An Evaluation of the Lineage of Brucella Isolates in Turkey by a Whole-Genome Single-Nucleotide Polymorphism Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of Bacterial Isolates
2.2. Brucella Isolation from Sample Materials and DNA Extraction
2.3. Whole-Genome Sequencing and Bioinformatics Procedure of the Raw Reads
2.4. Species and Antimicrobial Resistance Determination
2.5. Canonical SNP Assay
2.6. Core-Genome SNP Genotyping
3. Results
3.1. Origin of B. melitensis and B. abortus Isolates
3.2. Antimicrobial Resistance
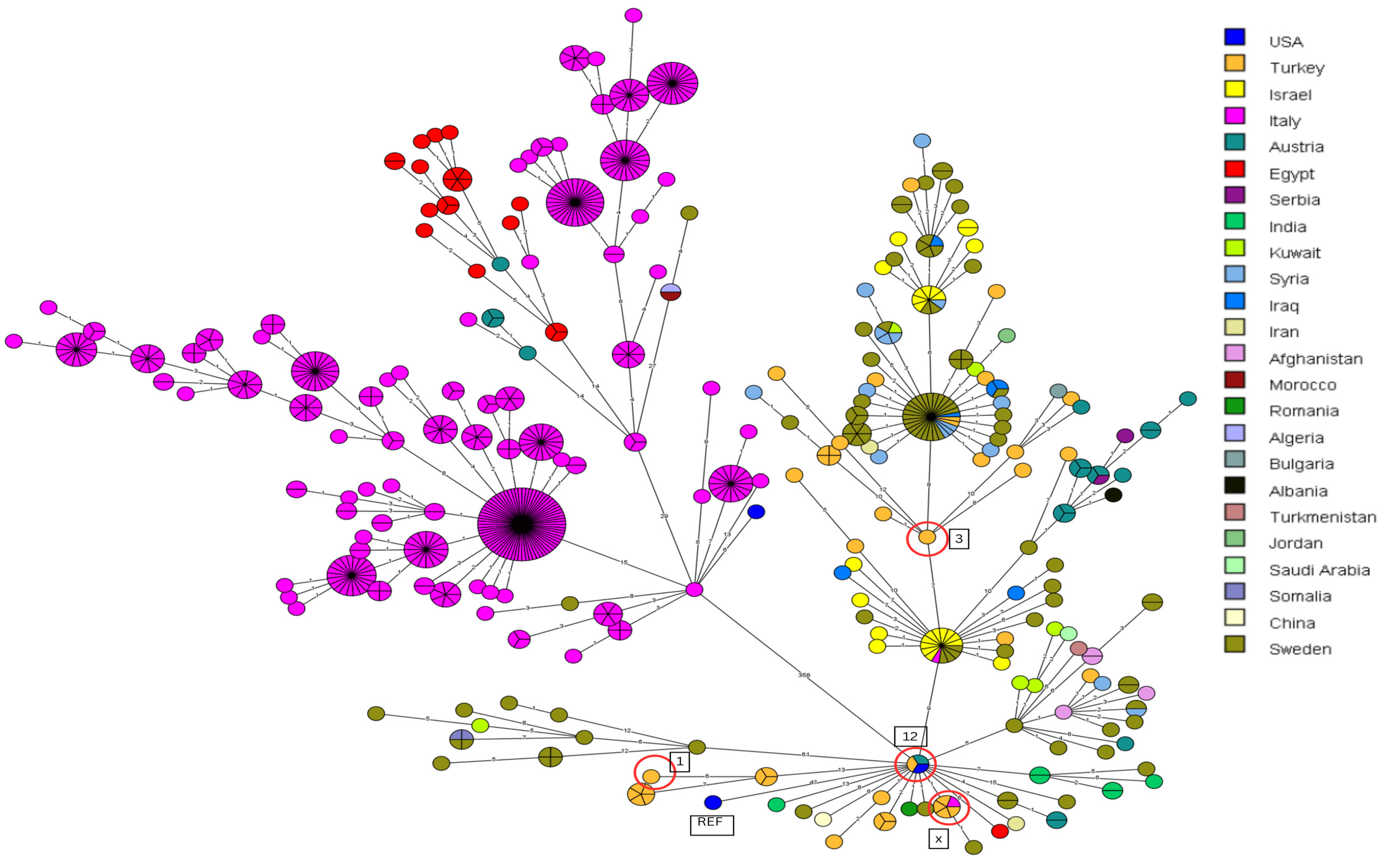
3.3. Canonical SNP Analysis
3.4. Core-Genome SNP Genotyping for B. melitensis
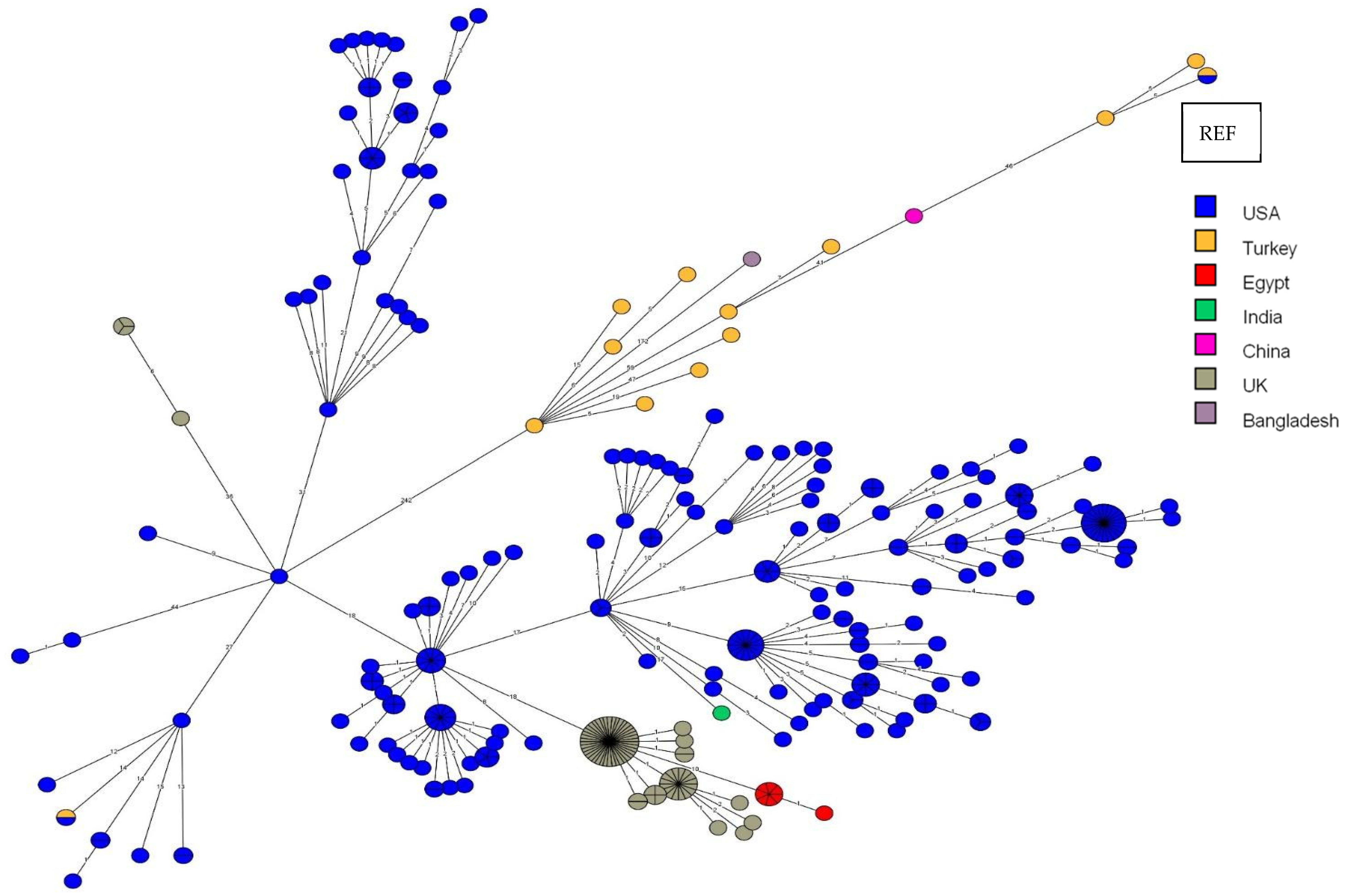
3.5. Core-Genome SNP Genotyping for B. abortus
4. Discussion
4.1. Comparison of B. melitensis Outbreak Strains to Strains from Other Countries
4.2. Comparison of B. abortus Outbreak Strains to Strains from Other Countries
4.3. Outlook
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- OIE. Brucellosis: Transmission and Spread. Available online: https://www.oie.int/en/disease/brucellosis/ (accessed on 22 September 2021).
- Occhialini, A.; Hofreuter, D.; Ufermann, C.M.; Al Dahouk, S.; Köhler, S. The Retrospective on Atypical Brucella Species Leads to Novel Definitions. Microorganisms 2022, 10, 813. [Google Scholar] [CrossRef]
- WOAH. Brucellosis (Infection with B. abortus, B. melitensis and B. suis). Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.01.04_BRUCELLOSIS.pdf (accessed on 23 October 2023).
- Holzer, K.; El-Diasty, M.; Wareth, G.; Abdel-Hamid, N.H.; Hamdy, M.E.R.; Moustafa, S.A.; Linde, J.; Bartusch, F.; Sayour, A.E.; Elbauomy, E.M.; et al. Tracking the Distribution of Brucella abortus in Egypt Based on Core Genome SNP Analysis and In Silico MLVA-16. Microorganisms 2021, 9, 1942. [Google Scholar] [CrossRef] [PubMed]
- Akar, K.; Tatar, F.; Schmoock, G.; Wareth, G.; Neubauer, H.; Erganiş, O. Tracking the diversity and Mediterranean lineage of Brucella melitensis isolates from different animal species in Turkey using MLVA-16 genotyping. Ger. J. Vet. Res. 2022, 2, 25–30. [Google Scholar] [CrossRef]
- Dadar, M.; Alamian, S.; Behrozikhah, A.M.; Yazdani, F.; Kalantari, A.; Etemadi, A.; Whatmore, A.M. Molecular identification of Brucella species and biovars associated with animal and human infection in Iran. Vet. Res. Forum 2019, 10, 315–321. [Google Scholar]
- Tuon, F.F.; Gondolfo, R.B.; Cerchiari, N. Human-to-human transmission of Brucella—A systematic review. Trop. Med. Int. Health 2017, 22, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Corbel, M.J. Brucellosis in Humans and Animals; World Health Organization: Geneva, Switzerland, 2006; ISBN 9789241547130. [Google Scholar]
- Akar, K.; Erganis, O. Evaluation of the genetic profiles of Brucella melitensis strain from Turkey using multilocus variable number tandem repeat analysis (MLVA) and multilocus sequence typing (MLST) techniques. Vet. Microbiol. 2022, 269, 109423. [Google Scholar] [CrossRef] [PubMed]
- Dorneles, E.M.S.; de Faria, A.P.P.; Pauletti, R.B.; Santana, J.A.; Caldeira, G.A.V.; Heinemann, M.B.; Titze-de-Almeida, R.; Lage, A.P. Genetic stability of Brucella abortus S19 and RB51 vaccine strains by multiple locus variable number tandem repeat analysis (MLVA16). Vaccine 2013, 31, 4856–4859. [Google Scholar] [CrossRef]
- Wareth, G.; El-Diasty, M.; Melzer, F.; Schmoock, G.; Moustafa, S.A.; El-Beskawy, M.; Khater, D.F.; Hamdy, M.E.R.; Zaki, H.M.; Ferreira, A.C.; et al. MLVA-16 Genotyping of Brucella abortus and Brucella melitensis Isolates from Different Animal Species in Egypt: Geographical Relatedness and the Mediterranean Lineage. Pathogens 2020, 9, 498. [Google Scholar] [CrossRef]
- Holzer, K.; Wareth, G.; El-Diasty, M.; Abdel-Hamid, N.H.; Hamdy, M.E.R.; Moustafa, S.A.; Linde, J.; Bartusch, F.; Abdel-Glil, M.Y.; Sayour, A.E.; et al. Tracking the distribution, genetic diversity, and lineage of Brucella melitensis recovered from humans and animals in Egypt based on core-genome SNP analysis and in silico MLVA-16. Transbound. Emerg. Dis. 2022, 69, 3952–3963. [Google Scholar] [CrossRef]
- Garofolo, G.; Di Giannatale, E.; Massis, F.D.E.; Zilli, K.; Ancora, M.; Cammà, C.; Calistri, P.; Foster, J.T. Investigating genetic diversity of Brucella abortus and Brucella melitensis in Italy with MLVA-16. Infect. Genet. Evol. 2013, 19, 59–70. [Google Scholar] [CrossRef]
- Alton, G.G.; Jones, L.M.; Angus, R.D.; Verger, J.M. Techniques for the Brucellosis Laboratory; Institute National de la Recherche Agronomique, INRA: Paris, France, 1988. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. Quast: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- García-Yoldi, D.; Marín, C.M.; De Miguel, M.J.; Muñoz, P.M.; Vizmanos, J.L.; López-Goñi, I. Multiplex PCR assay for the identification and differentiation of all Brucella species and the vaccine strains Brucella abortus S19 and RB51 and Brucella melitensis Rev1. Clin. Chem. 2006, 52, 779–781. [Google Scholar] [CrossRef] [PubMed][Green Version]
- López-Goñi, I.; García-Yoldi, D.; Marín, C.M.; De Miguel, M.J.; Muñoz, P.M.; Blasco, J.M.; Jacques, I.; Grayon, M.; Cloeckaert, A.; Ferreira, A.C.; et al. Evaluation of a multiplex PCR assay (Bruce-ladder) for molecular typing of all Brucella species, including the vaccine strains. J. Clin. Microbiol. 2008, 46, 3484–3487. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.I.; Her, M.; Kim, J.W.; Kim, J.Y.; Ko, K.Y.; Ha, Y.M.; Jung, S.C. Advanced multiplex PCR assay for differentiation of Brucella species. Appl. Environ. Microbiol. 2011, 77, 6726–6728. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, 10–1128. [Google Scholar] [CrossRef]
- Foster, J.T.; Walker, F.M.; Rannals, B.D.; Hussain, M.H.; Drees, K.P.; Tiller, R.V.; Hoffmaster, A.R.; Al-Rawahi, A.; Keim, P.; Saqib, M. African Lineage Brucella melitensis Isolates from Omani Livestock. Front. Microbiol. 2017, 8, 2702. [Google Scholar] [CrossRef]
- Yumuk, Z.; O’Callaghan, D. Brucellosis in Turkey—An overview. Int. J. Infect. Dis. 2012, 16, e228–e235. [Google Scholar] [CrossRef] [PubMed]
- Moriyón, I.; Blasco, J.M.; Letesson, J.J.; De Massis, F.; Moreno, E. Brucellosis and One Health: Inherited and Future Challenges. Microorganisms 2023, 11, 2070. [Google Scholar] [CrossRef]
- Kiliç, S.; Ivanov, I.N.; Durmaz, R.; Bayraktar, M.R.; Ayaslioglu, E.; Uyanik, M.H.; Aliskan, H.; Yasar, E.; Bayramoglu, G.; Arslantürk, A.; et al. Multiple-locus variable-number tandem-repeat analysis genotyping of human Brucella isolates from Turkey. J. Clin. Microbiol. 2011, 49, 3276–3283. [Google Scholar] [CrossRef]
- Zhu, X.; Zhao, Z.; Ma, S.; Guo, Z.; Wang, M.; Li, Z.; Liu, Z. Brucella melitensis, a latent “travel bacterium”, continual spread and expansion from Northern to Southern China and its relationship to worldwide lineages. Emerg. Microbes Infect. 2020, 9, 1618–1627. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, C.; Wei, K.; Zhao, Z.; Wang, M.; Li, D.; Wang, H.; Wei, Q.; Li, Z. Investigation of Genetic Relatedness of Brucella Strains in Countries Along the Silk Road. Front. Vet. Sci. 2020, 7, 539444. [Google Scholar] [CrossRef] [PubMed]
- Denk, A.; Demirdag, K.; Kalkan, A.; Ozden, M.; Cetinkaya, B.; Kilic, S.S. In vitro, activity of Brucella melitensis isolates to various antimicrobials in Turkey. Infect. Dis. 2015, 47, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Yücer, A.A.; Altıntaş, Ö. Turkeys animal selection and animal importation within the scopes of health and technical criteria. Bulg. J. Agric. Sci. 2021, 27, 972–979. [Google Scholar]
- De Massis, F.; Ali, R.M.; Serrani, S.; Toro, M.; Sferrella, A.; D’Aurelio, N.; Janowicz, A.; Zilli, K.; Romualdi, T.; Felicioni, E.; et al. Genetic Diversity of Brucella melitensis Isolated from Domestic Ruminants in Iraq. Microorganisms 2024, 12, 475. [Google Scholar] [CrossRef] [PubMed]
- Danış, D. Yeni Göç Hareketleri ve Türkiye. Available online: https://www.academia.edu/1427180/Yeni_G%C3%B6%C3%A7_Hareketleri_ve_T%C3%BCrkiye (accessed on 15 September 2004).
- Kavak, G. Körfez Savaşı Sonrası Irak’tan Türkiye’ye Göçler ve Sonuçları. Master’s Thesis, Marmara University, Istanbul, Turkey, 2011. [Google Scholar]
- Sacchini, L.; Wahab, T.; Di Giannatale, E.; Zilli, K.; Abass, A.; Garofolo, G.; Janowicz, A. Whole Genome Sequencing for Tracing Geographical Origin of Imported Cases of Human Brucellosis in Sweden. Microorganisms 2019, 7, 398. [Google Scholar] [CrossRef] [PubMed]
- Kay, G.L.; Sergeant, M.J.; Giuffra, V.; Bandiera, P.; Milanese, M.; Bramanti, B.; Bianucci, R.; Pallen, M.J. Recovery of a medieval Brucella melitensis genome using shotgun metagenomics. MBio 2014, 5, 10–1128. [Google Scholar] [CrossRef]
- Schaeffer, J.; Revilla-Fernández, S.; Hofer, E.; Posch, R.; Stoeger, A.; Leth, C.; Schmoll, F.; Djordjevic, V.; Lakicevic, B.; Matovic, K.; et al. Tracking the Origin of Austrian Human Brucellosis Cases Using Whole Genome Sequencing. Front. Med. 2021, 8, 635547. [Google Scholar] [CrossRef]
- Georgi, E.; Walter, M.C.; Pfalzgraf, M.-T.; Northoff, B.H.; Holdt, L.M.; Scholz, H.C.; Zoeller, L.; Zange, S.; Antwerpen, M.H. Whole genome sequencing of Brucella melitensis isolated from 57 patients in Germany reveals high diversity in strains from Middle East. PLoS ONE 2017, 12, e0175425. [Google Scholar] [CrossRef]
- Janowicz, A.; de Massis, F.; Ancora, M.; Cammà, C.; Patavino, C.; Battisti, A.; Prior, K.; Harmsen, D.; Scholz, H.; Zilli, K.; et al. Core Genome Multilocus Sequence Typing and Single Nucleotide Polymorphism Analysis in the Epidemiology of Brucella melitensis Infections. J. Clin. Microbiol. 2018, 56, 10–1128. [Google Scholar] [CrossRef]
- Dal, T.; Kara, S.S.; Cikman, A.; Balkan, C.E.; Acıkgoz, Z.C.; Zeybek, H.; Uslu, H.; Durmaz, R. Comparison of multiplex real-time polymerase chain reaction with serological tests and culture for diagnosing human brucellosis. J. Infect. Public Health 2019, 12, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Shevtsov, A.; Cloeckaert, A.; Berdimuratova, K.; Shevtsova, E.; Shustov, A.V.; Amirgazin, A.; Karibayev, T.; Kamalova, D.; Zygmunt, M.S.; Ramanculov, Y.; et al. Brucella abortus in Kazakhstan, population structure and comparison with worldwide genetic diversity. Front. Microbiol. 2023, 14, 1106994. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.R.; Neia, R.C.; Silva, S.B.; Williamson, C.H.D.; Gillece, J.D.; O’Callaghan, D.; Foster, J.T.; Oliveira, I.R.C.; Bueno Filho, J.S.S.; Lage, A.P.; et al. Comparison of Brucella abortus population structure based on genotyping methods with different levels of resolution. J. Microbiol. Methods 2023, 211, 106772. [Google Scholar] [CrossRef]
- Allen, A.R.; Milne, G.; Drees, K.; Presho, E.; Graham, J.; McAdam, P.; Jones, K.; Wright, L.; Skuce, R.; Whatmore, A.M.; et al. Genomic epizootiology of a Brucella abortus outbreak in Northern Ireland (1997–2012). Infect. Genet. Evol. 2020, 81, 104235. [Google Scholar] [CrossRef] [PubMed]
- Hikal, A.F.; Wareth, G.; Khan, A. Brucellosis: Why is it eradicated from domestic livestock in the United States but not in the Nile River Basin countries? Ger. J. Microbiol. 2023, 3, 19–25. [Google Scholar] [CrossRef]

| Strain ID | Species | Source of Isolation | Host | Geographical Area | Year of Isolation |
|---|---|---|---|---|---|
| bru1 | B. melitensis | Organ | Cattle | Mersin | 2021 |
| bru2 | B. melitensis | Organ | Cattle | Erzurum | 2018 |
| bru3 | B. melitensis | Organ | Cattle | Kars | 2012 |
| bru4 | B. melitensis | Organ | Sheep | Istanbul | 2019 |
| bru8 | B. melitensis | Organ | Cattle | Elazığ | 2019 |
| bru10 | B. melitensis | Organ | Cattle | Şanlıurfa | 2019 |
| bru12 | B. melitensis | Organ | Sheep | Izmir | 2019 |
| bru14 | B. melitensis | Organ | Cattle | Kars | 2020 |
| bru16 | B. melitensis | Organ | Cattle | Bayburt | 2020 |
| bru17 | B. melitensis | Organ | Goat | unknown | 2020 |
| bru18 | B. melitensis | Organ | Sheep | Edirne | 2021 |
| bru20 | B. melitensis | Organ | Cattle | Erzurum | 2021 |
| bru23 | B. melitensis | Organ | Human | Istanbul | 2022 |
| bru25 | B. melitensis | Organ | Sheep | Kırıkkale | 2022 |
| bru27 | B. melitensis | Organ | Sheep | Canakkale | 2022 |
| bru5 | B. abortus | Organ | Cattle | Aksaray | 2019 |
| bru6 | B. abortus | Organ | Cattle | Ankara | 2019 |
| bru7 | B. abortus | Organ | Buffalo | Düzce | 2019 |
| bru9 | B. abortus | Milk | Cattle | Kırklareli | 2019 |
| bru11 | B. abortus | Organ | Buffalo | Samsun | 2019 |
| bru13 | B. abortus | Organ | Sheep | Konya | 2019 |
| bru15 | B. abortus | Organ | Cattle | Erzincan | 2020 |
| bru19 | B. abortus | Organ | Cattle | Bursa | 2021 |
| bru21 | B. abortus | Organ | Sheep | Erzurum | 2018 |
| bru22 | B. abortus | Organ | Buffalo | Kocaeli | 2018 |
| bru24 | B. abortus | Organ | Cattle | Bilecik | 2022 |
| bru26 | B. abortus | Organ | Cattle | Adana | 2022 |
| bru28 | B. abortus | Organ | Cattle | Kars | 2022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akar, K.; Holzer, K.; Hoelzle, L.E.; Yıldız Öz, G.; Abdelmegid, S.; Baklan, E.A.; Eroğlu, B.; Atıl, E.; Moustafa, S.A.; Wareth, G.; et al. An Evaluation of the Lineage of Brucella Isolates in Turkey by a Whole-Genome Single-Nucleotide Polymorphism Analysis. Vet. Sci. 2024, 11, 316. https://doi.org/10.3390/vetsci11070316
Akar K, Holzer K, Hoelzle LE, Yıldız Öz G, Abdelmegid S, Baklan EA, Eroğlu B, Atıl E, Moustafa SA, Wareth G, et al. An Evaluation of the Lineage of Brucella Isolates in Turkey by a Whole-Genome Single-Nucleotide Polymorphism Analysis. Veterinary Sciences. 2024; 11(7):316. https://doi.org/10.3390/vetsci11070316
Chicago/Turabian StyleAkar, Kadir, Katharina Holzer, Ludwig E. Hoelzle, Gülseren Yıldız Öz, Shaimaa Abdelmegid, Emin Ayhan Baklan, Buket Eroğlu, Eray Atıl, Shawky A. Moustafa, Gamal Wareth, and et al. 2024. "An Evaluation of the Lineage of Brucella Isolates in Turkey by a Whole-Genome Single-Nucleotide Polymorphism Analysis" Veterinary Sciences 11, no. 7: 316. https://doi.org/10.3390/vetsci11070316
APA StyleAkar, K., Holzer, K., Hoelzle, L. E., Yıldız Öz, G., Abdelmegid, S., Baklan, E. A., Eroğlu, B., Atıl, E., Moustafa, S. A., Wareth, G., & Elkhayat, M. (2024). An Evaluation of the Lineage of Brucella Isolates in Turkey by a Whole-Genome Single-Nucleotide Polymorphism Analysis. Veterinary Sciences, 11(7), 316. https://doi.org/10.3390/vetsci11070316








