Machine Learning Prediction of Multidrug Resistance in Swine-Derived Campylobacter spp. Using United States Antimicrobial Resistance Surveillance Data (2013–2023)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
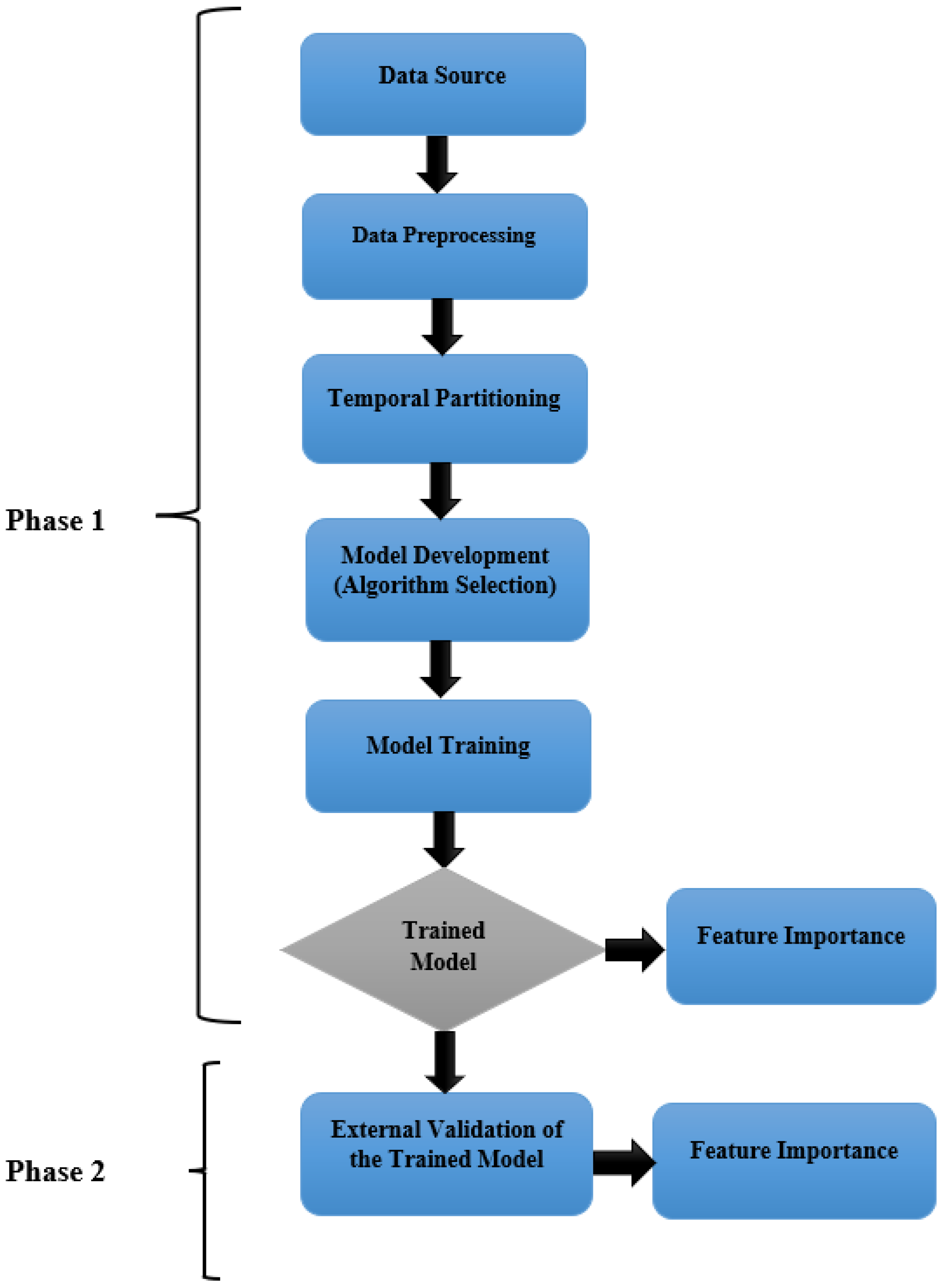
2.1. Study Design
2.2. Phase 1
2.2.1. Data Preprocessing and Temporal Partitioning
2.2.2. Algorithm Selection and Internal Validation
2.2.3. Model Development and Validation Using the Selected Machine Learning Algorithm
2.2.4. Feature Importance Analysis
2.3. Phase 2
External Validation of the Trained Model
3. Results
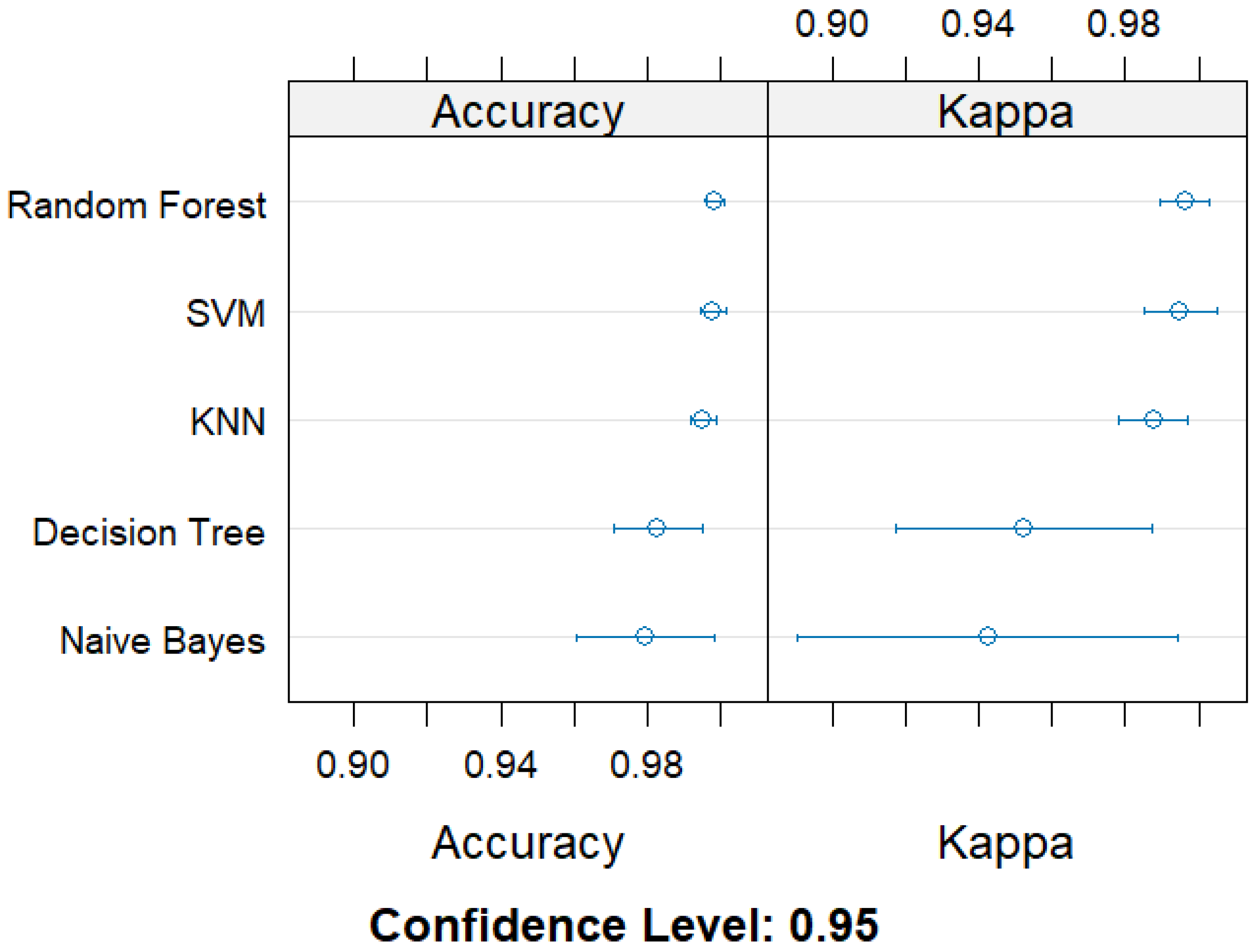
3.1. Performance Evaluation of Classification Machine Learning Algorithms for MDR Prediction in Swine-Derived Campylobacter
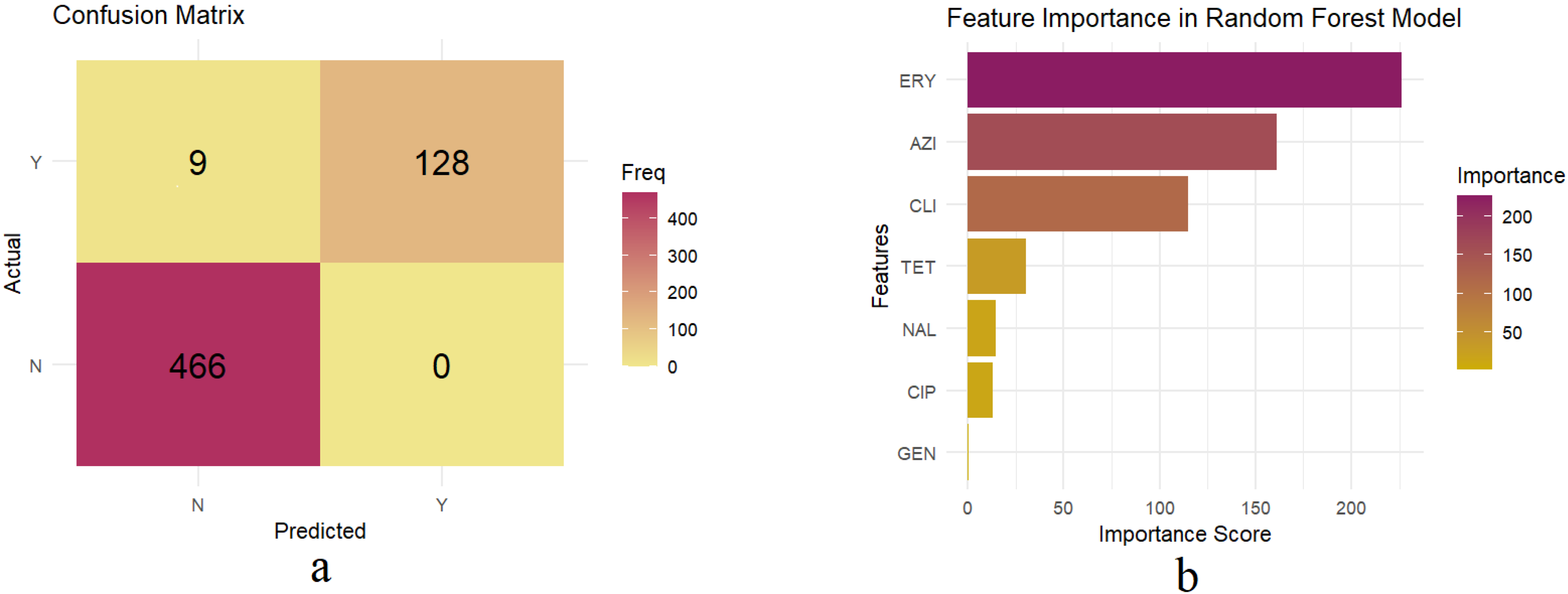
3.2. Development and Evaluation of a Random Forest Model to Predict Multidrug Resistance in Campylobacter from Swine
3.3. Important Features Predicting MDR in the Trained Random Forest Model
3.4. External Validation of the Trained Random Forest Model (Phase 2)
3.5. Important Features Predicting MDR in the External Validation Phase of the Trained Random Forest Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ranasinghe, S.; Fhogartaigh, C.N. Bacterial Gastroenteritis. Medicine 2021, 49, 687–693. [Google Scholar] [CrossRef]
- Chlebicz, A.; Śliżewska, K. Campylobacteriosis, Salmonellosis, Yersiniosis, and Listeriosis as Zoonotic Foodborne Diseases: A Review. Int. J. Environ. Res. Public Health 2018, 15, 863. [Google Scholar] [CrossRef]
- Ben Romdhane, R.; Merle, R. The Data Behind Risk Analysis of Campylobacter Jejuni and Campylobacter Coli Infections. Curr. Top. Microbiol. Immunol. 2021, 431, 25–58. [Google Scholar] [CrossRef]
- Francois Watkins, L.K.; Laughlin, M.E.; Joseph, L.A.; Chen, J.C.; Nichols, M.; Basler, C.; Breazu, R.; Bennett, C.; Koski, L.; Montgomery, M.P.; et al. Ongoing Outbreak of Extensively Drug-Resistant Campylobacter Jejuni Infections Associated with US Pet Store Puppies, 2016–2020. JAMA. Netw. Open 2021, 4, e2125203. [Google Scholar] [CrossRef] [PubMed]
- Habib, I.; Mohamed, M.-Y.I.; Lakshmi, G.B.; Al Marzooqi, H.M.; Afifi, H.S.; Shehata, M.G.; Khan, M.; Ghazawi, A.; Abdalla, A.; Anes, F. Quantitative Assessment and Genomic Profiling of Campylobacter Dynamics in Poultry Processing: A Case Study in the United Arab Emirates Integrated Abattoir System. Front. Microbiol. 2024, 15, 1439424. [Google Scholar] [CrossRef] [PubMed]
- Boes, J.; Nersting, L.; Nielsen, E.M.; Kranker, S.; Enøe, C.; Wachmann, H.C.; Baggesen, D.L. Prevalence and Diversity of Campylobacter Jejuni in Pig Herds on Farms with and without Cattle or Poultry. J. Food Prot. 2005, 68, 722–727. [Google Scholar] [CrossRef]
- Papadopoulos, D.; Petridou, E.; Papageorgiou, K.; Giantsis, I.A.; Delis, G.; Economou, V.; Frydas, I.; Papadopoulos, G.; Hatzistylianou, M.; Kritas, S.K. Phenotypic and Molecular Patterns of Resistance among Campylobacter Coli and Campylobacter Jejuni Isolates, from Pig Farms. Animals 2021, 11, 2394. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, C.A.; Young, S.; Li, C.; Hsu, C.-H.; Martin, G.; Zhao, S. Use of Whole-Genome Sequencing for Campylobacter Surveillance from NARMS Retail Poultry in the United States in 2015. Food Microbiol. 2018, 73, 122–128. [Google Scholar] [CrossRef]
- Sodagari, H.R.; Sohail, M.N.; Varga, C. Temporal, Regional, and Demographic Differences among Antimicrobial-Resistant Domestic Campylobacter Jejuni Human Infections across the United States, 2013–2019. Int. J. Antimicrob. Agents 2025, 65, 107467. [Google Scholar] [CrossRef]
- Ahmed, S.K.; Hussein, S.; Qurbani, K.; Ibrahim, R.H.; Fareeq, A.; Mahmood, K.A.; Mohamed, M.G. Antimicrobial Resistance: Impacts, Challenges, and Future Prospects. J. Med. Surg. Public Health 2024, 2, 100081. [Google Scholar] [CrossRef]
- Habib, I.; Ibrahim Mohamed, M.-Y.; Ghazawi, A.; Lakshmi, G.B.; Khan, M.; Li, D.; Sahibzada, S. Genomic Characterization of Molecular Markers Associated with Antimicrobial Resistance and Virulence of the Prevalent Campylobacter Coli Isolated from Retail Chicken Meat in the United Arab Emirates. Curr. Res. Food Sci. 2023, 6, 100434. [Google Scholar] [CrossRef]
- Sodagari, H.R.; Agrawal, I.; Sohail, M.N.; Yudhanto, S.; Varga, C. Monitoring Antimicrobial Resistance in Campylobacter Isolates of Chickens and Turkeys at the Slaughter Establishment Level across the United States, 2013–2021. Epidemiol. Infect. 2024, 152, e41. [Google Scholar] [CrossRef]
- Marin, C.; Lorenzo-Rebenaque, L.; Moreno-Moliner, J.; Sevilla-Navarro, S.; Montero, E.; Chinillac, M.C.; Jordá, J.; Vega, S. Multidrug-Resistant Campylobacer Jejuni on Swine Processing at a Slaughterhouse in Eastern Spain. Animals 2021, 11, 1339. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Palacios, P.; Galán-Sánchez, F.; Casimiro-Soriguer, C.S.; Jurado-Tarifa, E.; Arroyo, F.; Lara, M.; Chaves, J.A.; Dopazo, J.; Rodríguez-Iglesias, M.A. Genotypic Characterization and Antimicrobial Susceptibility of Human Campylobacter Jejuni Isolates in Southern Spain. Microbiol. Spectr. 2024, 12, e0102824. [Google Scholar] [CrossRef] [PubMed]
- Chavan, P.; Vashishth, R. Antimicrobial Resistance in Foodborne Pathogens: Consequences for Public Health and Future Approaches. Discov. Appl. Sci. 2025, 7, 623. [Google Scholar] [CrossRef]
- McClymont, H.; Lambert, S.B.; Barr, I.; Vardoulakis, S.; Bambrick, H.; Hu, W. Internet-Based Surveillance Systems and Infectious Diseases Prediction: An Updated Review of the Last 10 Years and Lessons from the COVID-19 Pandemic. J. Epidemiol. Glob. Health 2024, 14, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Jackson, S.A. Machine Learning and Complex Biological Data. Genome Biol. 2019, 20, 76. [Google Scholar] [CrossRef]
- Drouin, A.; Giguère, S.; Déraspe, M.; Marchand, M.; Tyers, M.; Loo, V.G.; Bourgault, A.-M.; Laviolette, F.; Corbeil, J. Predictive Computational Phenotyping and Biomarker Discovery Using Reference-Free Genome Comparisons. BMC Genom. 2016, 17, 754. [Google Scholar] [CrossRef]
- Hyun, J.C.; Kavvas, E.S.; Monk, J.M.; Palsson, B.O. Machine Learning with Random Subspace Ensembles Identifies Antimicrobial Resistance Determinants from Pan-Genomes of Three Pathogens. PLoS. Comput. Biol. 2020, 16, e1007608. [Google Scholar] [CrossRef]
- Naidenov, B.; Lim, A.; Willyerd, K.; Torres, N.J.; Johnson, W.L.; Hwang, H.J.; Hoyt, P.; Gustafson, J.E.; Chen, C. Pan-Genomic and Polymorphic Driven Prediction of Antibiotic Resistance in Elizabethkingia. Front. Microbiol. 2019, 10, 1446. [Google Scholar] [CrossRef]
- Parthasarathi, K.T.S.; Gaikwad, K.B.; Rajesh, S.; Rana, S.; Pandey, A.; Singh, H.; Sharma, J. A Machine Learning-Based Strategy to Elucidate the Identification of Antibiotic Resistance in Bacteria. Front. Antibiot. 2024, 3, 1405296. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Zhao, Q.; Xu, X.; Zhou, Y.; Huang, M. Early Prediction of Carbapenem-Resistant Gram-Negative Bacterial Carriage in Intensive Care Units Using Machine Learning. J. Glob. Antimicrob. Resist. 2022, 29, 225–231. [Google Scholar] [CrossRef]
- Moran, E.; Robinson, E.; Green, C.; Keeling, M.; Collyer, B. Towards Personalized Guidelines: Using Machine-Learning Algorithms to Guide Antimicrobial Selection. J. Antimicrob. Chemother. 2020, 75, 2677–2680. [Google Scholar] [CrossRef] [PubMed]
- Noman, S.M.; Zeeshan, M.; Arshad, J.; Deressa Amentie, M.; Shafiq, M.; Yuan, Y.; Zeng, M.; Li, X.; Xie, Q.; Jiao, X. Machine Learning Techniques for Antimicrobial Resistance Prediction of Pseudomonas Aeruginosa from Whole Genome Sequence Data. Comput. Intell. Neurosci. 2023, 2023, 5236168. [Google Scholar] [CrossRef]
- Chowdhury, A.S.; Call, D.R.; Broschat, S.L. Antimicrobial Resistance Prediction for Gram-Negative Bacteria via Game Theory-Based Feature Evaluation. Sci. Rep. 2019, 9, 14487. [Google Scholar] [CrossRef]
- Feucherolles, M.; Nennig, M.; Becker, S.L.; Martiny, D.; Losch, S.; Penny, C.; Cauchie, H.-M.; Ragimbeau, C. Combination of MALDI-TOF Mass Spectrometry and Machine Learning for Rapid Antimicrobial Resistance Screening: The Case of Campylobacter Spp. Front Microbiol. 2021, 12, 804484. [Google Scholar] [CrossRef]
- U.S. Department of Agriculture, Food Safety and Inspection Service. FSIS Laboratory Sampling Data: NARMS Cecal Sampling. Food Safety and Inspection Service. Available online: https://www.Fsis.Usda.Gov/Science-Data/Data-Sets-Visualizations/Laboratory-Sampling-Data (accessed on 1 August 2025).
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Singh, D.; Singh, B. Investigating the Impact of Data Normalization on Classification Performance. Appl. Soft. Comput. 2020, 97, 105524. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2025; Available online: https://www.r-project.org/ (accessed on 1 August 2025).
- Osisanwo, F.Y.; Akinsola, J.E.T.; Awodele, O.; Hinmikaiye, J.O.; Olakanmi, O.; Akinjobi, J. Supervised Machine Learning Algorithms: Classification and Comparison. Int. J. Comput. Trends Technol.-IJCTT 2017, 48. [Google Scholar] [CrossRef]
- Muhammad, I.; Yan, Z. Supervised Machine Learning Approaches: A Survey. ICTACT J. Soft Comput. 2015, 5, 946–952. [Google Scholar] [CrossRef]
- Radhoush, S.; Whitaker, B.M.; Nehrir, H. An Overview of Supervised Machine Learning Approaches for Applications in Active Distribution Networks. Energies 2023, 16, 5972. [Google Scholar] [CrossRef]
- Uddin, S.; Khan, A.; Hossain, M.E.; Moni, M.A. Comparing Different Supervised Machine Learning Algorithms for Disease Prediction. BMC Med. Inform. Decis. Mak. 2019, 19, 281. [Google Scholar] [CrossRef]
- ValizadehAslani, T.; Zhao, Z.; Sokhansanj, B.A.; Rosen, G.L. Amino Acid K-Mer Feature Extraction for Quantitative Antimicrobial Resistance (AMR) Prediction by Machine Learning and Model Interpretation for Biological Insights. Biology 2020, 9, 365. [Google Scholar] [CrossRef]
- Lu, J.; Chen, J.; Liu, C.; Zeng, Y.; Sun, Q.; Li, J.; Shen, Z.; Chen, S.; Zhang, R. Identification of Antibiotic Resistance and Virulence-Encoding Factors in Klebsiella Pneumoniae by Raman Spectroscopy and Deep Learning. Microb. Biotechnol. 2022, 15, 1270–1280. [Google Scholar] [CrossRef] [PubMed]
- Portelli, S.; Myung, Y.; Furnham, N.; Vedithi, S.C.; Pires, D.E.V.; Ascher, D.B. Prediction of Rifampicin Resistance beyond the RRDR Using Structure-Based Machine Learning Approaches. Sci. Rep. 2020, 10, 18120. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, D.; Chen, F.; Rao, G.; Zhang, Y. Machine Learning-Based Colistin Resistance Marker Screening and Phenotype Prediction in Escherichia Coli from Whole Genome Sequencing Data. J. Infect. 2024, 88, 191–193. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Chen, C.-H.; Lee, T.-Y.; Horng, J.-T.; Liu, T.-P.; Tseng, Y.-J.; Lu, J.-J. Rapid Detection of Heterogeneous Vancomycin-Intermediate Staphylococcus Aureus Based on Matrix-Assisted Laser Desorption Ionization Time-of-Flight: Using a Machine Learning Approach and Unbiased Validation. Front. Microbiol. 2018, 9, 2393. [Google Scholar] [CrossRef]
- Weis, C.; Cuénod, A.; Rieck, B.; Dubuis, O.; Graf, S.; Lang, C.; Oberle, M.; Brackmann, M.; Søgaard, K.K.; Osthoff, M.; et al. Direct Antimicrobial Resistance Prediction from Clinical MALDI-TOF Mass Spectra Using Machine Learning. Nat. Med. 2022, 28, 164–174. [Google Scholar] [CrossRef]
- Babiker, A.; Mustapha, M.M.; Pacey, M.P.; Shutt, K.A.; Ezeonwuka, C.D.; Ohm, S.L.; Cooper, V.S.; Marsh, J.W.; Doi, Y.; Harrison, L.H. Use of Online Tools for Antimicrobial Resistance Prediction by Whole-Genome Sequencing in Methicillin-Resistant Staphylococcus Aureus (MRSA) and Vancomycin-Resistant Enterococci (VRE). J. Glob. Antimicrob. Resist. 2019, 19, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Hendriksen, R.S.; Bortolaia, V.; Tate, H.; Tyson, G.H.; Aarestrup, F.M.; McDermott, P.F. Using Genomics to Track Global Antimicrobial Resistance. Front. Public Health 2019, 7, 242. [Google Scholar] [CrossRef]
- Dai, L.; Sahin, O.; Grover, M.; Zhang, Q. New and Alternative Strategies for the Prevention, Control, and Treatment of Antibiotic-Resistant Campylobacter. Transl. Res. 2020, 223, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, M.; Kamoun, S.; Hkimi, C.; Ghedira, K.; Béjaoui, A.; Maaroufi, A. Relationships between Virulence Genes and Antibiotic Resistance Phenotypes/Genotypes in Campylobacter Spp. Isolated from Layer Hens and Eggs in the North of Tunisia: Statistical and Computational Insights. Foods 2022, 11, 3554. [Google Scholar] [CrossRef] [PubMed]
- Painset, A.; Day, M.; Doumith, M.; Rigby, J.; Jenkins, C.; Grant, K.; Dallman, T.J.; Godbole, G.; Swift, C. Comparison of Phenotypic and WGS-Derived Antimicrobial Resistance Profiles of Campylobacter Jejuni and Campylobacter Coli Isolated from Cases of Diarrhoeal Disease in England and Wales, 2015-16. J. Antimicrob. Chemother. 2020, 75, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Pyörälä, S.; Baptiste, K.E.; Catry, B.; van Duijkeren, E.; Greko, C.; Moreno, M.A.; Pomba, M.C.M.F.; Rantala, M.; Ružauskas, M.; Sanders, P.; et al. Macrolides and Lincosamides in Cattle and Pigs: Use and Development of Antimicrobial Resistance. Vet. J. 2014, 200, 230–239. [Google Scholar] [CrossRef]
- de la Lastra, J.M.P.; Wardell, S.J.T.; Pal, T.; de la Fuente-Nunez, C.; Pletzer, D. From Data to Decisions: Leveraging Artificial Intelligence and Machine Learning in Combating Antimicrobial Resistance—A Comprehensive Review. J. Med. Syst. 2024, 48, 71. [Google Scholar] [CrossRef]

| Metric | Value |
|---|---|
| Accuracy | 99.73% |
| 95% Confidence Interval | 99.42–99.90% |
| No Information Rate (NIR) | 76.54% |
| p-Value [Accuracy > NIR] | p < 2 × 10−16 |
| Kappa | 0.9925 |
| McNemar’s Test p-Value | 0.0412 |
| Sensitivity | 98.86% |
| Specificity | 100.00% |
| Positive Predictive Value | 100.00% |
| Negative Predictive Value | 99.65% |
| Prevalence | 23.46% |
| Detection Rate | 23.19% |
| Detection Prevalence | 23.19% |
| Balanced Accuracy | 99.43% |
| Metric | Value |
|---|---|
| Accuracy | 98.51% |
| 95% Confidence Interval | 97.19–99.32% |
| No Information Rate (NIR) | 77.28% |
| p-Value [Acc > NIR] | p < 2.2 × 10−16 |
| Kappa | 0.9565 |
| McNemar’s Test p-Value | 0.007661 |
| Sensitivity | 93.43% |
| Specificity | 100% |
| Positive Predictive Value | 100% |
| Negative Predictive Value | 98.11 |
| Prevalence | 22.72% |
| Detection Rate | 21.23% |
| Detection Prevalence | 21.%23 |
| Balanced Accuracy | 96.72% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sodagari, H.R.; Ghasemi, M.; Varga, C.; Habib, I. Machine Learning Prediction of Multidrug Resistance in Swine-Derived Campylobacter spp. Using United States Antimicrobial Resistance Surveillance Data (2013–2023). Vet. Sci. 2025, 12, 937. https://doi.org/10.3390/vetsci12100937
Sodagari HR, Ghasemi M, Varga C, Habib I. Machine Learning Prediction of Multidrug Resistance in Swine-Derived Campylobacter spp. Using United States Antimicrobial Resistance Surveillance Data (2013–2023). Veterinary Sciences. 2025; 12(10):937. https://doi.org/10.3390/vetsci12100937
Chicago/Turabian StyleSodagari, Hamid Reza, Maryam Ghasemi, Csaba Varga, and Ihab Habib. 2025. "Machine Learning Prediction of Multidrug Resistance in Swine-Derived Campylobacter spp. Using United States Antimicrobial Resistance Surveillance Data (2013–2023)" Veterinary Sciences 12, no. 10: 937. https://doi.org/10.3390/vetsci12100937
APA StyleSodagari, H. R., Ghasemi, M., Varga, C., & Habib, I. (2025). Machine Learning Prediction of Multidrug Resistance in Swine-Derived Campylobacter spp. Using United States Antimicrobial Resistance Surveillance Data (2013–2023). Veterinary Sciences, 12(10), 937. https://doi.org/10.3390/vetsci12100937







