Evidence Supporting Oral Hygiene Management by Owners through a Genetic Analysis of Dental Plaque Bacteria in Dogs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Recruitment of Dogs and Owners
2.2. Periodontal Examination of Dogs
2.3. Dog Owner’s Evaluation of the Dog’s Oral Condition
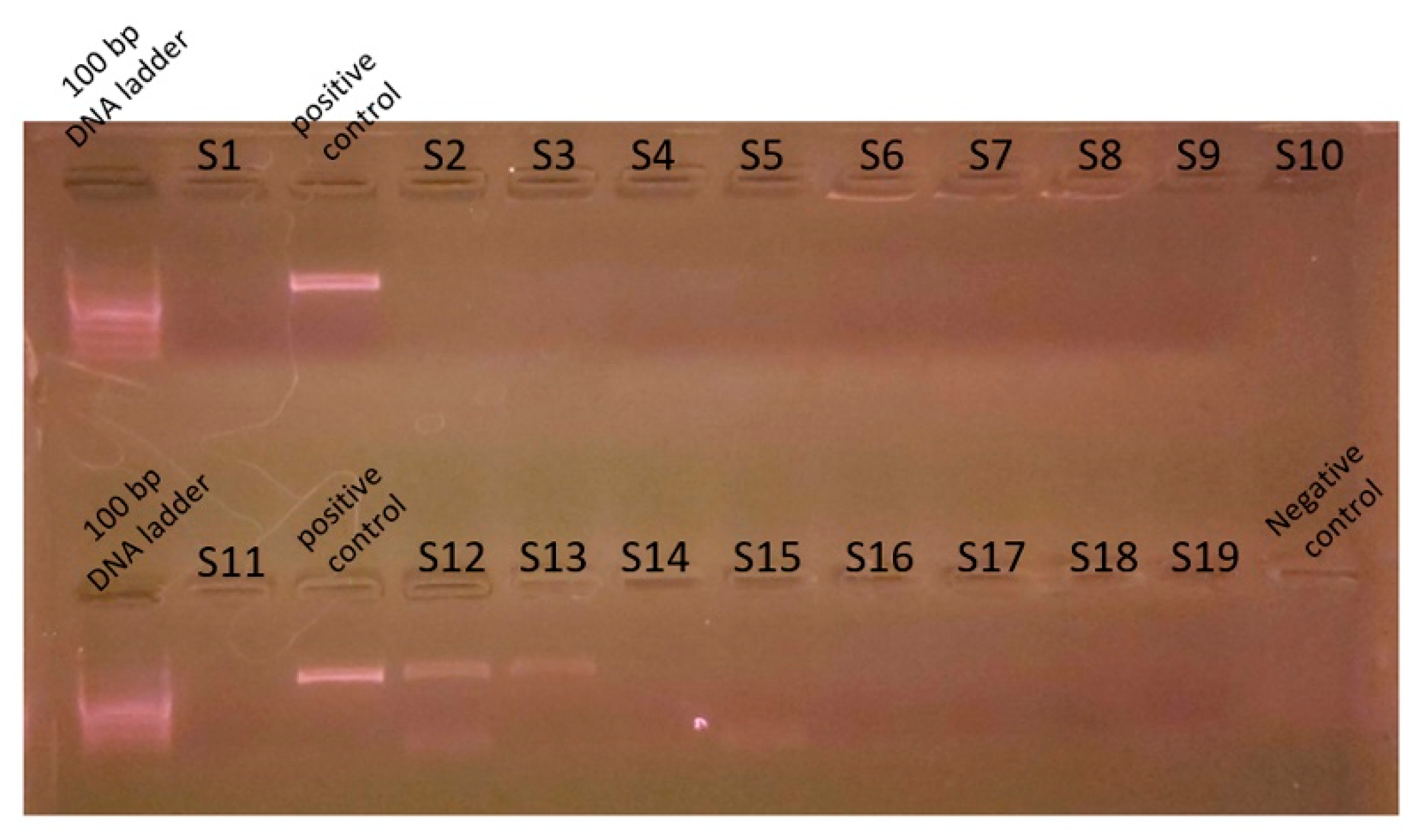
2.4. Dental Plaque Bacterial Culture and Polymerase Chain Reaction Analysis
2.5. Metagenome Amplicon Sequencing
2.6. Statistical Analysis
3. Results
3.1. Owner’s Awareness of Dog’s Oral Condition and the Degree of Tooth Brushing Practice
3.2. Genetic Analysis of Dental Plaque Bacteria of Dogs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- National Awareness Survey on Animal Protection. 2022 Animal Welfare Policy Division, Department of Agriculture and Life Policy, Food Industry Policy Office, Ministry of Agriculture, Food and Rural Affairs. Available online: https://www.korea.kr/briefing/pressReleaseView.do?newsId=156550504 (accessed on 2 February 2023).
- Hwang, W.K.; Son, K.P. 2021 Companion Animal Report in Korea. Available online: https://www.kbfg.com/kbresearch/report/reportView.do?reportId=2000160 (accessed on 3 March 2023).
- National Institute of Dental and Craniofacial Research. Dental Caries (Tooth Decay) in Adults (Ages 20 to 64 Years). Available online: https://www.nidcr.nih.gov/research/data-statistics/dental-caries/adults (accessed on 25 January 2023).
- Hale, F.A. Dental caries in the dog. Can. Vet. J. 2009, 50, 1301–1304. [Google Scholar] [CrossRef] [PubMed]
- Hale, F.A. Dental caries in the dog. J. Vet. Dent. 1998, 15, 79–83. [Google Scholar] [CrossRef]
- Berning, J.A.; Snyder, C.J.; Hetzel, S.; Sarment, D.P. Analysis of the Anatomic Relationship between the Mandibular First Molar Roots and Mandibular Canal Using Cone-Beam Computed-Tomography in 101 Dogs. Front. Vet. Sci. 2020, 2019, 485. [Google Scholar] [CrossRef] [PubMed]
- Nyvad, B.; Takahashi, N. Integrated hypothesis of dental caries and periodontal diseases. J. Oral. Microbiol. 2020, 12, 1710953. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.F.; Darke, B.H.; Keary, G.T. Dental caries in domesticated dogs. J. Am. Vet. Med. Assoc. 1962, 140, 433–436. [Google Scholar] [PubMed]
- Kwon, D.; Bae, K.; Kim, H.; Kim, S.H.; Lee, D.; Lee, J.H. Treponema denticola as a prognostic biomarker for periodontitis in dogs. PLoS ONE 2022, 17, e0262859. [Google Scholar] [CrossRef] [PubMed]
- Lavy, E.; Golani, Y.; Friedman, M.; Abram, T. Comparison of the distribution of oral cavity bacteria in various dog populations. Isr. J. Vet. Med. 2009, 64, 78–83. [Google Scholar]
- Kazuko, T.; Kazuhiko, H.; Masatomo, H. S. mutans from Dog Oral Cavity Was Transmitted from Human. Poster Discussion Session. Continental European and Israeli Divisions Meeting (28 September 2007, Thessaloniki, Greece). Available online: https://iadr.abstractarchives.com/abstract/israel07-96560/s-mutans-from-dog-oral-cavity-was-transmitted-from-human (accessed on 25 January 2023).
- Kim, S.S. Veterinary Dentistry as a Case Report. J. Vet. Clin. 2007, 24, 505–516. [Google Scholar]
- Cho, S.E.; Kwon, S.Y.; Kim, S.Y.; Park, S.Y.; Son, N.R. Evaluation of Canine Oral Health Status and Owners’ Awareness about Oral Care for Companion Dogs. Korean J. Clin. Dent. Hyg. 2018, 6, 17–27. [Google Scholar]
- Ramfjord, S.P. The Periodontal Disease Index (PDI). J. Periodontol. 1967, 38, 602–610. [Google Scholar] [CrossRef]
- Löe, H.; Silness, J. Periodontal Disease in Pregnancy. I. Prevalence and Severity. Acta Odontol. Scand. 1963, 21, 533–551. [Google Scholar] [CrossRef]
- Korea Development Institute. Innovation of the Company as Seen through Petconomy. 2022. Available online: https://eiec.kdi.re.kr/material/pageoneView.do?idx=1521 (accessed on 22 March 2023).
- High Frequency Disease Statistics. 2021. Healthcare Bigdata Hub. Available online: https://opendata.hira.or.kr/op/opc/olapHifrqSickInfo.do (accessed on 13 February 2023).
- Lemos, J.A.; Palmer, S.R.; Zeng, L.; Wen, Z.T.; Kajfasz, I.A.; Freires, I.A.; Abranches, J.; Brady, L.J. The Biology of Streptococcus mutans. Microbiol. Spectr. 2019, 7, 10–128. [Google Scholar] [CrossRef]
- Pal, M. Why do Dogs Lick You? Forbes Advisor. 2023. Available online: https://www.forbes.com/advisor/pet-insurance/pet-care/why-do-dogs-lick-you/ (accessed on 22 January 2023).
- Stull, J.W.; Brophy, J.; Weese, J.S. Reducing the risk of pet-associated zoonotic infections. CMAJ 2018, 187, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Bastos, V.A.; Freitas-Fernandes, L.B.; da Silva Fidalgo, T.K.; Martins, C.; Mattos, C.T.; de Souza, I.P.R.; Maia, L.C. Mother-to-child transmission of Streptococcus mutans: A systematic review and meta-analysis. J. Dent. 2015, 43, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 1986, 50, 353–380. [Google Scholar] [CrossRef] [PubMed]
- Hamada, S.; Slade, H.D. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbial. Rev. 1980, 44, 331–384. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Nomura, R.; Nakagawa, I.; Hamada, S.; Ooshima, T. Demonstration of Streptococcus mutans with a cell wall polysaccharide specific to a new serotype, k, in the human oral cavity. J. Clin. Microbiol. 2004, 42, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Ozak, K.; Seki, M.; Kawato, T.; Tanaka, H.; Nakano, Y.; Yamashita, Y. Analysis of loci required for determination of serotype antigenicity in Streptococcus mutans and its clinical utilization. J. Clin. Microbial. 2003, 41, 4107–4112. [Google Scholar] [CrossRef]
- Hirasawa, M.; Takada, K. A new selective medium for Streptococcus mutans and the distribution of S. mutans and S. sobrinus and their serotypes in dental plaque. Caries. Res. 2003, 37, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Jeong, T.S.; Chung, J.; Kim, S. The comparison of Streptococcus mutans isolated from occlusal surfaces of caries and non-caries teeth. J. Korean Acad. Pediatr. Dent. 2001, 28, 19–141. [Google Scholar]
- Kim, S.Y.; Bae, I.K.; Lee, J.H.; Shin, J.H.; Kim, J.B. Molecular epidemiology and characterization of Streptococcus mutans strains in Korea. J. Korean. Acad. Oral. Health 2020, 44, 34–40. [Google Scholar] [CrossRef]
- Huis in’t Veld, J.H.J.; van Palenstein-Helderman, W.H.; Dirks, O.B. Streptococcus mutans and dental caries in humans: A bacteriological and immunological study. Antonie Van Leeuwenhoek 1979, 45, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Holst, E.; Rollof, J.; Larsson, L.; Nielsen, J.P. Characterization and distribution of Pasteurella species recovered from infected humans. J. Clin. Microbiol. 1992, 30, 2984–2987. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Ochiai, Y.; Morishima, T.; Arashima, Y.; Kumasaka, K.; Kawano, K.Y. Pasteurella canis osteomyelitis and cutaneous abscess after a domestic dog bite. J. Am. Acad. Dermatol. 2002, 46, S151–S152. [Google Scholar] [CrossRef] [PubMed]
- Kelesidis, T.; Tsiodras, S. Staphylococcus intermedius is not only a zoonotic pathogen, but may also cause skin abscesses in humans after exposure to saliva. Int. J. Infect. Dis. 2010, 14, e838–e841. [Google Scholar] [CrossRef]
- Qin, S.; Wen, X.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- Savini, V.; Catavitello, C.; Talia, M.; Manna, A.; Pompetti, F.; Favaro, M.; Fontana, C.; Febbo, F.; Balbinot, A.; Berardino, F.D.; et al. Multidrug-resistant Escherichia fergusonii: A case of acute cystitis. J. Clin. Microbiol. 2008, 46, 1551–1552. [Google Scholar] [CrossRef][Green Version]
- Casolari, C.; Fabio, U. Isolation of Pasteurella multocida from Human Clinical Specimens: First Report in Italy. Eur. J. Epidemiol. 1988, 4, 389–390. [Google Scholar] [CrossRef]
- Yamasaki, Y.; Nomura, R.; Nakano, K.; Naka, S.; Matsumoto-Nakano, M.; Asai, F.; Ooshima, T. Distribution of periodontopathic bacterial species in dogs and their owners. Arch. Oral. Biol. 2012, 57, 1183–1188. [Google Scholar] [CrossRef]
- Rutland, B.E.; Weese, J.S.; Bolin, C.; Au, J.; Malani, A.N. Human-to-Dog Transmission of Methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 2009, 15, 1328–1330. [Google Scholar] [CrossRef]
- Weese, J.S.; Dick, H.; Willey, B.M.; McGeer, A.; Kreiswirth, B.N.; Innis, B.; Low, D.E. Suspected transmission of methicillin-resistant Staphylococcus aureus between domestic pets and humans in veterinary clinics and in the household. Vet. Microbiol. 2006, 115, 148–155. [Google Scholar] [CrossRef]
- Marcano, R.; Rojo, M.Á.; Cordoba-Diaz, D.; Garrosa, M. Pathological and Therapeutic Approach to Endotoxin-Secreting Bacteria Involved in Periodontal Disease. Toxins 2021, 13, 533. [Google Scholar] [CrossRef] [PubMed]
- Sedghi, L.; DiMassa, V.; Harrington, A.; Lynch, S.V.; Kapila, Y.L. The oral microbiome: Role of key organisms and complex networks in oral health and disease. Periodontol. 2000 2021, 87, 107–131. [Google Scholar] [CrossRef]
- Raspa, F.; Schiavone, A.; Pattono, D.; Galaverna, D.; Cavallini, D.; Vinassa, M.; Bergero, D.; Dalmasso, A.; Bottero, M.T.; Emanuela Valle, E. Pet feeding habits and the microbiological contamination of dog food bowls: Effect of feed type, cleaning method and bowl material. BMC Vet. Res. 2023, 19, 261. [Google Scholar] [CrossRef] [PubMed]
- Muca, E.; Buonaiuto, G.; Lamanna, M.; Silvestrelli, S.; Ghiaccio, F.; Federiconi, A.; Vettori, J.D.M.; Colleluori, R.; Fusaro, I.; Raspa, F.; et al. Reaching a Wider Audience: Instagram’s Role in Dairy Cow Nutrition Education and Engagement. Animals 2023, 13, 3503. [Google Scholar] [CrossRef] [PubMed]

| Category | Response | Unit | p Value | |
|---|---|---|---|---|
| Gender | Male | 9 (47.36%) | >0.05 * | |
| Female | 10 (52.64%) | |||
| Tendency to lick people frequently | Yes | n = 15 (78.95%) | 0.000 * | |
| No | n = 4 (21.05%) | |||
| Oral hygiene of dogs | Tooth brushing at least once a week (N = 11, 57.89%) | CI | 0.74 ± 0.41 〒 | CI; 0.479 * GI; 0.840 * |
| GI | 0.54 ± 0.36 〒 | |||
| Tooth brushing less than once a month (N = 8, 42.10%) | CI | 0.83 ± 0.51 〒 | ||
| GI | 0.61 ± 0.47 〒 | |||
| Correlation between dog’s clinical diagnosis vs. the owner’s awareness of the oral condition | yes (n = 12, 63.2%) | 0.000 * | ||
| no (n = 7, 36.8%) | ||||
| Tooth brushing frequency vs. the dog owner’s awareness of the dog’s oral condition | 0.422 # | |||
| Group | Kingdom | Phylum | Class | Order | Family | Genus | Species | Accession No. | SI | S2 | S4 | S5 | S8 | S9 | S12 | S13 | S15 | S18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASV1 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus intermedius | NR_036829.1 | 23,919 | 53,961 | 0 | 40,926 | 38,350 | 0 | 27,799 | 55,779 | 27,311 | 46 |
| ASV2 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | Pseudomonas aeruginosa | NR_113599.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 54,196 |
| ASV3 | Bacteria | Actinomycetota | Actinomycetes | Micrococcales | Micrococcaceae | Micrococcus | Micrococcus aloeverae | NR_134088.1 | 0 | 0 | 0 | 0 | 0 | 54,304 | 0 | 0 | 0 | 0 |
| ASV4 | Bacteria | Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Pasteurella | Pasteurella canis | NR_042882.1 | 658 | 603 | 11,342 | 13,574 | 15,373 | 0 | 0 | 0 | 0 | 0 |
| ASV5 | Bacteria | Bacillota | Bacilli | Bacillales | Bacillaceae | Bacillus | Bacillus clarus | NR_180213.1 | 0 | 0 | 35,677 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV6 | Bacteria | Proteobacteria | Gammaproteobacteria | Enterobacterales | Enterobacteriaceae | Escherichia | Escherichia fergusonii | NR_114079.1 | 29,916 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 2 |
| ASV7 | Bacteria | Proteobacteria | Gammaproteobacteria | Moraxellales | Moraxellaceae | Acinetobacter | Acinetobacter geminorum | NR_181169.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 21,760 | 0 |
| ASV8 | Bacteria | Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Pasteurella | Pasteurella multocida | NR_115138.1 | 0 | 0 | 0 | 0 | 0 | 0 | 12,118 | 0 | 0 | 0 |
| ASV9 | Bacteria | Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Pasteurella | Pasteurella multocida | NR_115138.1 | 0 | 0 | 0 | 0 | 0 | 0 | 1632 | 0 | 0 | 0 |
| ASV10 | Bacteria | Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Pasteurella | Pasteurella stomatis | NR_042888.1 | 0 | 0 | 6178 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV11 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus intermedius | NR_036829.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5558 | 0 |
| ASV12 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus intermedius | NR_036829.1 | 541 | 1314 | 0 | 1006 | 966 | 0 | 766 | 0 | 718 | 0 |
| ASV13 | Bacteria | Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Pasteurella | Pasteurella multocida | NR_115138.1 | 0 | 0 | 0 | 0 | 0 | 0 | 2171 | 0 | 0 | 0 |
| ASV14 | Bacteria | Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Pasteurella | Pasteurella multocida | NR_115137.1 | 0 | 0 | 0 | 0 | 0 | 1 | 2076 | 0 | 0 | 5 |
| ASV15 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | Pseudomonas aeruginosa | NR_113599.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1275 |
| ASV16 | Bacteria | Actinomycetota | Actinomycetes | Micrococcales | Micrococcaceae | Micrococcus | Micrococcus aloeverae | NR_134088.1 | 0 | 0 | 0 | 0 | 0 | 1414 | 0 | 0 | 0 | 0 |
| ASV17 | Bacteria | Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Pasteurella | Pasteurella stomatis | NR_042888.1 | 0 | 0 | 1105 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV18 | Bacteria | Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Pasteurella | Pasteurella canis | NR_042882.1 | 0 | 0 | 0 | 370 | 401 | 0 | 0 | 0 | 0 | 0 |
| ASV19 | Bacteria | Proteobacteria | Gammaproteobacteria | Enterobacterales | Enterobacteriaceae | Escherichia | Escherichia fergusonii | NR_114079.1 | 780 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV20 | Bacteria | Proteobacteria | Gammaproteobacteria | _Moraxellales | Moraxellaceae | Acinetobacter | Acinetobacter geminorum | NR_181169.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 533 | 0 |
| ASV21 | Bacteria | Proteobacteria | Gammaproteobacteria | Enterobacterales | Erwiniaceae | Kalamiella | Kalamiella piersonii | NR_181783.1 | 0 | 0 | 454 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV22 | Bacteria | Bacillota | Bacilli | Bacillales | Bacillaceae | Bacillus | Bacillus clarus | NR_180213.1 | 0 | 0 | 346 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV23 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus cohnii | NR_036902.1 | 0 | 0 | 298 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV24 | Bacteria | Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Pasteurella | Pasteurella multocida | NR_115138.1 | 0 | 0 | 0 | 0 | 0 | 0 | 308 | 0 | 0 | 0 |
| ASV25 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus simulans | NR_036906.1 | 0 | 0 | 0 | 0 | 266 | 0 | 0 | 0 | 0 | 0 |
| ASV26 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus agnetis | NR_117863.1 | 0 | 0 | 0 | 0 | 255 | 0 | 0 | 0 | 0 | 0 |
| ASV27 | Bacteria | Bacillota | Bacilli | Bacillales | Bacillaceae | Bacillus | Bacillus clarus | NR_180213.1 | 0 | 0 | 187 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV28 | Bacteria | Bacillota | Bacilli | Lactobacillales | Enterococcaceae | Enterococcus | Enterococcus hirae | NR_114783.2 | 0 | 0 | 0 | 0 | 242 | 0 | 0 | 0 | 0 | 0 |
| ASV29 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | Pseudomonas aeruginosa | NR_113599.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 145 |
| ASV30 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus ureilyticus | NR_037046.1 | 0 | 0 | 117 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV31 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | Pseudomonas aeruginosa | NR_113599.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 94 |
| ASV32 | Bacteria | Proteobacteria | Gammaproteobacteria | Enterobacterales | Erwiniaceae | Kalamiella | Kalamiella piersonii | NR_181783.1 | 0 | 0 | 99 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV33 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus canis | NR_181183.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 63 | 0 | 0 |
| ASV34 | Bacteria | Bacillota | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | Streptococcus mutans | NR_114726.1 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 3 | 0 | 0 |
| ASV35 | Bacteria | Proteobacteria | Alphaproteobacteria | Hyphomicrobiales | Phyllobacteriaceae | Phyllobacterium | Phyllobacterium zundukense | NR_181634.1 | 0 | 2 | 0 | 0 | 0 | 39 | 0 | 0 | 0 | 30 |
| ASV36 | Bacteria | Actinomycetota | Actinomycetes | Micrococcales | Micrococcaceae | Micrococcus | Micrococcus aloeverae | NR_134088.1 | 0 | 0 | 0 | 0 | 0 | 68 | 0 | 0 | 0 | 0 |
| ASV37 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus cohnii | NR_036902.1 | 0 | 0 | 59 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV38 | Bacteria | Proteobacteria | Gammaproteobacteria | Enterobacterales | Enterobacteriaceae | Escherichia | Escherichia fergusonii | NR_114079.1 | 66 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV39 | Bacteria | Bacteroidota | Chitinophagia | Chitinophagales | Chitinophagaceae | Hydrotalea | Hydrotalea flava | NR_117026.1 | 0 | 0 | 0 | 0 | 0 | 25 | 0 | 0 | 0 | 17 |
| ASV40 | Bacteria | Proteobacteria | Alphaproteobacteria | Hyphomicrobiales | Phyllobacteriaceae | Mesorhizobium | Mesorhizobium terrae | NR_180479.1 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 7 |
| ASV41 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus lugdunensis | NR_024668.1 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV42 | Bacteria | Proteobacteria | Betaproteobacteria | Neisseriales | Neisseriaceae | Neisseria | Neisseria zoodegmatis | NR_043459.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 |
| ASV43 | Bacteria | Bacillota | Bacilli | Lactobacillales | Enterococcaceae | Enterococcus | Enterococcus lactis | NR_117562.1 | 0 | 0 | 0 | 0 | 16 | 0 | 0 | 0 | 0 | 0 |
| ASV44 | Bacteria | Proteobacteria | Betaproteobacteria | Neisseriales | Neisseriaceae | Neisseria | Neisseria animaloris | NR_043458.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 |
| ASV45 | Bacteria | - | - | - | - | - | - | - | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 |
| ASV46 | Bacteria | Cyanobacteria | - | Oscillatoriales | - | Potamosiphon | Potamosiphon australiensis | NR_177904.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
| ASV47 | Bacteria | Actinomycetota | Actinomycetes | Micrococcales | Micrococcaceae | Micrococcus | Micrococcus luteus | NR_075062.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 |
| ASV48 | Bacteria | Cyanobacteria | - | Oscillatoriales | - | Potamosiphon | Potamosiphon australiensis | NR_177904.1 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 |
| ASV49 | Bacteria | Bacillota | Bacilli | Bacillales | Bacillaceae | Calidifontibacillus | Calidifontibacillus erzurumensis | NR_180225.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| ASV50 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | Pseudomonas aeruginosa | NR_113599.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| ASV51 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus muscae | NR_104762.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| ASV52 | Bacteria | Proteobacteria | Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | Campylobacter | Campylobacter massiliensis | NR_181373.1 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 |
| ASV53 | Bacteria | Proteobacteria | Alphaproteobacteria | Hyphomicrobiales | Nitrobacteraceae | Bradyrhizobium | Bradyrhizobium australafricanum | NR_180493.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| ASV54 | Bacteria | Proteobacteria | Alphaproteobacteria | Hyphomicrobiales | Xanthobacteraceae | Labrys | Labrys wisconsinensis | NR_116004.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| ASV55 | Bacteria | Bacteroidota | Chitinophagia | Chitinophagales | Chitinophagaceae | Chitinophaga | Chitinophaga vietnamensis | NR_180543.1 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| ASV56 | Bacteria | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | Variovorax | Variovorax boronicumulans | NR_114214.1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 |
| ASV57 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus intermedius | NR_036829.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| ASV58 | Bacteria | Proteobacteria | Gammaproteobacteria | Enterobacterales | Erwiniaceae | Pantoea | [Curtobacterium] plantarum | NR_104943.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| ASV59 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | Pseudomonas nicosulfuronedens | NR_180597.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| ASV60 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus agnetis | NR_117863.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| ASV61 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | Pseudomonas aeruginosa | NR_113599.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| ASV62 | Bacteria | - | - | - | - | - | - | - | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| ASV63 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus durrellii | NR_181502.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| ASV64 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus chromogenes | NR_036901.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ASV65 | Bacteria | Bacillota | Bacilli | Bacillales | Bacillaceae | Halalkalibacterium | Halalkalibacterium halodurans | NR_025446.1 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV66 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus simulans | NR_036906.1 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| ASV67 | Bacteria | Bacillota | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | Staphylococcus lutrae | NR_036791.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ASV68 | Bacteria | Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Pasteurella | Pasteurella canis | NR_042882.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| ASV69 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | Pseudomonas glycinis | NR_181729.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ASV70 | Bacteria | - | - | - | - | - | - | - | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV71 | Bacteria | Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | Brevundimonas | Brevundimonas poindexterae | NR_114709.1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| ASV72 | Bacteria | Actinomycetota | Actinomycetes | Corynebacteriales | Mycobacteriaceae | Mycobacterium | Mycobacterium numidiamassiliense | NR_179524.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ASV73 | Bacteria | Bacillota | Bacilli | Lactobacillales | Lactobacillaceae | Lacticaseibacillus | Lacticaseibacillus baoqingensis | NR_180279.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| ASV74 | Bacteria | Bacillota | Bacilli | Bacillales | Bacillaceae | Bacillus | Bacillus rhizoplanae | NR_181926.1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV75 | Bacteria | Bacillota | Bacilli | Bacillales | Bacillaceae | Bacillus | Bacillus clarus | NR_180213.1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ASV76 | Bacteria | Proteobacteria | Proteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | Pseudomonas migulae | NR_114223.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.s.; Kim, M.; Cho, I.-H.; Sim, Y.-M.; Hwang, Y.S. Evidence Supporting Oral Hygiene Management by Owners through a Genetic Analysis of Dental Plaque Bacteria in Dogs. Vet. Sci. 2024, 11, 96. https://doi.org/10.3390/vetsci11020096
Yu Js, Kim M, Cho I-H, Sim Y-M, Hwang YS. Evidence Supporting Oral Hygiene Management by Owners through a Genetic Analysis of Dental Plaque Bacteria in Dogs. Veterinary Sciences. 2024; 11(2):96. https://doi.org/10.3390/vetsci11020096
Chicago/Turabian StyleYu, Jeong suk, Minhee Kim, Il-Hoon Cho, Yu-Min Sim, and Young Sun Hwang. 2024. "Evidence Supporting Oral Hygiene Management by Owners through a Genetic Analysis of Dental Plaque Bacteria in Dogs" Veterinary Sciences 11, no. 2: 96. https://doi.org/10.3390/vetsci11020096
APA StyleYu, J. s., Kim, M., Cho, I.-H., Sim, Y.-M., & Hwang, Y. S. (2024). Evidence Supporting Oral Hygiene Management by Owners through a Genetic Analysis of Dental Plaque Bacteria in Dogs. Veterinary Sciences, 11(2), 96. https://doi.org/10.3390/vetsci11020096





