Transcriptomic Signatures of the Foetal Liver and Late Prenatal Development in Vitrified Rabbit Embryos
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Embryo Recovery
2.3. Vitrification and Warming Procedure
2.4. Embryo Transfer by Laparoscopy
2.5. Foetal Growth by Ultrasound Examination
2.6. Foetal and Offspring Phenotypic Determinations
2.7. RNA Isolation, Sequencing, and Analysis of Foetal Liver
2.8. Statistical Analysis
2.8.1. Phenotypical Parameter Analysis
2.8.2. Differential Gene Expression Analysis in Foetal Liver
3. Result
3.1. Prenatal Survival and Development of Fresh and Vitrified Embryos
3.2. Postnatal Survival Rate and Body Weight at Birth
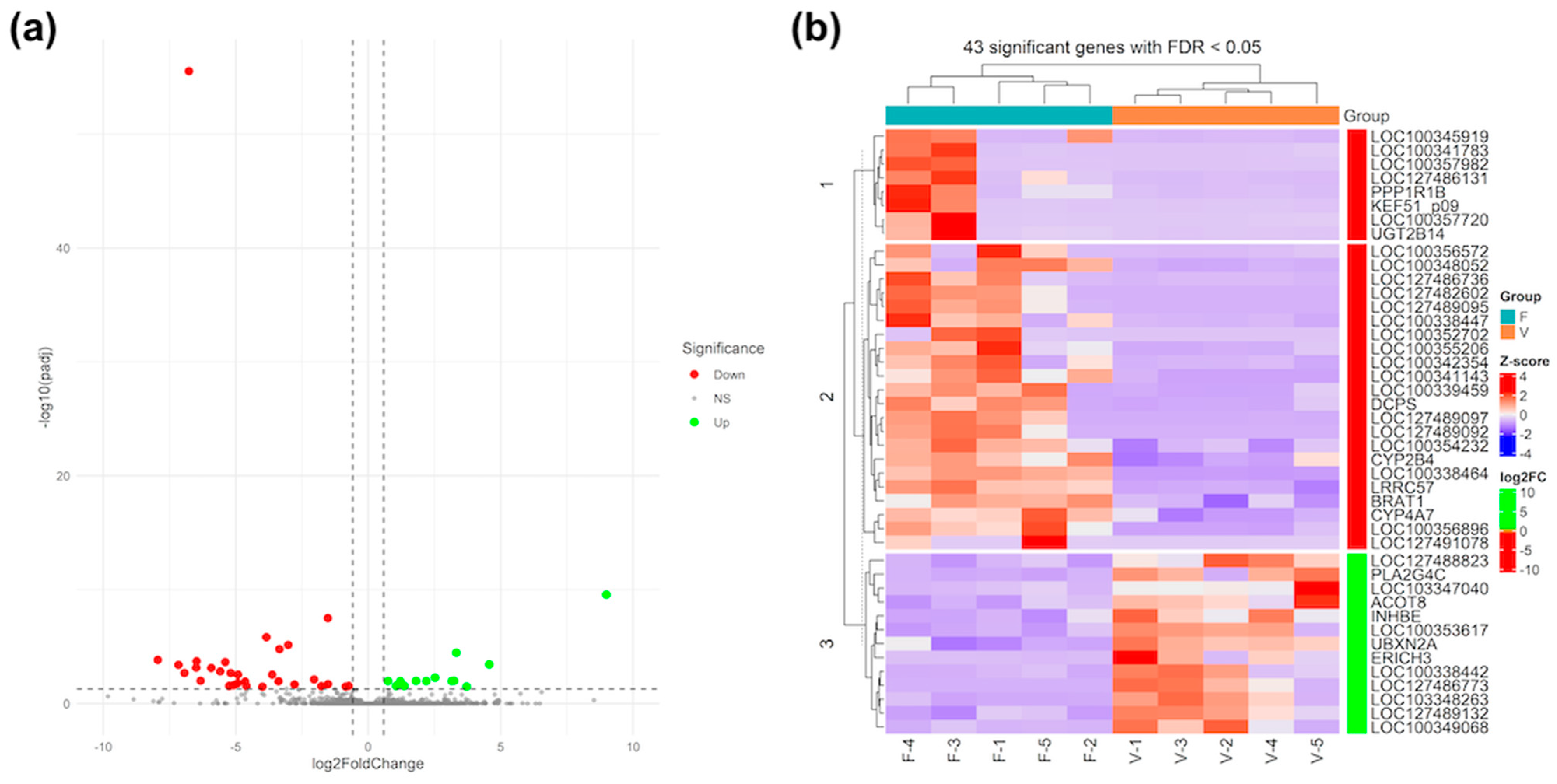
3.3. Differential Gene Expression and Functional Analysis in Foetal Liver
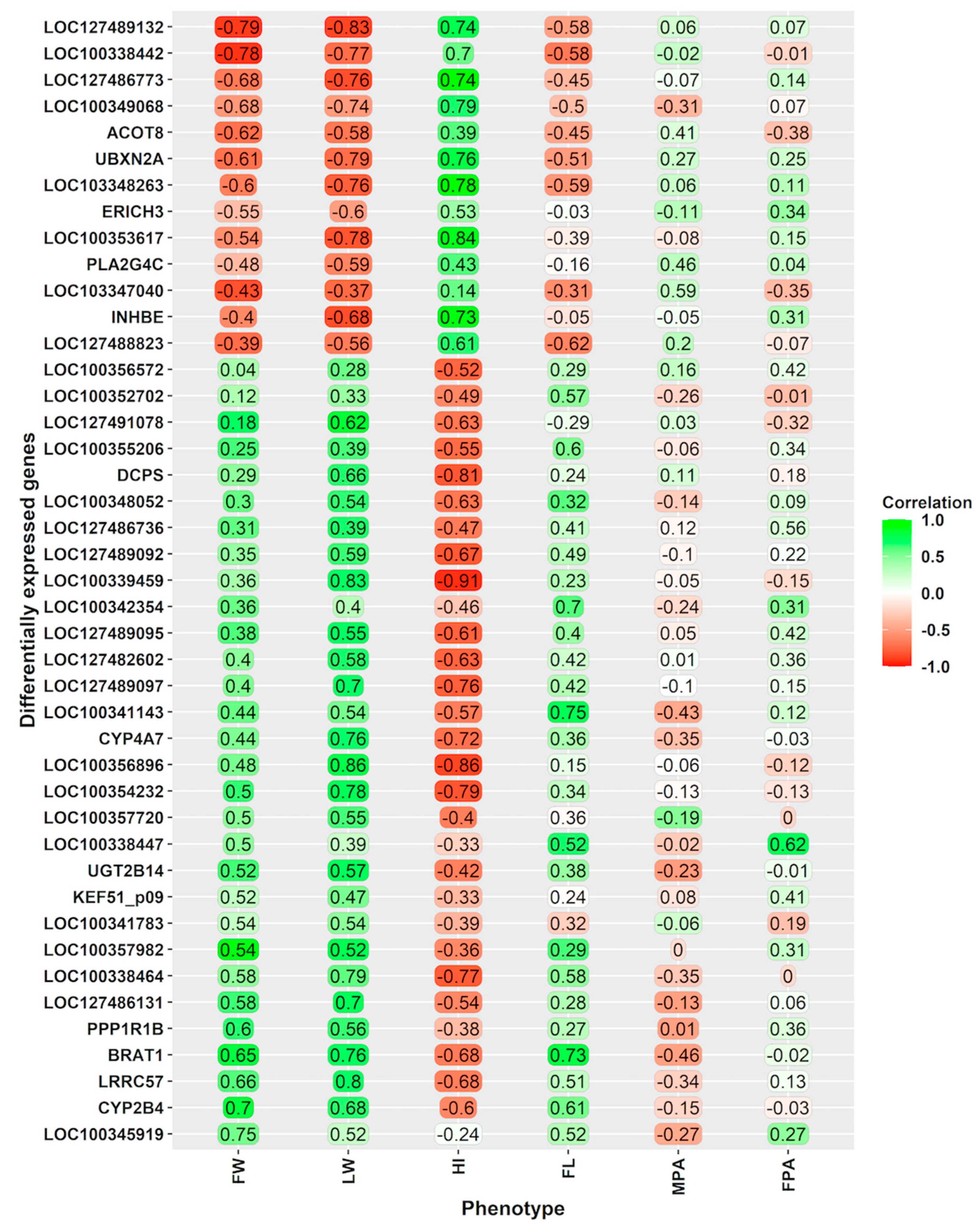
3.4. Relationships of the Gene Expression with Phenotypical Traits, Including Foetal-Placental Measurements
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duranthon, V.; Chavatte-Palmer, P. Long term effects of ART: What do animals tell us? Mol. Reprod. Dev. 2018, 85, 348–368. [Google Scholar] [CrossRef]
- Feuer, S.; Rinaudo, P. From Embryos to Adults: A DOHaD Perspective on In Vitro Fertilization and Other Assisted Reproductive Technologies. Healthcare 2016, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Novakovic, B.; Lewis, S.; Halliday, J.; Kennedy, J.; Burgner, D.P.; Czajko, A.; Kim, B.; Sexton-Oates, A.; Juonala, M.; Hammarberg, K.; et al. Assisted reproductive technologies are associated with limited epigenetic variation at birth that largely resolves by adulthood. Nat. Commun. 2019, 10, 3922. [Google Scholar] [CrossRef]
- Ramos-Ibeas, P.; Heras, S.; Gómez-Redondo, I.; Planells, B.; Fernández-González, R.; Pericuesta, E.; Laguna-Barraza, R.; Pérez-Cerezales, S.; Gutiérrez-Adán, A. Embryo responses to stress induced by assisted reproductive technologies. Mol. Reprod. Dev. 2019, 86, 1292–1306. [Google Scholar] [CrossRef]
- García-Martínez, S.; Sánchez Hurtado, M.; Gutiérrez, S.; Sánchez Margallo, F.M.; Romar, R.; Latorre, R.; Coy, P.; López Albors, O. Mimicking Physiological O2 Tension in the Female Reproductive Tract Improves Assisted Reproduction Outcomes in Pig. Mol. Hum. Reprod. 2018, 24, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.Y.B.; Mingels, R.; Morgan, H.; Macklon, N.; Cheong, Y.C. In Vivo Oxygen, Temperature and pH Dynamics in the Female Reproductive Tract and Their Importance in Human Conception: A Systematic Review. Hum. Reprod. Update 2018, 24, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Kohda, T. Effects of embryonic manipulation and epigenetics. J. Hum. Genet. 2013, 58, 416–420. [Google Scholar] [CrossRef]
- Canovas, S.; Ross, P.J.; Kelsey, G.; Coy, P. DNA Methylation in Embryo Development: Epigenetic Impact of ART (Assisted Reproductive Technologies). BioEssays 2017, 39, 1700106. [Google Scholar] [CrossRef]
- Canovas, S.; Ivanova, E.; Romar, R.; García-Martínez, S.; Soriano-Úbeda, C.; García-Vázquez, F.A.; Saadeh, H.; Andrews, S.; Kelsey, G.; Coy, P. DNA methylation and gene expression changes derived from assisted reproductive technologies can be decreased by reproductive fluids. eLife 2017, 6, e23670. [Google Scholar] [CrossRef]
- Chen, W.; Peng, Y.; Ma, X.; Kong, S.; Tan, S.; Wei, Y.; Zhao, Y.; Zhang, W.; Wang, Y.; Yan, L.; et al. Integrated multi-omics reveal epigenomic disturbance of assisted reproductive technologies in human offspring. EBioMedicine 2020, 61, 103076. [Google Scholar] [CrossRef]
- Ivanova, E.; Canovas, S.; Garcia-Martínez, S.; Romar, R.; Lopes, J.S.; Rizos, D.; Sanchez-Calabuig, M.J.; Krueger, F.; Andrews, S.; Perez-Sanz, F.; et al. DNA methylation changes during preimplantation development reveal inter-species differences and reprogramming events at imprinted genes. Clin. Epigenetics 2020, 12, 64. [Google Scholar] [CrossRef] [PubMed]
- García-Domínguez, X.; Diretto, G.; Peñaranda, D.S.; Frusciante, S.; García-Carpintero, V.; Cañizares, J.; Vicente, J.S.; Marco-Jiménez, F. Early Embryo Exposure to Assisted Reproductive Manipulation Induced Subtle Changes in Liver Epigenetics with No Apparent Negative Health Consequences in Rabbit. Int. J. Mol. Sci. 2021, 22, 9716. [Google Scholar] [CrossRef] [PubMed]
- Fleming, T.P.; Watkins, A.J.; Velazquez, M.A.; Mathers, J.C.; Prentice, A.M.; Stephenson, J.; Barker, M.; Saffery, R.; Yajnik, C.S.; Eckert, J.J.; et al. Origins of lifetime health around the time of conception: Causes and consequences. Lancet 2018, 391, 1842–1852. [Google Scholar] [CrossRef] [PubMed]
- Roseboom, T.J. Developmental plasticity and its relevance to assisted human reproduction. Hum. Reprod. 2018, 33, 546–552. [Google Scholar] [CrossRef]
- Avilés, M.; Gutiérrez-Adán, A.; Coy, P. Oviductal secretions: Will they be key factors for the future ARTs? Mol. Hum. Reprod. 2010, 16, 896–906. [Google Scholar] [CrossRef]
- Li, S.; Winuthayanon, W. Oviduct: Roles in fertilization and early embryo development. J. Endocrinol. 2017, 232, R1–R26. [Google Scholar] [CrossRef] [PubMed]
- Rizos, D.; Maillo, V.; Sánchez-Calabuig, M.J.; Lonergan, P. The Consequences of maternal-embryonic cross talk during the periconception period on subsequent embryonic development. Adv. Exp. Med. Biol. 2017, 1014, 69–86. [Google Scholar] [PubMed]
- Garcia-Domínguez, X.; Marco-Jiménez, F.; Peñaranda, D.S.; Diretto, G.; Garcia-Carpintero, V.; Cañizares, J.; Vicente, J.S. Long-term and transgenerational phenotypic, transcriptional and metabolic effects in rabbit males born following vitrified embryo transfer. Sci. Rep. 2020, 10, 11313. [Google Scholar] [CrossRef] [PubMed]
- Lavara, R.; Baselga, M.; Marco-Jiménez, F.; Vicente, J.S. Embryo vitrification in rabbits: Consequences for progeny growth. Theriogenology 2015, 84, 674–680. [Google Scholar] [CrossRef]
- Saenz-De-Juano, M.D.; Marco-Jimenez, F.; Schmaltz-Panneau, B.; Jimenez-Trigos, E.; Viudes-De-Castro, M.P.; Penaranda, D.S.; Jouneau, L.; Lecardonnel, J.; Lavara, R.; Naturil-Alfonso, C.; et al. Vitrification alters rabbit foetal placenta at transcriptomic and proteomic level. Reproduction 2014, 147, 789–801. [Google Scholar] [CrossRef]
- Vicente, J.S.; Saenz-de-Juano, M.D.; Jiménez-Trigos, E.; Viudes-de-Castro, M.P.; Peñaranda, D.S.; Marco-Jiménez, F. Rabbit morula vitrification reduces early foetal growth and increases losses throughout gestation. Cryobiology 2013, 67, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Marco-Jiménez, F.; Garcia-Domínguez, X.; Domínguez-Martínez, M.; Viudes-de-Castro, M.P.; Diretto, G.; Peñaranda, D.S.; Vicente, J.S. Effect of embryo vitrification on the steroid biosynthesis of liver tissue in rabbit offspring. Int. J. Mol. Sci. 2020, 21, 8642. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Le, F.; Wang, N.; Li, L.; Liu, X.Z.; Zheng, Y.M.; Lou, H.Y.; Xu, X.R.; Chen, Y.L.; Zhu, X.M.; et al. Alteration of fatty acid metabolism in the liver, adipose tissue, and testis of male mice conceived through assisted reproductive technologies: Fatty acid metabolism in ART mice. Lipids Health Dis. 2013, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Feuer, S.K.; Donjacour, A.; Simbulan, R.K.; Lin, W.; Liu, X.; Maltepe, E.; Rinaudo, P.F. Sexually dimorphic effect of In Vitro Fertilization (IVF) on adult mouse fat and liver metabolomes. Endocrinology 2014, 155, 4554–4567. [Google Scholar] [CrossRef]
- Liang, Y.U.; Jiang, X.C.; Liu, R.; Liang, G.; Beyer, T.P.; Gao, H.; Ryan, T.P.; Shuyu, D.L.; Eacho, P.I.; Cao, G. Liver X receptors (LXRs) regulate apolipoprotein AIV-Implications of the antiatherosclerotic effect of LXR agonists. Mol. Endocrinol. 2004, 18, 2000–2010. [Google Scholar] [CrossRef]
- Belva, F.; Bonduelle, M.; Provyn, S.; Painter, R.C.; Tournaye, H.; Roelants, M.; De Schepper, J. Metabolic Syndrome and Its Components in Young Adults Conceived by ICSI. Int. J. Endocrinol. 2018, 2018, 8170518. [Google Scholar] [CrossRef]
- Bessenfelder, U.; Brem, G. Laparoscopic embryo transfer in rabbits. Reproduction 1993, 99, 53–56. [Google Scholar] [CrossRef]
- García-Dominguez, X.; Marco-Jimenez, F.; Viudes-de-Castro, M.P.; Vicente, J.S. Minimally invasive embryo transfer and embryo vitrification at the optimal embryo stage in rabbit model. J. Vis. Exp. 2019, 147, e58055. [Google Scholar] [CrossRef]
- Juárez, J.D.; Marco-Jiménez, F.; Vicente, J.S. Evaluation of foetal growth, litter size and reproductive performance in rabbit after 18 generations of selection for growth rate using cryopreserved embryos. Livest. Sci. 2021, 253, 104702. [Google Scholar] [CrossRef]
- Juárez, J.D.; Marco-Jiménez, F.; Vicente, J.S. Effects of rederivation by embryo vitrification on performance in a rabbit paternal line. Front. Anim. Sci. 2022, 3, 909446. [Google Scholar]
- Overbey, E.G.; Saravia-Butler, A.M.; Zhang, Z.; Rathi, K.S.; Fogle, H.; da Silveira, W.A.; Barker, R.J.; Bass, J.J.; Beheshti, A.; Berrios, D.C.; et al. NASA GeneLab RNA-seq consensus pipeline: Standardized processing of short-read RNA-seq data. iScience 2021, 24, 102361. [Google Scholar] [CrossRef]
- Andrews, S. FASTQC. A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 June 2024).
- Krueger, F. TrimGalore: A Wrapper around Cutadapt and FastQC to Consistently Apply Adapter and Quality Trimming to FastQ Files, with Extra Functionality for RRBS Data. 2021. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 10 June 2024).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- McKay, J.A.; Xie, L.; Adriaens, M.; Evelo, C.T.; Ford, D.; Mathers, J.C. Organ-Specific Gene Expression Changes in the Fetal Liver and Placenta in Response to Maternal Folate Depletion. Nutrients 2016, 8, 661. [Google Scholar] [CrossRef]
- Marco-Jiménez, F.; Baselga, M.; Vicente, J.S. Successful re-establishment of a rabbit population from embryos vitrified 15 years ago: The importance of biobanks in livestock conservation. PLoS ONE 2018, 13, e0199234. [Google Scholar]
- Mocé, M.L.; Blasco, A.; Santacreu, M.A. In vivo development of vitrified rabbit embryos: Effects on prenatal survival and placental development. Theriogenology 2010, 73, 704–710. [Google Scholar] [CrossRef]
- Mushahary, D.; Gautam, P.; Sundaram, C.S.; Sirdeshmukh, R. Expanded protein expression profile of human placenta using two-dimentional gel electrophoresis. Placenta 2013, 34, 193–196. [Google Scholar] [CrossRef]
- Landers, K.A.; Mortimer, R.H.; Richard, K. Transthyretin and the human placenta. Placenta 2013, 34, 513–517. [Google Scholar] [CrossRef]
- Leese, H.J.; Guerif, F.; Allgar, V.; Brison, D.R.; Lundin, K.; Sturmey, R.G. Biological optimization, the Goldilocks principle, and how much is lagom in the preimplantation embryo. Mol. Reprod. Dev. 2016, 83, 748–754. [Google Scholar] [CrossRef]
- Marco-Jiménez, F.; Lavara, R.; Jiménez-Trigos, E.; Vicente, J.S. In vivo development of vitrified rabbit embryos: Effects of vitrification device, recipient genotype, and asynchrony. Theriogenology 2013, 79, 1124–1129. [Google Scholar] [CrossRef]
- Loughran, P.A.; Roman, L.J.; Aitken, A.E.; Miller, R.T.; Masters, B.S. Identification of unique amino acids that modulate CYP4A7 activity. Biochemistry 2000, 39, 15110–15120. [Google Scholar] [CrossRef]
- Yoon, M.; Madden, M.C.; Barton, H.A. Developmental Expression of Aldehyde Dehydrogenase in Rat: A Comparison of Liver and Lung Development. Toxicol. Sci. 2006, 89, 386–398. [Google Scholar] [CrossRef]
- Naganuma, T.; Takagi, S.; Kanetake, T.; Kitamura, T.; Hattori, S.; Miyakawa, T.; Sassa, T.; Kihara, A. Disruption of the Sjögren-Larsson Syndrome Gene Aldh3a2 in Mice Increases Keratinocyte Growth and Retards Skin Barrier Recovery. J. Biol. Chem. 2016, 291, 11676–11688. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, R.; Delgado, M.E.; Gloe, V.; Schuetz, K.; Plazzo, A.P.; Franke, B.; San Phan, T.; Fleming, J.; Mayans, O.; Brunner, T. Liver receptor homolog-1 (NR5A2) orchestrates hepatic inflammation and TNF-induced cell death. Cell Rep. 2023, 42, 113513. [Google Scholar] [CrossRef]
- Miles, H.L.; Hofman, P.L.; Peek, J.; Harris, M.; Wilson, D.; Robinson, E.M.; Gluckman, P.D.; Cutfield, W.S. In vitro fertilization improves childhood growth and metabolism. J. Clin. Endocrinol. Metab. 2007, 92, 3441–3445. [Google Scholar] [CrossRef]
- Guo, X.Y.; Liu, X.M.; Jin, L.; Wang, T.T.; Ullah, K.; Sheng, J.Z.; Huang, H.F. Cardiovascular and metabolic profiles of ofspring conceived by assisted reproductive technologies: A systematic review and meta-analysis. Fertil. Steril. 2017, 107, 622–631. [Google Scholar] [CrossRef]
- Sawado, A.; Ezoe, K.; Miki, T.; Ohata, K.; Amagai, A.; Shimazaki, K.; Okimura, T.; Kato, K. Fatty acid supplementation during warming improves pregnancy outcomes after frozen blastocyst transfers: A propensity score-matched study. Sci. Rep. 2024, 14, 9343. [Google Scholar] [CrossRef]
- Zhang, Y.; O’Leary, M.N.; Peri, S.; Wang, M.; Zha, J.; Melov, S.; Kappes, D.J.; Feng, Q.; Rhodes, J.; Amieux, P.S.; et al. Ribosomal Proteins Rpl22 and Rpl22l1 Control Morphogenesis by Regulating Pre-mRNA Splicing. Cell Rep. 2017, 18, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.E. Peptidyl-prolyl isomerases: A new twist to transcription. EMBO Rep. 2002, 3, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Dolan, J.; Walshe, K.; Alsbury, S.; Hokamp, K.; O’Keeffe, S.; Okafuji, T.; Miller, S.F.; Tear, G.; Mitchell, K.J. The extracellular Leucine-Rich Repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genom. 2007, 8, 320. [Google Scholar] [CrossRef]
- Smith, R.D.; Lupashin, V.V. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr. Res. 2008, 343, 2024–2031. [Google Scholar] [CrossRef] [PubMed]
- Shears, S.B.; Gokhale, N.A.; Wang, H.; Zaremba, A. Diphosphoinositol polyphosphates: What are the mechanisms? Adv. Enzyme. Regul. 2011, 51, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Hanes, S.D. Prolyl isomerases in gene transcription. Biochim. Biophys. Acta 2015, 1850, 2017–2034. [Google Scholar] [CrossRef] [PubMed]
- Meunier, E.; Broz, P. Interferon-inducible GTPases in cell autonomous and innate immunity. Cell. Microbiol. 2016, 18, 168–180. [Google Scholar] [CrossRef]
- Li, N.; Kong, M.; Zeng, S.; Hao, C.; Li, M.; Li, L.; Xu, Z.; Zhu, M.; Xu, Y. Brahma related gene 1 (Brg1) contributes to liver regeneration by epigenetically activating the Wnt/β-catenin pathway in mice. FASEB J. 2019, 33, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Fahl, S.P.; Sertori, R.; Zhang, Y.; Contreras, A.V.; Harris, B.; Wang, M.; Perrigoue, J.; Balachandran, S.; Kennedy, B.K.; David, L.; et al. Loss of Ribosomal Protein Paralog Rpl22-like1 Blocks Lymphoid Development without Affecting Protein Synthesis. J. Immunol. 2022, 208, 870–880. [Google Scholar] [CrossRef]
- Zhao, W.; Zhou, Y.; Li, C.; Bi, Y.; Wang, K.; Ye, M.; Li, H. Molecular basis for protein histidine N1-specific methylation of the “His-x-His” motifs by METTL9. Cell Insight 2023, 2, 100090. [Google Scholar] [CrossRef]
- Onda, H.; Yoshikawa, J. Alpha1-acid glycoprotein as a hepatocyte-specific mitosis-inhibiting protein in regenerating rat liver. Gan 1975, 66, 227–235. [Google Scholar] [PubMed]
- Ray, B.K.; Gao, X.; Ray, A. Regulation of rabbit alpha 1-acid glycoprotein gene expression in acute-phase liver. Identification of inducible and constitutive proteins like CCAAT-enhancer binding protein that interact with the 5′-proximal promoter elements. Eur. J. Biochem. 1993, 216, 127–136. [Google Scholar] [CrossRef] [PubMed]
- So, E.Y.; Ouchi, T. BRAT1 deficiency causes increased glucose metabolism and mitochondrial malfunction. BMC Cancer 2014, 14, 548. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xun, Y.; Rong, S.; Yan, L.; Sorelle, J.A.; Li, X.; Tang, M.; Keller, K.; Ludwig, S.; Moresco, E.M.; et al. Loss of immunity-related GTPase GM4951 leads to nonalcoholic fatty liver disease without obesity. Nat. Commun. 2022, 13, 4136. [Google Scholar] [CrossRef]
- Colantuono, R.; D’Acunto, E.; Melis, D.; Vajro, P.; Freeze, H.H.; Mandato, C. Liver Involvement in Congenital Disorders of Glycosylation: A Systematic Review. J. Pediatr. Gastroenterol. Nutr. 2021, 73, 444–454. [Google Scholar] [CrossRef]

| Group | n | Survival | Foetal Weight (g) | Placenta Weight (g) | |||
|---|---|---|---|---|---|---|---|
| Body | Liver | Hepatosomatic Index 1 | Foetal | Maternal | |||
| Fresh | 49 | 0.84 ± 0.053 a | 13.3 ± 0.43 a | 1.28 ± 0.063 a | 11.0 ± 0.46 | 2.7 ± 0.12 | 1.32 ± 0.103 |
| Vitrified | 65 | 0.66 ± 0.067 b | 11.4 ± 0.43 b | 1.06 ± 0.064 b | 11.8 ± 0.45 | 2.8 ± 0.12 | 1.19 ± 0.106 |
| Group | Embryos Transferred | Survival at Birth | Birth Weight 1 (g) (n) | Live Birth Weight 1 (g) (n) |
|---|---|---|---|---|
| Vitrified | 146 | 0.47 ± 0.055 b | 59.2 ± 2.55 (68) | 56.4 3.12 (64) |
| Fresh | 81 | 0.71 ± 0.058 a | 49.8 ± 2.90 (56) | 52.6 3.51 (51) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vicente, J.S.; Valdés-Hernández, J.; Marco-Jiménez, F. Transcriptomic Signatures of the Foetal Liver and Late Prenatal Development in Vitrified Rabbit Embryos. Vet. Sci. 2024, 11, 347. https://doi.org/10.3390/vetsci11080347
Vicente JS, Valdés-Hernández J, Marco-Jiménez F. Transcriptomic Signatures of the Foetal Liver and Late Prenatal Development in Vitrified Rabbit Embryos. Veterinary Sciences. 2024; 11(8):347. https://doi.org/10.3390/vetsci11080347
Chicago/Turabian StyleVicente, José Salvador, Jesús Valdés-Hernández, and Francisco Marco-Jiménez. 2024. "Transcriptomic Signatures of the Foetal Liver and Late Prenatal Development in Vitrified Rabbit Embryos" Veterinary Sciences 11, no. 8: 347. https://doi.org/10.3390/vetsci11080347
APA StyleVicente, J. S., Valdés-Hernández, J., & Marco-Jiménez, F. (2024). Transcriptomic Signatures of the Foetal Liver and Late Prenatal Development in Vitrified Rabbit Embryos. Veterinary Sciences, 11(8), 347. https://doi.org/10.3390/vetsci11080347






