Characterization and Function Analysis of miRNA Editing during Fat Deposition in Chinese Indigenous Ningxiang Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Filtration and Comparison of Sequencing Data
2.3. Identification and Characterization of miRNA Editing Sites
2.4. Screening of Differential miRNA Editing Sites
2.5. Prediction of Target Genes and KEGG Functional Enrichment Analysis
3. Results
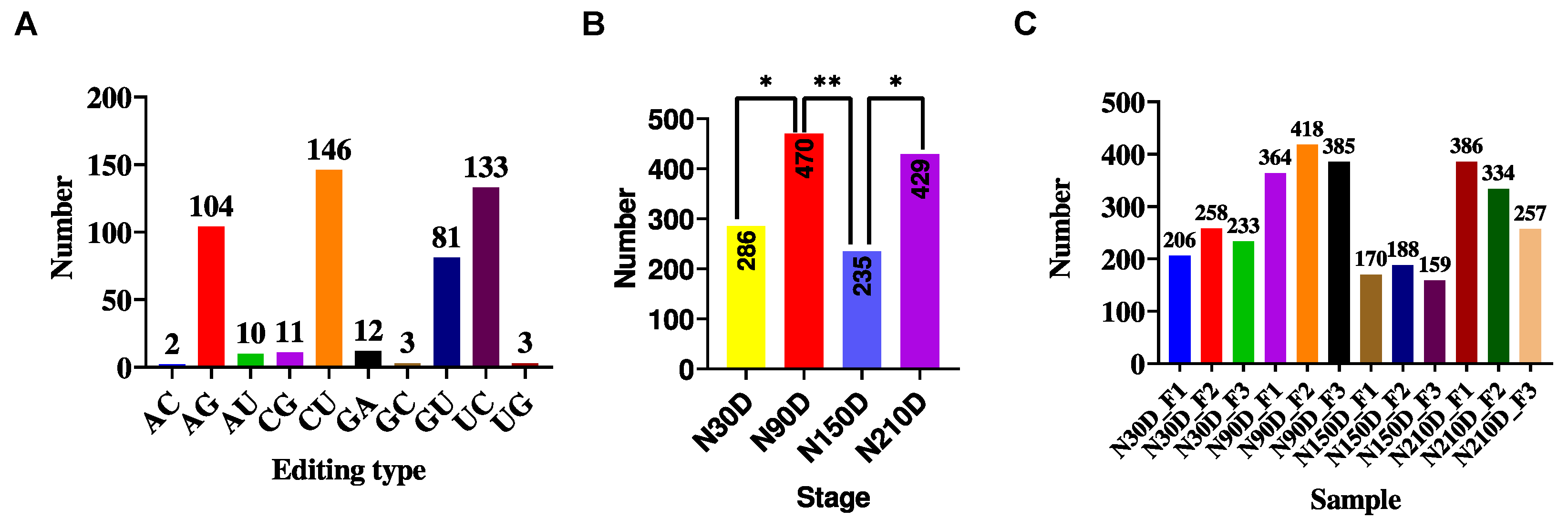
3.1. Identification of miRNA Editing Sites
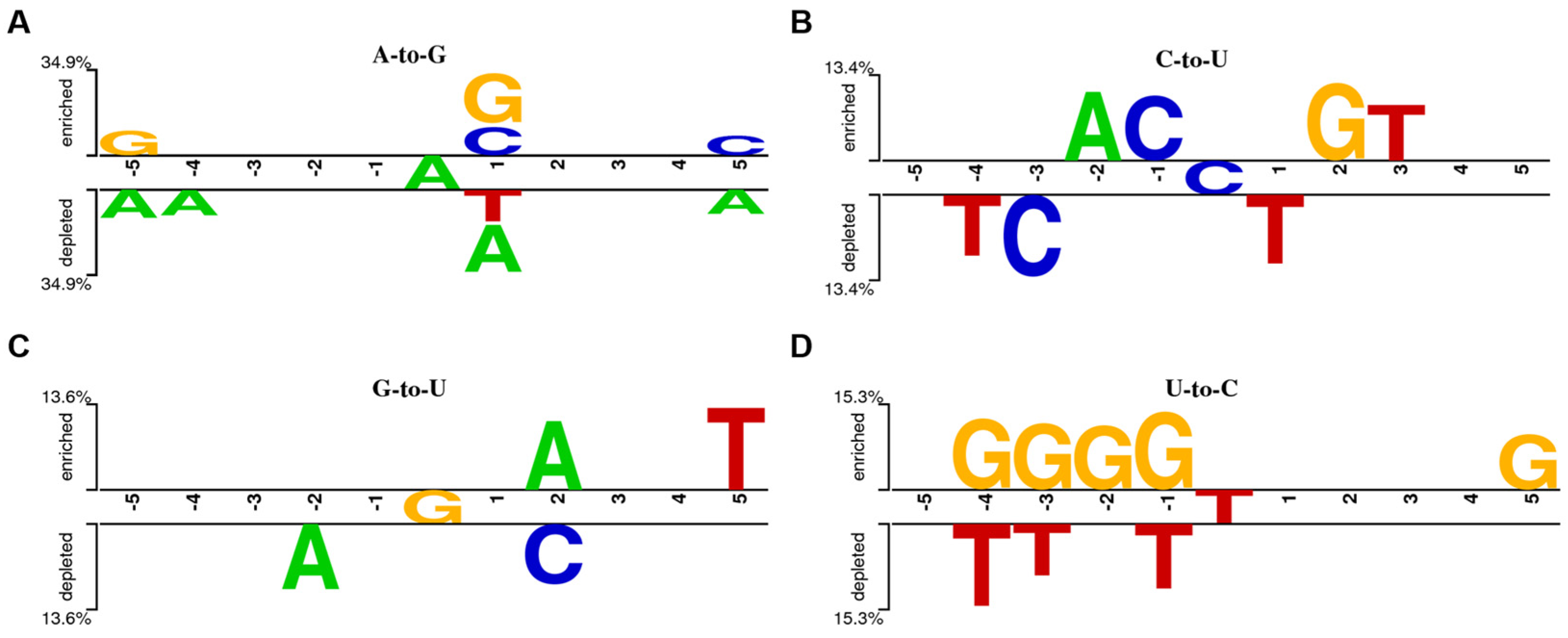
3.2. miRNA Editing Characteristic Analysis
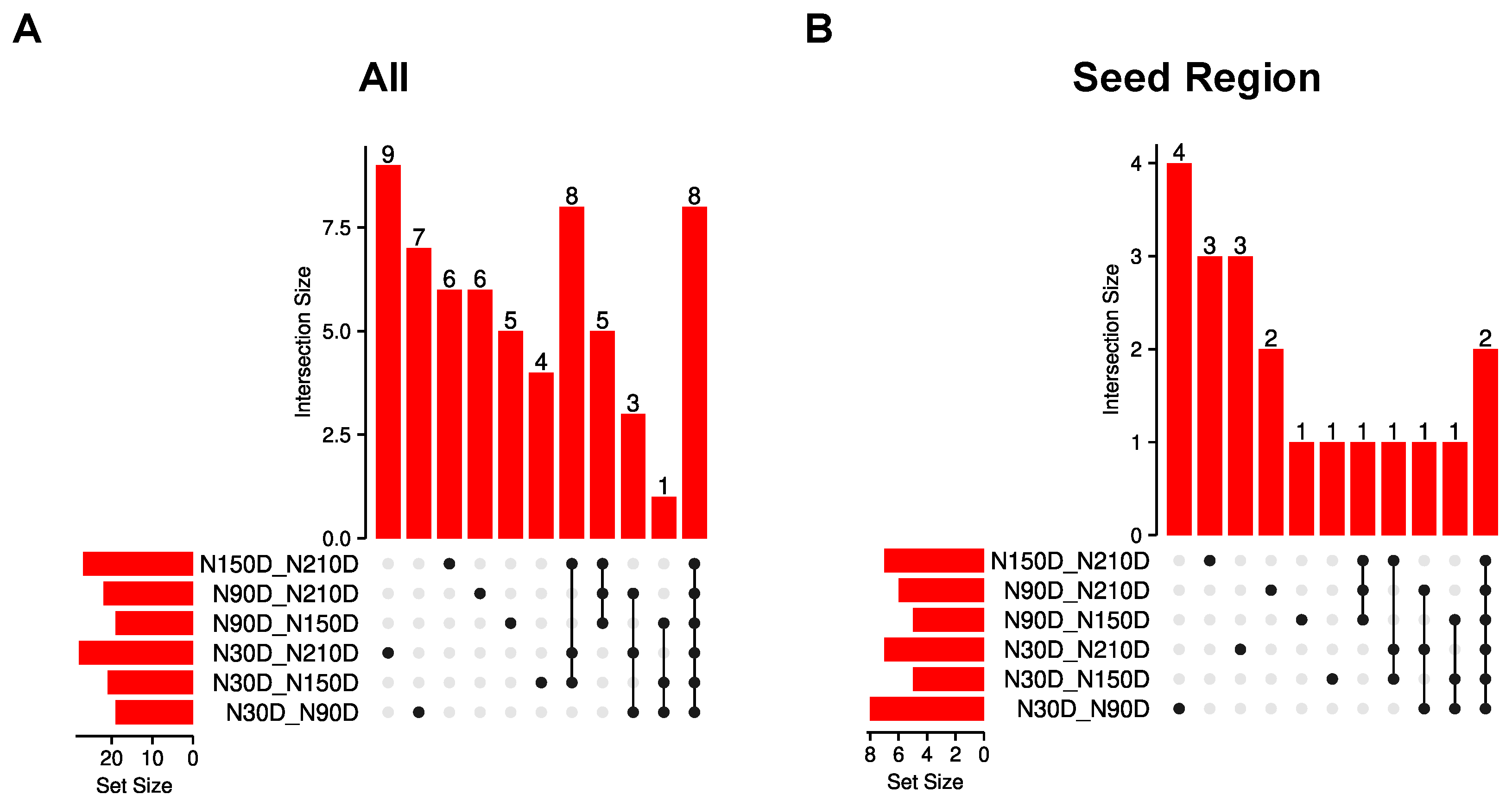
3.3. Identification of Differential Editing Sites
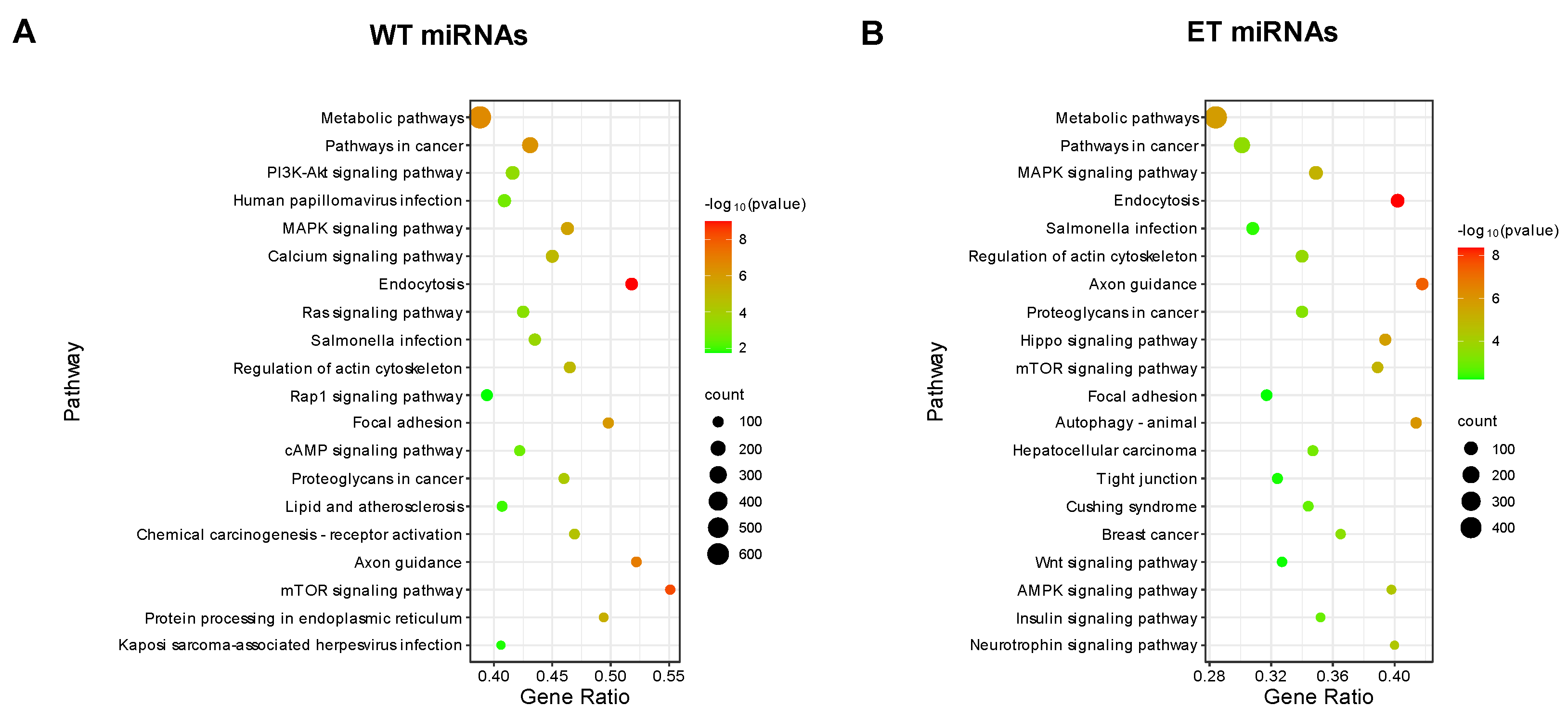
3.4. Target Gene Prediction and Functional Enrichment Analysis for A-to-G Editing Site Host
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, B.; Gao, H.; Yang, F.; Li, Y.; Yang, Q.; Liao, Y.; Guo, H.; Xu, K.; Tang, Z.; Gao, N.; et al. Comparative Characterization of Volatile Compounds of Ningxiang Pig, Duroc and Their Crosses (Duroc × Ningxiang) by Using SPME-GC-MS. Foods 2023, 12, 1059. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Wang, Z.; Li, J.; Yang, H.; Yin, Y.; Tan, B.; Chen, J. Comparative Microbial Profiles of Colonic Digesta between Ningxiang Pig and Large White Pig. Animals 2021, 11, 1862. [Google Scholar] [CrossRef] [PubMed]
- Benne, R.; Van den Burg, J.; Brakenhoff, J.P.; Sloof, P.; Van Boom, J.H.; Tromp, M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 1986, 46, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Sandhoff, R.; Sandhoff, K. Emerging concepts of ganglioside metabolism. FEBS Lett. 2018, 592, 3835–3864. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Vesely, C.; Tauber, S.; Sedlazeck, F.J.; Tajaddod, M.; Haeseler, A.V.; Jantsch, M.F. ADAR2 induces reproducible changes in sequence and abundance of mature microRNAs in the mouse brain. Nucleic Acids Res. 2014, 42, 12155–12168. [Google Scholar] [CrossRef]
- Feng, H.; Liu, T.; Yousuf, S.; Zhang, X.; Huang, W.; Li, A.; Xie, L.; Miao, X. Identification of potential miRNA-mRNA regulatory network and the key miRNAs in intramuscular and subcutaneous adipose. Front. Vet. Sci. 2022, 9, 976603. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, W.; Bai, Y.; Yang, W.; Ling, Y.; Fang, M. ssc-miR-7134-3p regulates fat accumulation in castrated male pigs by targeting MARK4 gene. Int. J. Biol. Sci. 2017, 13, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xie, Y.; Chen, W.; Zhang, Y.; Zeng, Y. miR-34a Regulates Lipid Droplet Deposition in 3T3-L1 and C2C12 Cells by Targeting LEF1. Cells 2022, 12, 167. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Chao, M.; Zhao, T.; Li, R.; Zhang, Z.; Yan, W.; Pang, W. miR-503 targets MafK to inhibit subcutaneous preadipocyte adipogenesis causing a decrease of backfat thickness in Guanzhong Black pigs. Meat Sci. 2023, 198, 109116. [Google Scholar] [CrossRef]
- Luciano, D.J.; Mirsky, H.; Vendetti, N.J.; Maas, S. RNA editing of a miRNA precursor. RNA 2004, 10, 1174–1177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, X.; Cui, Y.; Zhang, X. Suppression of RNA editing by miR-17 inhibits the stemness of melanoma stem cells. Mol. Ther. Nucleic Acids 2022, 27, 439–455. [Google Scholar] [CrossRef] [PubMed]
- Meadows, S.; Seidler, A.; Wall, M.; Page, J.; Taylor, C.; Flinn, B.; Turner, R.; Santanam, N. Altered Regulation of adipomiR Editing with Aging. Int. J. Mol. Sci. 2020, 21, 6899. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yang, J.; He, J.; Gong, Y.; Xiao, Y.; Zeng, Q.; Xu, K.; Duan, Y.; He, J.; Ma, H. Spatiotemporal Regulation and Functional Analysis of Circular RNAs in Skeletal Muscle and Subcutaneous Fat during Pig Growth. Biology 2021, 10, 841. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.; Tomaru, Y.; Nishibu, T.; Ukekawa, R.; Funakoshi, T.; Kurokawa, T.; Suzuki, H.; Hayashizaki, Y.; et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010, 20, 1398–1410. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010, 24, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Alon, S.; Eisenberg, E. Identifying RNA editing sites in miRNAs by deep sequencing. Methods Mol. Biol. 2013, 1038, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.F.M. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef]
- Marceca, G.P.; Distefano, R.; Tomasello, L.; Lagana, A.; Russo, F.; Calore, F.; Romano, G.; Bagnoli, M.; Gasparini, P.; Ferro, A.; et al. MiREDiBase, a manually curated database of validated and putative editing events in microRNAs. Sci. Data 2021, 8, 199. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014, 47, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Pesole, G. REDItools: High-throughput RNA editing detection made easy. Bioinformatics 2013, 29, 1813–1814. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, P.; Li, L.; Che, D.; Li, T.; Li, H.; Li, Q.; Jia, H.; Tao, S.; Hua, J.; et al. miRNA editing landscape reveals miR-34c regulated spermatogenesis through structure and target change in pig and mouse. Biochem. Biophys. Res. Commun. 2018, 502, 486–492. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Lei, W.; Ge, C.; Du, P.; Wang, L.; Li, F. Large-scale detection and analysis of adenosine-to-inosine RNA editing during development in Plutella xylostella. Mol. Genet. Genom. 2015, 290, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Zamyatnin, A.J.; Lyamzaev, K.G.; Zinovkin, R.A. A-to-I RNA editing: A contribution to diversity of the transcriptome and an organism’s development. Biochemistry 2010, 75, 1316–1323. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhao, C.; Xu, Y.; Huang, K.; Wang, Y.; Wang, X.; Zhou, X.; Pang, W.; Yang, G.; Yu, T. Adipose-specific BMP and activin membrane-bound inhibitor (BAMBI) deletion promotes adipogenesis by accelerating ROS production. J. Biol. Chem. 2021, 296, 100037. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chen, X.; Zhao, C.; Wang, X.; Cheng, Y.; Xi, F.; Yao, X.; Zhang, L.; Yang, G.; Yu, T. MiR-99b-5p Attenuates Adipogenesis by Targeting SCD1 and Lpin1 in 3T3-L1 Cells. J. Agric. Food Chem. 2021, 69, 2564–2575. [Google Scholar] [CrossRef] [PubMed]
- Ekdahl, Y.; Farahani, H.S.; Behm, M.; Lagergren, J.; Ohman, M. A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res. 2012, 22, 1477–1487. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, X.; Zhang, L.; Wang, L.; He, J.; Ma, H.; Wang, L. Preliminary identification and analysis of differential RNA editing between higher and lower backfat thickness pigs using DNA-seq and RNA-seq data. Anim. Genet. 2022, 53, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Pinto, Y.; Buchumenski, I.; Levanon, E.Y.; Eisenberg, E. Human cancer tissues exhibit reduced A-to-I editing of miRNAs coupled with elevated editing of their targets. Nucleic Acids Res. 2018, 46, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Li, H.; Yang, J.; Shen, X.; Song, C.; Yang, Z.; Wang, X.; Huang, Y.; Lan, X.; Lei, C.; et al. circRNA Profiling Reveals an Abundant circFUT10 that Promotes Adipocyte Proliferation and Inhibits Adipocyte Differentiation via Sponging let-7. Mol. Ther. Nucleic Acids 2020, 20, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ji, B.; Song, R.; Wang, S.; Li, T.; Zhang, X.; Chen, K.; Li, T.; Li, J. Accurate detection for a wide range of mutation and editing sites of microRNAs from small RNA high-throughput sequencing profiles. Nucleic Acids Res. 2016, 44, e123. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, A.Y.; Lee, H.W.; Son, Y.H.; Lee, G.Y.; Lee, J.W.; Lee, Y.S.; Kim, J.B. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem. Biophys. Res. Commun. 2010, 392, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, X.; Li, Y.; Jia, L.; Zhai, L.; Wei, W.; Zhang, L.; Jiang, H.; Bai, Y. Exercise-Induced Browning of White Adipose Tissue and Improving Skeletal Muscle Insulin Sensitivity in Obese/Non-obese Growing Mice: Do Not Neglect Exosomal miR-27a. Front. Nutr. 2022, 9, 940673. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, D.; Peng, C.; Gao, R.; Li, X.; Zhang, L.; Lv, Q.; Xiao, X.; Li, Q. MicroRNA-27a, downregulated in human obesity, exerts an antiapoptotic function in adipocytes. Endocr. J. 2023, 70, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ma, X.F.; Dong, M.Z.; Tan, J.; Zhang, J.; Zhuang, L.K.; Liu, S.S.; Xin, Y.N. MiR-30b-5p regulates the lipid metabolism by targeting PPARGC1A in Huh-7 cell line. Lipids Health Dis. 2020, 19, 76. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Xu, H.; Fang, J.; Zhang, C.; Song, J.; Zhang, X.; Hao, B.; Yin, B.; Xia, G. miR-15a regulates the preadipocyte differentiation by targeting ABAT gene in Yanbian yellow cattle. Anim. Biotechnol. 2023, 34, 2343–2352. [Google Scholar] [CrossRef]
- Wu, Q.; Yang, H.; Tai, R.; Li, C.; Xia, T.; Liu, Y.; Sun, C. Lnc-hipk1 inhibits mouse adipocyte apoptosis as a sponge of miR-497. Biofactors 2022, 48, 135–147. [Google Scholar] [CrossRef]
- Chu, M.; Zhao, Y.; Yu, S.; Hao, Y.; Zhang, P.; Feng, Y.; Zhang, H.; Ma, D.; Liu, J.; Cheng, M.; et al. miR-15b negatively correlates with lipid metabolism in mammary epithelial cells. Am. J. Physiol. Cell Physiol. 2018, 314, C43–C52. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.F.; Xiong, Y.; Peng, Y.; Gao, Y.; Qin, J.; Chu, G.Y.; Pang, W.J.; Yang, G.S. miR-425-5p Inhibits Differentiation and Proliferation in Porcine Intramuscular Preadipocytes. Int. J. Mol. Sci. 2017, 18, 2101. [Google Scholar] [CrossRef]
- Diniz, T.A.; de Lima, J.E.; Teixeira, A.A.; Biondo, L.A.; Da, R.L.; Valadao, I.C.; Silveira, L.S.; Cabral-Santos, C.; de Souza, C.O.; Rosa, N.J. Aerobic training improves NAFLD markers and insulin resistance through AMPK-PPAR-alpha signaling in obese mice. Life Sci. 2021, 266, 118868. [Google Scholar] [CrossRef] [PubMed]
- Neopane, K.; Kozlov, N.; Negoita, F.; Murray-Segal, L.; Brink, R.; Hoque, A.; Ovens, A.J.; Tjin, G.; McAloon, L.M.; Yu, D.; et al. Blocking AMPK beta1 myristoylation enhances AMPK activity and protects mice from high-fat diet-induced obesity and hepatic steatosis. Cell Rep. 2022, 41, 111862. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.W.; Park, H.S.; Choi, G.H.; Kim, D.; Lee, T. beta-aminoisobutyric acid attenuates LPS-induced inflammation and insulin resistance in adipocytes through AMPK-mediated pathway. J. Biomed. Sci. 2018, 25, 27. [Google Scholar] [CrossRef] [PubMed]
- Frosig, C.; Jensen, T.E.; Jeppesen, J.; Pehmoller, C.; Treebak, J.T.; Maarbjerg, S.J.; Kristensen, J.M.; Sylow, L.; Alsted, T.J.; Schjerling, P.; et al. AMPK and insulin action--responses to ageing and high fat diet. PLoS ONE 2013, 8, e62338. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Sun, X.; Lin, Z.; Yang, Y.; Zhang, M.; Xu, Z.; Liu, P.; Liu, Z.; Huang, H. Gentiopicroside targets PAQR3 to activate the PI3K/AKT signaling pathway and ameliorate disordered glucose and lipid metabolism. Acta Pharm. Sin. B 2022, 12, 2887–2904. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; You, W.; Chen, W.; Zhou, Y.; Nong, Q.; Valencak, T.G.; Wang, Y.; Shan, T. Single-cell RNA sequencing and lipidomics reveal cell and lipid dynamics of fat infiltration in skeletal muscle. J. Cachexia Sarcopenia Muscle 2021, 12, 109–129. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhang, Y.; Liu, Y.; Zhu, D.; Yu, J.; Li, G.; Sun, Z.; Wang, W.; Jiang, H.; Hong, Z. MiR-27a promotes insulin resistance and mediates glucose metabolism by targeting PPAR-gamma-mediated PI3K/AKT signaling. Aging 2019, 11, 7510–7524. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Wang, J. Wnt/beta-Catenin Signaling and Obesity. Front. Physiol. 2018, 9, 792. [Google Scholar] [CrossRef] [PubMed]
- Warnefors, M.; Liechti, A.; Halbert, J.; Valloton, D.; Kaessmann, H. Conserved microRNA editing in mammalian evolution, development and disease. Genome Biol. 2014, 15, R83. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.; Liang, X.; Wu, X.; Liu, J.; Yang, S.; Tao, C.; Zhang, J.; Tian, J.; Zhao, J.; et al. Stearoyl-CoA Desaturase is Essential for Porcine Adipocyte Differentiation. Int. J. Mol. Sci. 2020, 21, 2446. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chai, J.; Wang, Y.; Gu, Y.; Long, K.; Li, M.; Jin, L. LncPLAAT3-AS Regulates PLAAT3-Mediated Adipocyte Differentiation and Lipogenesis in Pigs through miR-503-5p. Genes 2023, 14, 161. [Google Scholar] [CrossRef] [PubMed]
- Lan, Q.; Liufu, S.; Liu, X.; Ai, N.; Xu, X.; Li, X.; Yu, Z.; Yin, Y.; Liu, M.; Ma, H. Comprehensive analysis of transcriptomic and metabolomic profiles uncovered the age-induced dynamic development pattern of subcutaneous fat in Ningxiang pig. Gene 2023, 880, 147624. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, X.; Yang, Z.; Zhang, Q.; Hao, W.; Pang, Y.; Zhang, D.; Liu, D. Transcriptional Regulation Associated with Subcutaneous Adipogenesis in Porcine ACSL1 Gene. Biomolecules 2023, 13, 1057. [Google Scholar] [CrossRef]
- Ma, H.; Zhang, S.; Zhang, K.; Zhan, H.; Peng, X.; Xie, S.; Li, X.; Zhao, S.; Ma, Y. Identifying Selection Signatures for Backfat Thickness in Yorkshire Pigs Highlights New Regions Affecting Fat Metabolism. Genes 2019, 10, 254. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Zheng, M.; Zhang, J.; Ye, Y.; Duan, M.; Chamba, Y.; Wang, Z.; Shang, P. Transcriptomics-Based Study of Differentially Expressed Genes Related to Fat Deposition in Tibetan and Yorkshire Pigs. Front. Vet. Sci. 2022, 9, 919904. [Google Scholar] [CrossRef] [PubMed]
- Renaville, B.; Prandi, A.; Fan, B.; Sepulcri, A.; Rothschild, M.F.; Piasentier, E. Candidate gene marker associations with fatty acid profiles in heavy pigs. Meat Sci. 2013, 93, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Zhang, H.; Hua, Z.; Zhu, Z.; Tao, J.; Xiao, H.; Zhang, L.; Bi, Y.; Wang, H. ACSL4 Directs Intramuscular Adipogenesis and Fatty Acid Composition in Pigs. Animals 2022, 12, 119. [Google Scholar] [CrossRef] [PubMed]
- Ballester, M.; Ramayo-Caldas, Y.; Revilla, M.; Corominas, J.; Castello, A.; Estelle, J.; Fernandez, A.I.; Folch, J.M. Integration of liver gene co-expression networks and eGWAs analyses highlighted candidate regulators implicated in lipid metabolism in pigs. Sci. Rep. 2017, 7, 46539. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Onteru, S.K.; Rothschild, M.F. The GGT1 and IGFBP5 genes are associated with fat deposition traits in the pig (Brief Report). Arch. Anim. Breed. 2009, 52, 337–339. [Google Scholar] [CrossRef]
- Shang, P.; Li, W.; Liu, G.; Zhang, J.; Li, M.; Wu, L.; Wang, K.; Chamba, Y. Identification of lncRNAs and Genes Responsible for Fatness and Fatty Acid Composition Traits between the Tibetan and Yorkshire Pigs. Int. J. Genom. 2019, 2019, 5070975. [Google Scholar] [CrossRef] [PubMed]
- Dione, N.; Lacroix, S.; Taschler, U.; Deschenes, T.; Abolghasemi, A.; Leblanc, N.; Di Marzo, V.; Silvestri, C. Mgll Knockout Mouse Resistance to Diet-Induced Dysmetabolism Is Associated with Altered Gut Microbiota. Cells 2020, 9, 2705. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, L.M.; Pereira, V.; Noronha, N.Y.; de Souza, P.M.; Wolf, L.S.; de Oliveira, C.C.; Placa, J.R.; Noma, I.; Da, S.R.G.; de Souza, V.; et al. The influence of serum selenium in differential epigenetic and transcriptional regulation of CPT1B gene in women with obesity. J. Trace Elem. Med. Biol. 2023, 83, 127376. [Google Scholar] [CrossRef] [PubMed]

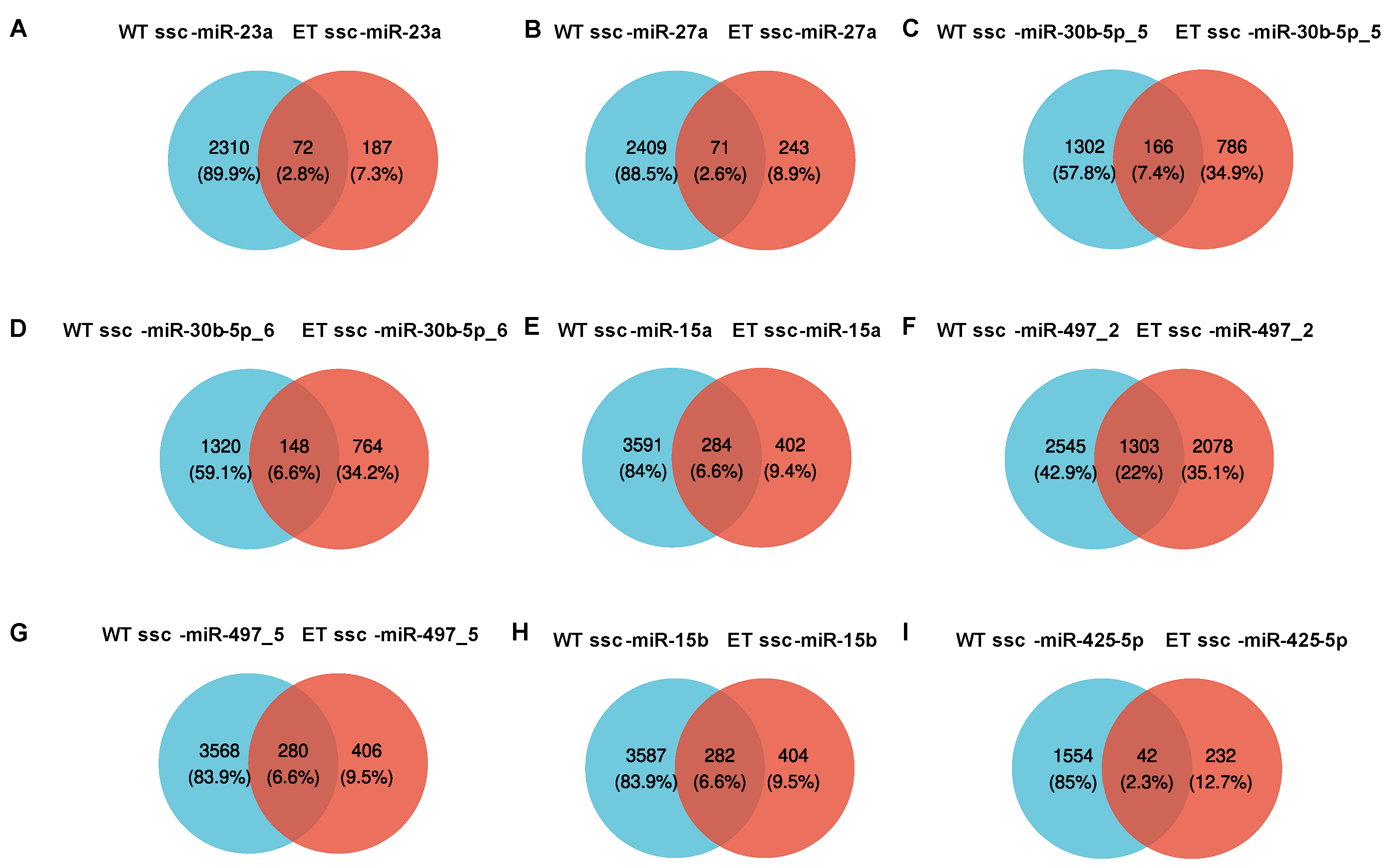
| Chromosome | Position | Mature miRNA Name | Position in miRNA | Editing Type |
|---|---|---|---|---|
| 2 | 65308161 | ssc-miR-23a | 4 | A-to-G |
| 2 | 65308350 | ssc-miR-27a | 4 | A-to-G |
| 4 | 6952809 | ssc-miR-30b-5p | 5 | A-to-G |
| 4 | 6952808 | ssc-miR-30b-5p | 5 | A-to-G |
| 11 | 17757478 | ssc-miR-15a | 5 | A-to-G |
| 12 | 52422400 | ssc-miR-497 | 2 | A-to-G |
| 12 | 52422397 | ssc-miR-497 | 5 | A-to-G |
| 13 | 100083195 | ssc-miR-15b | 5 | A-to-G |
| 13 | 31655056 | ssc-miR-425-5p | 7 | A-to-G |
| 2 | 65308351 | ssc-miR-27a | 5 | C-to-U |
| 12 | 46211541 | ssc-miR-423-5p | 8 | C-to-U |
| 14 | 6520933 | ssc-miR-320 | 6 | C-to-U |
| 1 | 224065570 | ssc-miR-204 | 2 | U-to-C |
| 2 | 150580147 | ssc-miR-145-5p | 7 | U-to-C |
| 2 | 65308354 | ssc-miR-27a | 8 | U-to-C |
| 6 | 58332107 | ssc-let-7e | 6 | U-to-C |
| 12 | 45088852 | ssc-miR-451 | 8 | U-to-C |
| 12 | 43337029 | ssc-miR-193a-5p | 5 | U-to-C |
| 13 | 189138833 | ssc-miR-155-5p | 2 | U-to-C |
| 15 | 120453426 | ssc-miR-26b-5p | 7 | U-to-C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, J.; Yang, F.; Li, Y.; Gao, N.; Zeng, Q.; Ma, H.; He, J.; Zhang, Y. Characterization and Function Analysis of miRNA Editing during Fat Deposition in Chinese Indigenous Ningxiang Pigs. Vet. Sci. 2024, 11, 183. https://doi.org/10.3390/vetsci11040183
Lv J, Yang F, Li Y, Gao N, Zeng Q, Ma H, He J, Zhang Y. Characterization and Function Analysis of miRNA Editing during Fat Deposition in Chinese Indigenous Ningxiang Pigs. Veterinary Sciences. 2024; 11(4):183. https://doi.org/10.3390/vetsci11040183
Chicago/Turabian StyleLv, Jiayu, Fang Yang, Yiyang Li, Ning Gao, Qinghua Zeng, Haiming Ma, Jun He, and Yuebo Zhang. 2024. "Characterization and Function Analysis of miRNA Editing during Fat Deposition in Chinese Indigenous Ningxiang Pigs" Veterinary Sciences 11, no. 4: 183. https://doi.org/10.3390/vetsci11040183
APA StyleLv, J., Yang, F., Li, Y., Gao, N., Zeng, Q., Ma, H., He, J., & Zhang, Y. (2024). Characterization and Function Analysis of miRNA Editing during Fat Deposition in Chinese Indigenous Ningxiang Pigs. Veterinary Sciences, 11(4), 183. https://doi.org/10.3390/vetsci11040183





