Screening of Neutralizing Antibodies against FaeG Protein of Enterotoxigenic Escherichia coli
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Median Lethal Dose and Minimum Lethal Dose Test of Isolated ETEC Strains
2.2. Immunization of Mice
2.3. Expression and Fluorescent Labeling of F4ac FaeG Protein in ETEC
2.4. Indirect Enzyme-Linked Immunosorbent Assay (iELISA)
2.5. Fluorescence-Activated Cell Sorting
2.6. Amplification of Variable Region and Construction of Expression Plasmids
2.7. Expression and Analyses of Anti-rFaeG IgG mAbs
2.8. Adhesion Assay of ETEC
2.9. Anti-rFaeG IgG mAb Neutralization Assay in Mice
3. Results
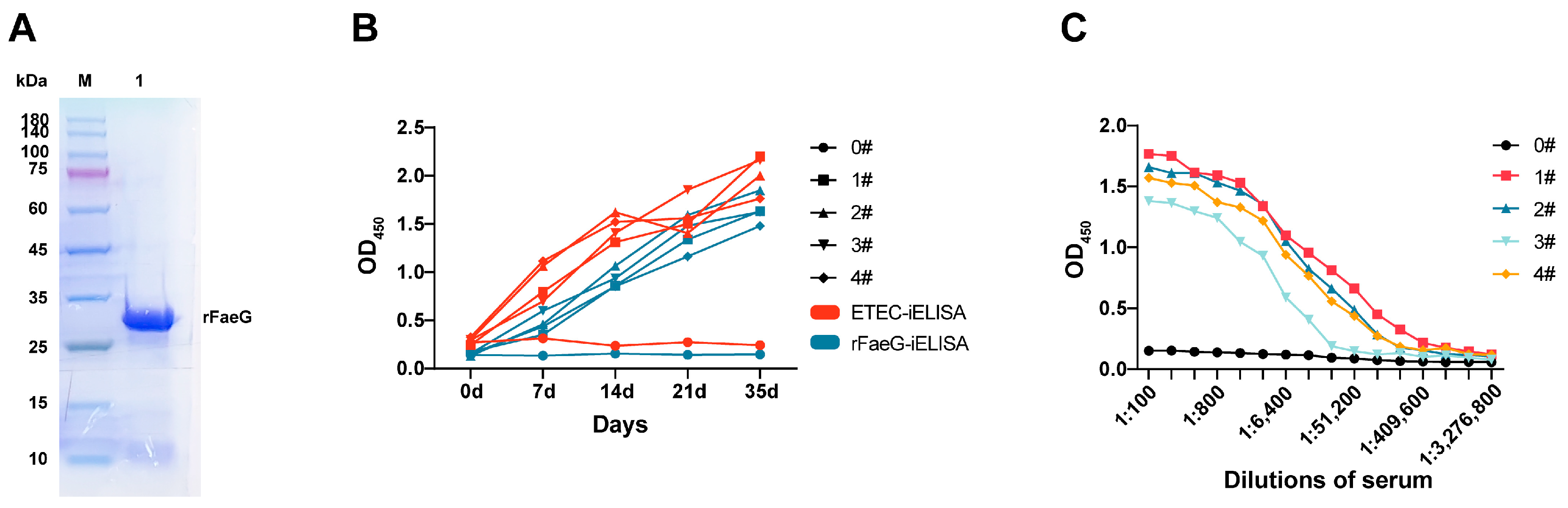
3.1. Soluble Expression and Immune Response of rFaeG Protein in Mice
3.2. Sorting of rFaeG-Specific Mouse Memory B Cells
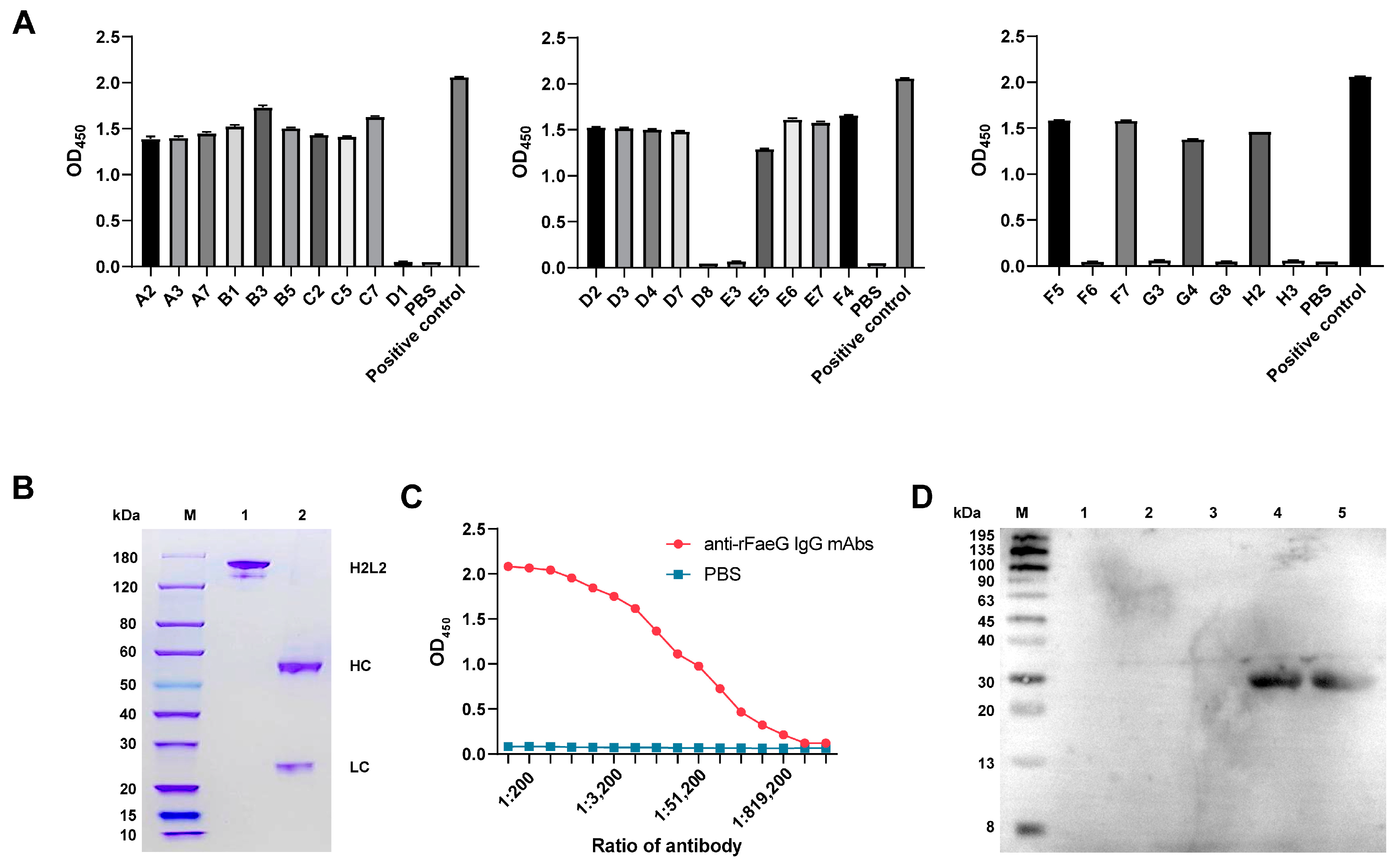
3.3. Preparation of Anti-rFaeG IgG mAbs
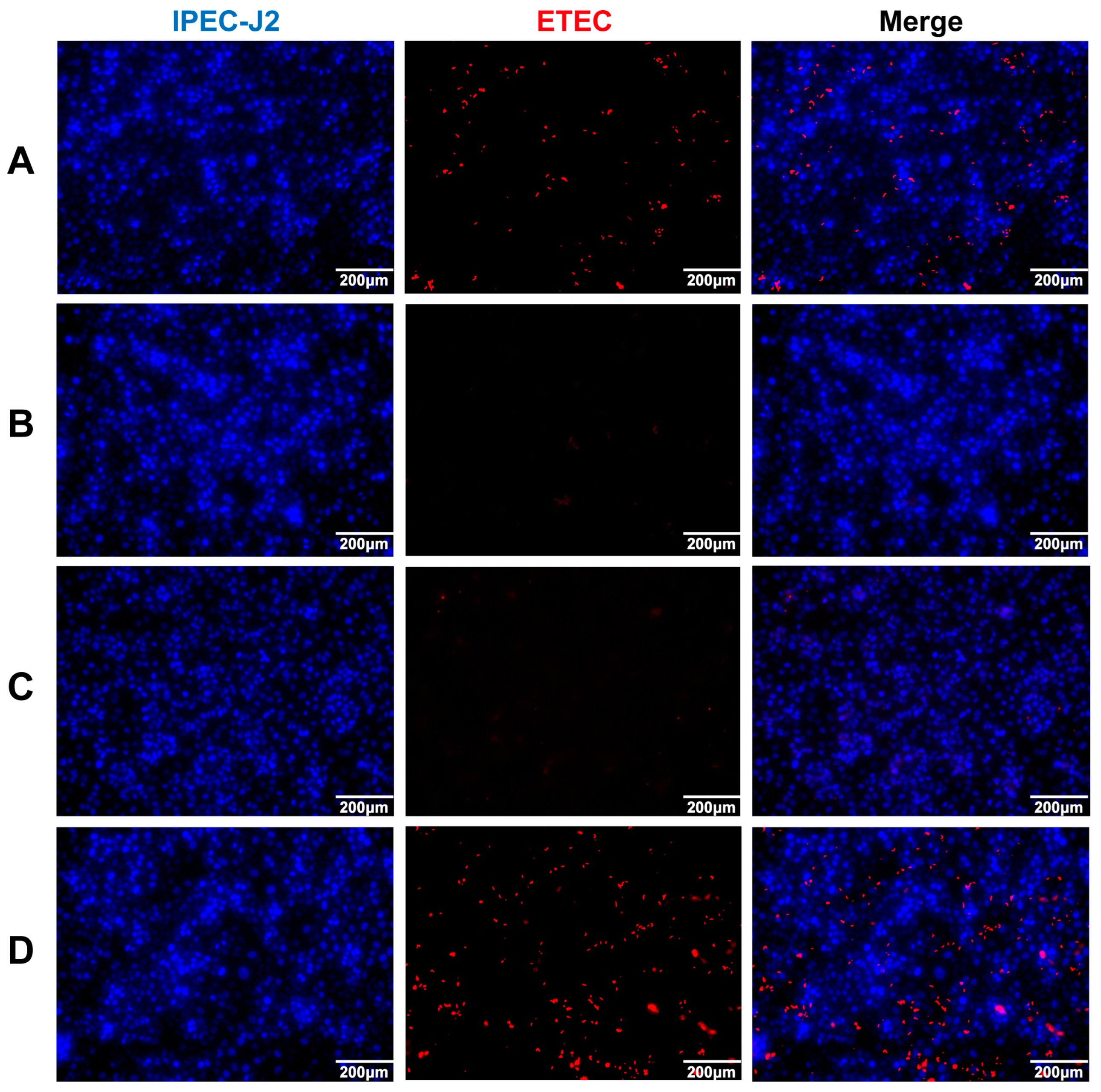
3.4. Neutralizing Activity of Anti-rFaeG IgG mAbs In Vitro
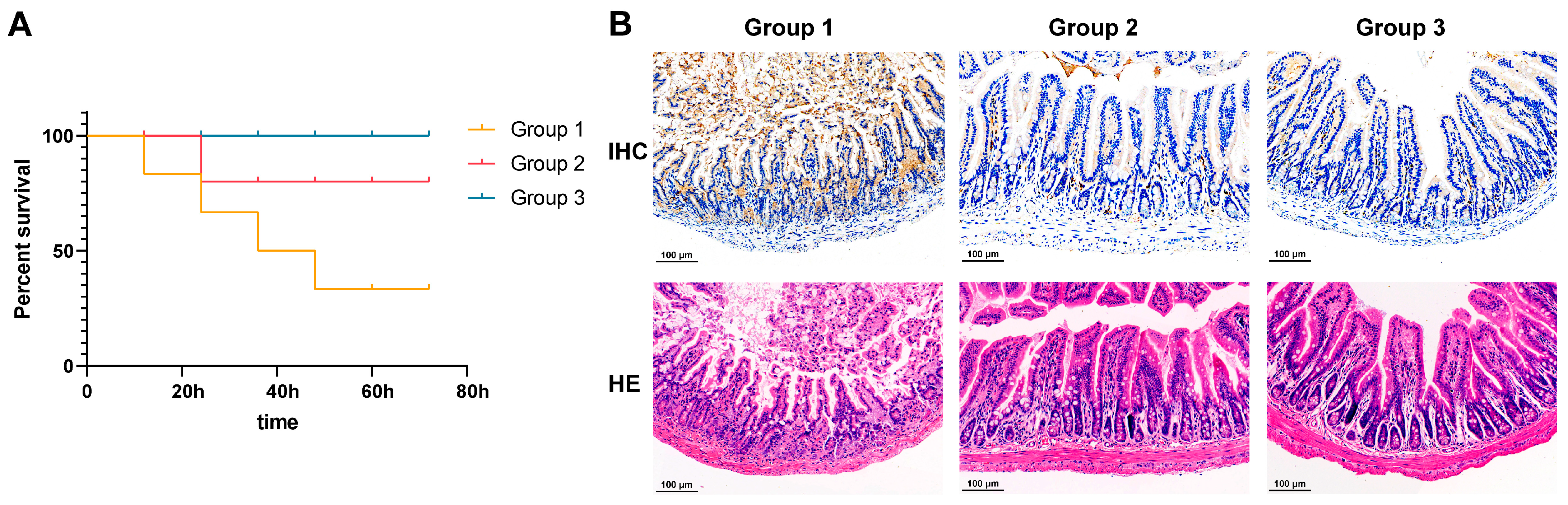
3.5. Neutralizing Activity of Anti-rFaeG IgG mAbs In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rhouma, M.; Fairbrother, J.M.; Beaudry, F.; Letellier, A. Post weaning diarrhea in pigs: Risk factors and non-colistin-based control strategies. Acta Vet. Scand. 2017, 59, 31. [Google Scholar] [CrossRef] [PubMed]
- Fairbrother, J.M.; Nadeau, E.; Gyles, C.L. Escherichia coli in postweaning diarrhea in pigs: An update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 2005, 6, 17–39. [Google Scholar] [CrossRef]
- Nagy, B.; Fekete, P.Z. Enterotoxigenic Escherichia coli in veterinary medicine. Int. J. Med. Microbiol. 2005, 295, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Kopic, S.; Geibel, J.P. Toxin mediated diarrhea in the 21 century: The pathophysiology of intestinal ion transport in the course of ETEC, V. cholerae and rotavirus infection. Toxins 2010, 2, 2132–2157. [Google Scholar] [CrossRef]
- Calderon Toledo, C.; von Mentzer, A.; Agramont, J.; Thorell, K.; Zhou, Y.; Szabo, M.; Colque, P.; Kuhn, I.; Gutiérrez-Cortez, S.; Joffré, E. Circulation of enterotoxigenic Escherichia coli (ETEC) isolates expressing CS23 from the environment to clinical settings. mSystems 2023, 8, e0014123. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, L.; Ali, Z.B.; Nygren, E.; Wang, Z.; Karlsson, S.; Zhu, B.; Quiding-Järbrink, M.; Sjöling, Å. Alkaline pH Is a signal for optimal production and secretion of the heat labile toxin, LT in enterotoxigenic Escherichia coli (ETEC). PLoS ONE 2013, 8, e74069. [Google Scholar] [CrossRef]
- Xia, L.; Dai, L.; Yu, Q.; Yang, Q.; Jin, L.Z.; Zhao, X. Intestinal receptors for adhesive fimbriae of enterotoxigenic Escherichia coli (ETEC) K88 in swine—A review. Appl. Microbiol. Biotechnol. 2000, 54, 311–318. [Google Scholar] [CrossRef]
- Duan, Q.; Wu, W.; Pang, S.; Pan, Z.; Zhang, W.; Zhu, G. Coimmunization with Two Enterotoxigenic Escherichia coli (ETEC) Fimbrial Multiepitope Fusion Antigens Induces the Production of Neutralizing Antibodies against Five ETEC Fimbriae (F4, F5, F6, F18, and F41). Appl. Environ. Microbiol. 2020, 86, e00217-20. [Google Scholar] [CrossRef]
- Xia, P.; Song, Y.; Zou, Y.; Yang, Y.; Zhu, G. F4+ enterotoxigenic Escherichia coli (ETEC) adhesion mediated by the major fimbrial subunit FaeG. J. Basic Microbiol. 2015, 55, 1118–1124. [Google Scholar] [CrossRef]
- Verdonck, F.; Cox, E.; Goddeeris, B.M. F4 fimbriae expressed by porcine enterotoxigenic Escherichia coli, an example of an eccentric fimbrial system? J. Mol. Microbiol. Biotechnol. 2004, 7, 155–169. [Google Scholar] [CrossRef]
- Lin, J.; Mateo, K.S.; Zhao, M.; Erickson, A.K.; Garcia, N.; He, D.; Moxley, R.A.; Francis, D.H. Protection of piglets against enteric colibacillosis by intranasal immunization with K88ac (F4ac) fimbriae and heat labile enterotoxin of Escherichia coli. Vet. Microbiol. 2013, 162, 731–739. [Google Scholar] [CrossRef]
- Melkebeek, V.; Sonck, E.; Verdonck, F.; Goddeeris, B.M.; Cox, E. Optimized FaeG expression and a thermolabile enterotoxin DNA adjuvant enhance priming of an intestinal immune response by an FaeG DNA vaccine in pigs. Clin. Vaccine Immunol. 2007, 14, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Foged, N.T.; Klemm, P.; Elling, F.; Jorsal, S.E.; Zeuthen, J. Monoclonal antibodies to K88ab, K88ac and K88ad fimbriae from enterotoxigenic Escherichia coli. Microb. Pathog. 1986, 1, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Xu, Q.; Serafino, M.A.; Zhang, Q.; Wang, C.; Yu, Y. Comprehensive analysis of circular RNAs in porcine small intestine epithelial cells associated with susceptibility to Escherichia coli F4ac diarrhea. BMC Genom. 2023, 24, 211. [Google Scholar] [CrossRef]
- Heo, J.M.; Opapeju, F.O.; Pluske, J.R.; Kim, J.C.; Hampson, D.J.; Nyachoti, C.M. Gastrointestinal health and function in weaned pigs: A review of feeding strategies to control post-weaning diarrhoea without using in-feed antimicrobial compounds. J. Anim. Physiol. Anim. Nutr. 2013, 97, 207–237. [Google Scholar] [CrossRef]
- Smith, M.G.; Jordan, D.; Chapman, T.A.; Chin, J.J.; Barton, M.D.; Do, T.N.; Fahy, V.A.; Fairbrother, J.M.; Trott, D.J. Antimicrobial resistance and virulence gene profiles in multi-drug resistant enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhoea. Vet. Microbiol. 2010, 145, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Butaye, P.; van Duijkeren, E.; Prescott, J.F.; Schwarz, S. Antimicrobial resistance in bacteria from animals and the environment. Vet. Microbiol. 2014, 171, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.; Barros, M.M.; Araujo, D.; Campos, A.M.; Oliveira, R.; Silva, S.; Almeida, C. Swine enteric colibacillosis: Current treatment avenues and future directions. Front. Vet. Sci. 2022, 9, 981207. [Google Scholar] [CrossRef]
- Yang, W.; Li, J.; Yao, Z.; Li, M. A review on the alternatives to antibiotics and the treatment of antibiotic pollution: Current development and future prospects. Sci. Total Environ. 2024, 926, 171757. [Google Scholar] [CrossRef]
- Castelli, M.S.; McGonigle, P.; Hornby, P.J. The pharmacology and therapeutic applications of monoclonal antibodies. Pharmacol. Res. Perspect. 2019, 7, e00535. [Google Scholar] [CrossRef]
- Canzoneri, R.; Lacunza, E.; Abba, M.C. Genomics and bioinformatics as pillars of precision medicine in oncology. Medicina 2019, 79, 587–592. [Google Scholar]
- Robinson, M.P.; Jung, J.; Lopez-Barbosa, N.; Chang, M.; Li, M.; Jaroentomeechai, T.; Cox, E.C.; Zheng, X.; Berkmen, M.; DeLisa, M.P. Isolation of full-length IgG antibodies from combinatorial libraries expressed in the cytoplasm of Escherichia coli. Nat. Commun. 2023, 14, 3514. [Google Scholar] [CrossRef]
- Reinhart, D.; Damjanovic, L.; Kaisermayer, C.; Kunert, R. Benchmarking of commercially available CHO cell culture media for antibody production. Appl. Microbiol. Biotechnol. 2015, 99, 4645–4657. [Google Scholar] [CrossRef]
- Song, X.; Yang, Y.; Li, J.; He, M.; Zou, Y.; Jia, R.; Li, L.; Hang, J.; Cui, M.; Bai, L.; et al. Tannins extract from Galla Chinensis can protect mice from infection by Enterotoxigenic Escherichia coli O101. BMC Complement. Med. Ther. 2021, 21, 84. [Google Scholar] [CrossRef] [PubMed]
- Hui Xian, T.; Parasuraman, S.; Ravichandran, M.; Prabhakaran, G. Dual-Use Vaccine for Diarrhoeal Diseases: Cross-Protective Immunogenicity of a Cold-Chain-Free, Live-Attenuated, Oral Cholera Vaccine against Enterotoxigenic Escherichia coli (ETEC) Challenge in BALB/c Mice. Vaccines 2022, 10, 2161. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Peng, K.; She, Y.; Fu, F.; Shi, Q.; Lin, Y.; Xu, C. Preparation of novel trivalent vaccine against enterotoxigenic Escherichia coli for preventing newborn piglet diarrhea. Am. J. Vet. Res. 2023, 84, 2. [Google Scholar] [CrossRef]
- Kettleborough, C.A.; Saldanha, J.; Ansell, K.H.; Bendig, M.M. Optimization of primers for cloning libraries of mouse immunoglobulin genes using the polymerase chain reaction. Eur. J. Immunol. 1993, 23, 206–211. [Google Scholar] [CrossRef]
- Xiao, K.; Yang, Y.; Zhang, Y.; Lv, Q.; Huang, F.; Wang, D.; Zhao, J.; Liu, Y. Long-chain PUFA ameliorate enterotoxigenic Escherichia coli-induced intestinal inflammation and cell injury by modulating pyroptosis and necroptosis signaling pathways in porcine intestinal epithelial cells. Br. J. Nutr. 2022, 128, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Yang, X.; Huang, N.; Wu, M.; Sun, H.; He, Q.; Lang, Q.; Zou, X.; Liu, Z.; Wang, J.; et al. Generation of fully human anti-GPC3 antibodies with high-affinity recognition of GPC3 positive tumors. Investig. New Drugs 2021, 39, 615–626. [Google Scholar] [CrossRef]
- Doki, T.; Takano, T.; Hohdatsu, T. Development of a mouse-feline chimeric antibody against feline tumor necrosis factor-alpha. J. Vet. Med. Sci. 2016, 78, 1447–1455. [Google Scholar] [CrossRef]
- Virdi, V.; Coddens, A.; De Buck, S.; Millet, S.; Goddeeris, B.M.; Cox, E.; De Greve, H.; Depicker, A. Orally fed seeds producing designer IgAs protect weaned piglets against enterotoxigenic Escherichia coli infection. Proc. Natl. Acad. Sci. USA 2013, 110, 11809–11814. [Google Scholar] [CrossRef] [PubMed]
- Yahong, H.; Liang, W.; Pan, A.; Zhou, Z.; Wang, Q.; Huang, C.; Chen, J.; Zhang, D. Protective immune response of bacterially-derived recombinant FaeG in piglets. J. Microbiol. 2006, 44, 548–555. [Google Scholar] [PubMed]
- Verdonck, F.; Cox, E.; Van der Stede, Y.; Goddeeris, B.M. Oral immunization of piglets with recombinant F4 fimbrial adhesin FaeG monomers induces a mucosal and systemic F4-specific immune response. Vaccine 2004, 22, 4291–4299. [Google Scholar] [CrossRef]
- Van den Broeck, W.; Cox, E.; Goddeeris, B.M. Receptor-specific binding of purified F4 to isolated villi. Vet. Microbiol. 1999, 68, 255–263. [Google Scholar] [CrossRef]
- Liang, W.; Huang, Y.; Yang, X.; Zhou, Z.; Pan, A.; Qian, B.; Huang, C.; Chen, J.; Zhang, D. Oral immunization of mice with plant-derived fimbrial adhesin FaeG induces systemic and mucosal K88ad enterotoxigenic Escherichia coli-specific immune responses. FEMS Immunol. Med. Microbiol. 2006, 46, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Moxley, R.A.; Zhang, W. Mapping the Neutralizing Epitopes of Enterotoxigenic Escherichia coli K88 (F4) Fimbrial Adhesin and Major Subunit FaeG. Appl. Environ. Microbiol. 2019, 85, e00329-19. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Hu, S.; Guan, Y.X.; Yao, S.J. Coexpression of chaperonin GroEL/GroES markedly enhanced soluble and functional expression of recombinant human interferon-gamma in Escherichia coli. Appl. Microbiol. Biotechnol. 2012, 93, 1065–1074. [Google Scholar] [CrossRef]
- Verma, R.; Khanna, P.; Chawla, S. Whole-cell inactivated leptospirosis vaccine: Future prospects. Hum. Vaccin. Immunother. 2013, 9, 763–765. [Google Scholar] [CrossRef]
- Sanchez, R.M.; Sierra, A.P.; Obregon Fuentes, A.M.; Gonzalez, I.R.; Gil, A.B.; Suarez, M.B.; Silveira, J.R.; Gastón, B.D. Reactogenecity and immunogenecity of Cuban trivalent inactivated vaccine against human leptospirosis in different vaccination schedules. Rev. Cubana Med. Trop. 2002, 54, 37–43. [Google Scholar]
- Bell, J.; Gray, D. Antigen-capturing cells can masquerade as memory B cells. J. Exp. Med. 2003, 197, 1233–1244. [Google Scholar] [CrossRef]
- Starkie, D.O.; Compson, J.E.; Rapecki, S.; Lightwood, D.J. Generation of Recombinant Monoclonal Antibodies from Immunised Mice and Rabbits via Flow Cytometry and Sorting of Antigen-Specific IgG+ Memory B Cells. PLoS ONE 2016, 11, e0152282. [Google Scholar] [CrossRef] [PubMed]
- Prashar, P.; Swain, S.; Adhikari, N.; Aryan, P.; Singh, A.; Kwatra, M.; Prabhakar, B. A novel high-throughput single B-cell cloning platform for isolation and characterization of high-affinity and potent SARS-CoV-2 neutralizing antibodies. Antivir. Res. 2022, 203, 105349. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.; Garman, L.; Wrammert, J.; Zheng, N.Y.; Capra, J.D.; Ahmed, R.; Wilson, P.C. Corrigendum: Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 2014, 9, 2903. [Google Scholar] [CrossRef] [PubMed]
- Weitkamp, J.H.; Kallewaard, N.; Kusuhara, K.; Feigelstock, D.; Feng, N.; Greenberg, H.B.; Crowe, J.E., Jr. Generation of recombinant human monoclonal antibodies to rotavirus from single antigen-specific B cells selected with fluorescent virus-like particles. J. Immunol. Methods 2003, 275, 223–237. [Google Scholar] [CrossRef]
- Jiang, Q.; Zhang, H.; Xie, Y.; Wang, Y. Recombinant expression of porcine lactoferrin peptide LF-6 with intein technology and its immunomodulatory function in ETEC K88-infected mice. Int. Immunopharmacol. 2016, 39, 181–191. [Google Scholar] [CrossRef]

| Primers | Sequence (5′-3′) a |
|---|---|
| VH1-F | (CCGATCCAGCCTCCGGAC)MAGCTTCAGGAGTCRGGACC |
| VH2-F | (CCGATCCAGCCTCCGGAC)CAGCTGAAGSASTCAGGACC |
| VH3-F | (CCGATCCAGCCTCCGGAC)MWGSKGGTGGAGTCTGGGGGA |
| VH4-F | (CCGATCCAGCCTCCGGAC)ARSSTGGWGGAATCTGGAGGA |
| VH5-F | (CCGATCCAGCCTCCGGAC)ARGSTGRTSGAGTCTGGAGG |
| VH6-F | (CCGATCCAGCCTCCGGAC)CARSYGCAGCARYCTGGG |
| VH7-F | (CCGATCCAGCCTCCGGAC)CAGYTGSWGCARTCTGGA |
| VH8-F | (CCGATCCAGCCTCCGGAC)CAGCTGCAGCAGTCWGTG |
| VH9-F | (CCGATCCAGCCTCCGGAC)MASYTGSWGGWGWCTGGAGG |
| VH10-F | (CCGATCCAGCCTCCGGAC)CAGMTSCAGCAGYCTGG |
| 1st reverse γ | CKYGGTSYTGCTGGCYGGGTG |
| 2nd reverse γ | GCTCAGGGAARTAGCCCTTGAC |
| Vκ1-F | (CCGATCCAGCCTCCGGAC)CAGACRTCMAGATRAYCCAGWCTMCA |
| Vκ2-F | (CCGATCCAGCCTCCGGAC)CARAMATTKTGCTGACYCARTYTCC |
| Vκ3-F | (CCGATCCAGCCTCCGGAC)CAGATRYTKTGATGACCCAAACTCCA |
| Vκ4-F | (CCGATCCAGCCTCCGGAC)CASRAAWTSTTCTCWYMCAGTGTCC |
| 1st reverse κ | CTAACACTCATTCCTGTTGAAGC |
| 2nd reverse κ | TGGGAAGATGGATACAGTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Lu, S.; Zhou, S.; Li, Z.; Guan, S.; Chen, H.; Song, Y. Screening of Neutralizing Antibodies against FaeG Protein of Enterotoxigenic Escherichia coli. Vet. Sci. 2024, 11, 419. https://doi.org/10.3390/vetsci11090419
Tian Y, Lu S, Zhou S, Li Z, Guan S, Chen H, Song Y. Screening of Neutralizing Antibodies against FaeG Protein of Enterotoxigenic Escherichia coli. Veterinary Sciences. 2024; 11(9):419. https://doi.org/10.3390/vetsci11090419
Chicago/Turabian StyleTian, Yang, Sijia Lu, Saisai Zhou, Zhen Li, Shuaiyin Guan, Huanchun Chen, and Yunfeng Song. 2024. "Screening of Neutralizing Antibodies against FaeG Protein of Enterotoxigenic Escherichia coli" Veterinary Sciences 11, no. 9: 419. https://doi.org/10.3390/vetsci11090419
APA StyleTian, Y., Lu, S., Zhou, S., Li, Z., Guan, S., Chen, H., & Song, Y. (2024). Screening of Neutralizing Antibodies against FaeG Protein of Enterotoxigenic Escherichia coli. Veterinary Sciences, 11(9), 419. https://doi.org/10.3390/vetsci11090419






