Investigation of Prototheca bovis Infection and Its Correlation with Dairy Herd Improvement Data from a Dairy Farm in Central China
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Approval and Sample Collection
2.2. Materials
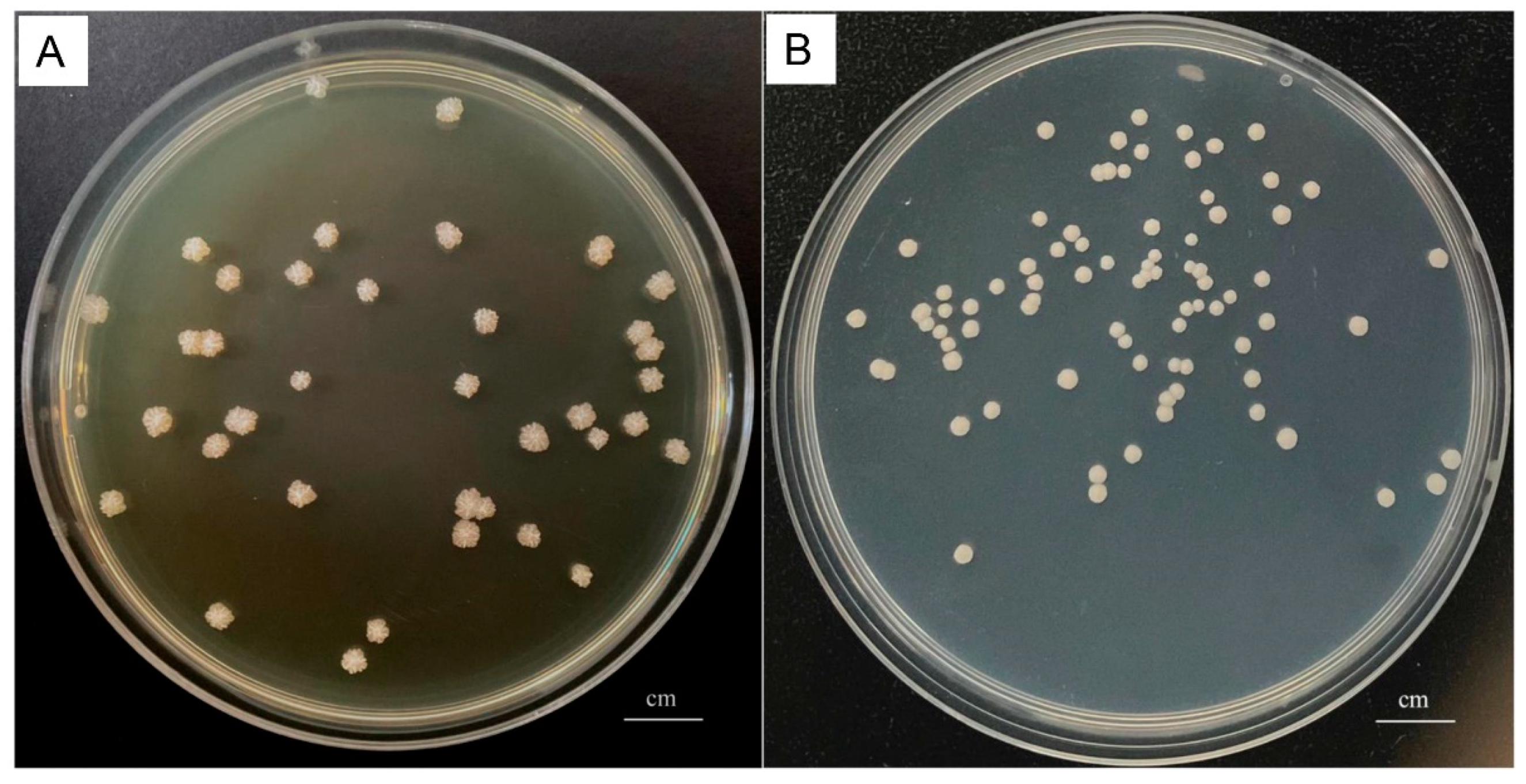
2.3. Isolation and Colony Counting of P. bovis
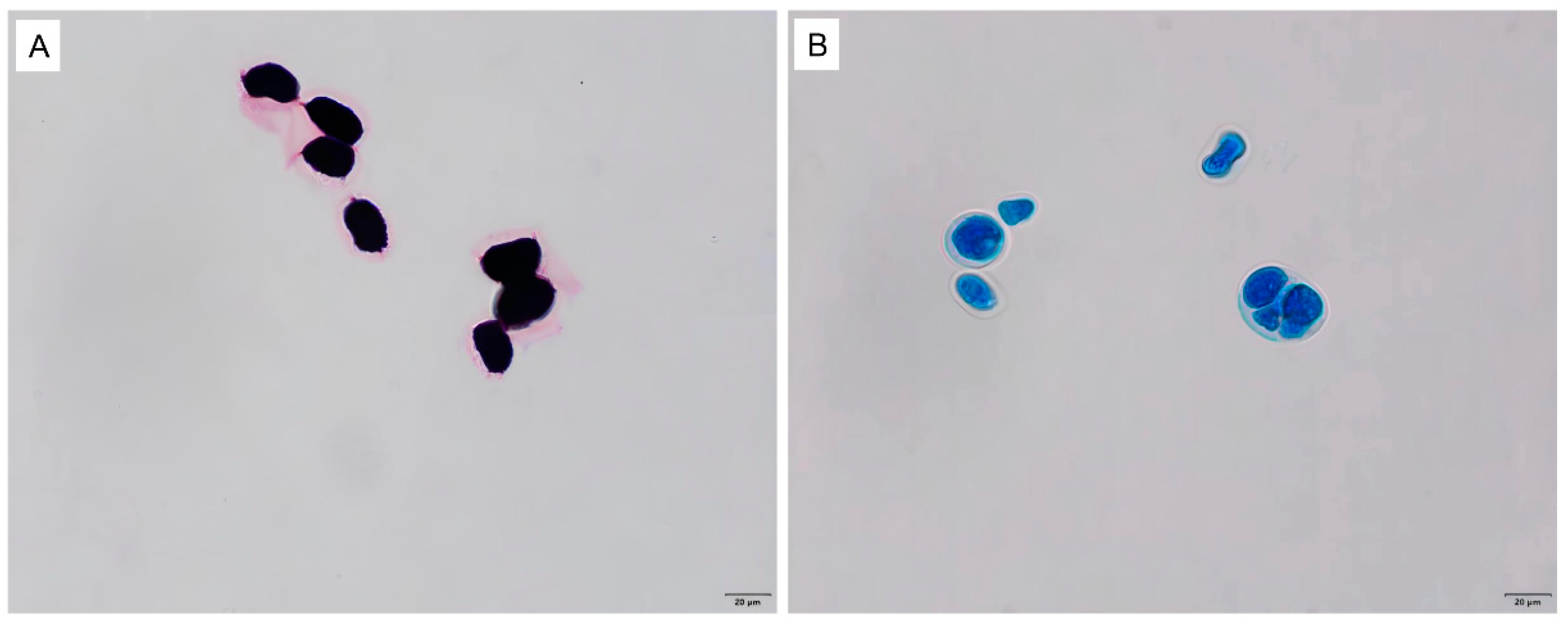
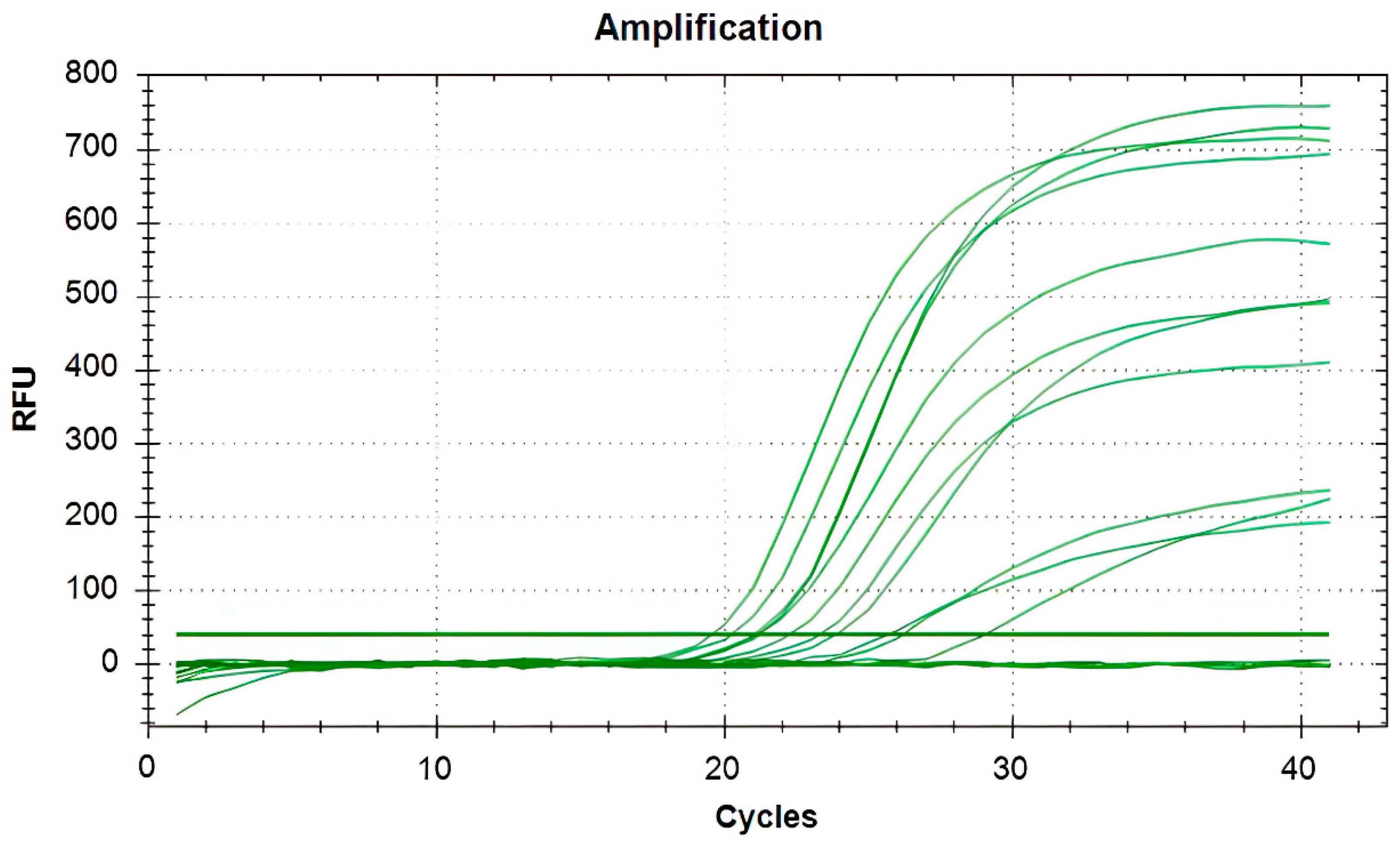
2.4. Identification of Microorganisms
2.5. Statistical Analyses
3. Results
3.1. Isolation and Identification of Bacteria and P. bovis
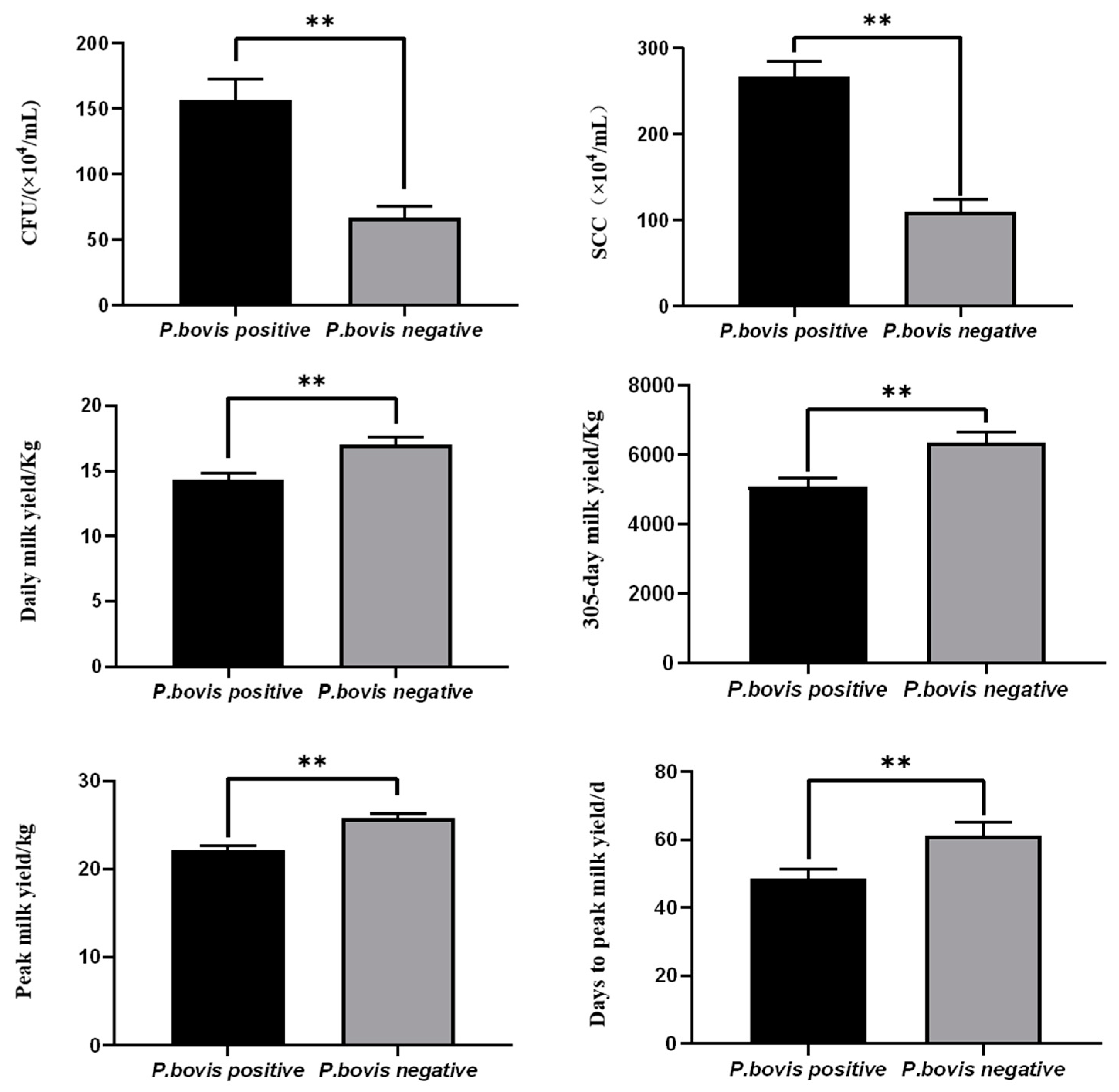
3.2. Effects of P. bovis Infection on Milk Production
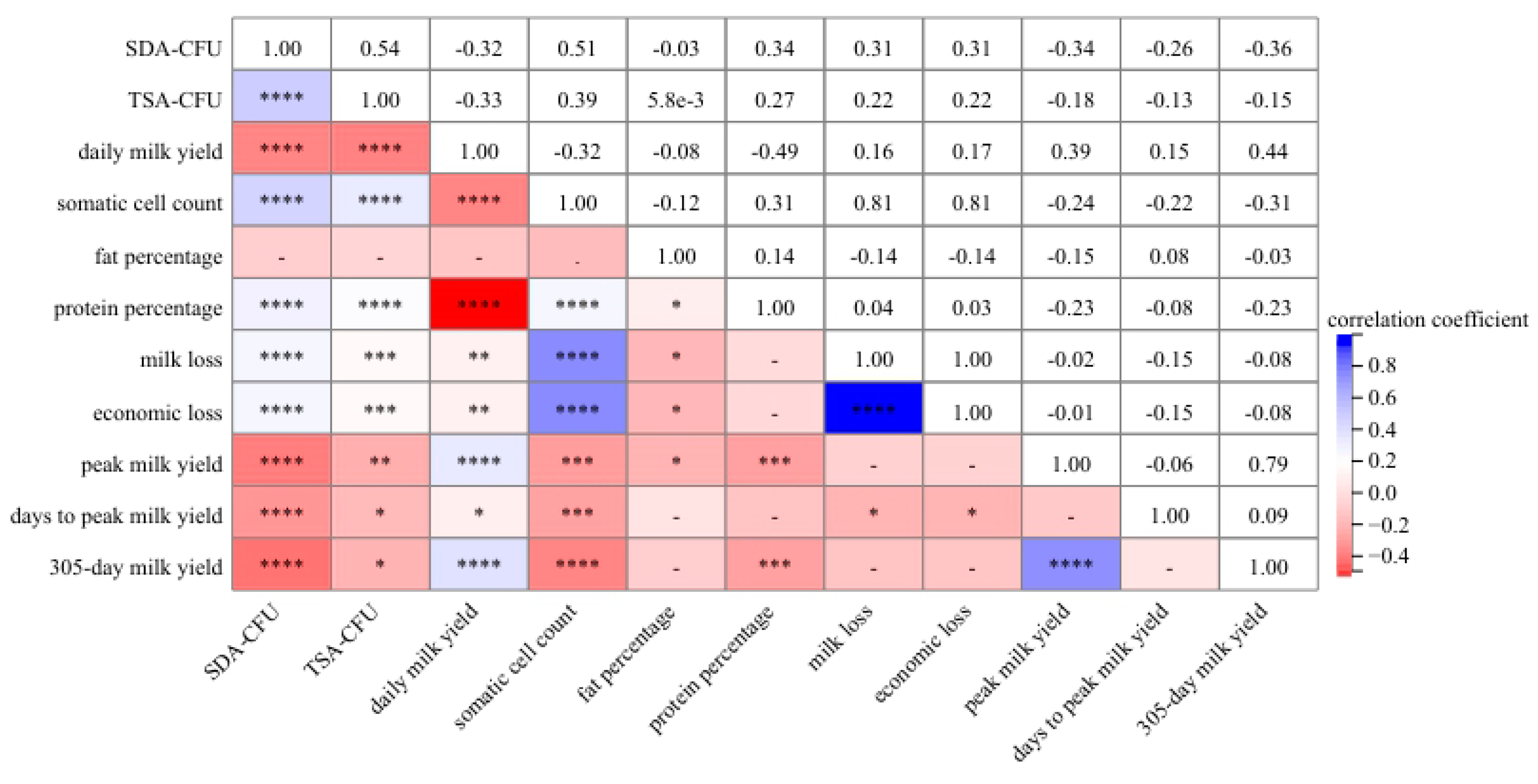
3.3. Correlation Analysis between P. bovis Infection and DHI Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kano, R. Emergence of Fungal-Like Organisms: Prototheca. Mycopathologia 2020, 185, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Jagielski, T.; Bakuła, Z.; Gawor, J.; Maciszewski, K.; Kusber, W.H.; Dyląg, M.; Nowakowska, J.; Gromadka, R.; Karnkowska, A. The genus Prototheca (Trebouxiophyceae, Chlorophyta) revisited: Implications from molecular taxonomic studies. Algal Res. 2019, 43, 101639. [Google Scholar] [CrossRef]
- Huilca-Ibarra, M.P.; Vasco-Julio, D.; Ledesma, Y.; Guerrero-Freire, S.; Zurita, J.; Castillejo, P.; Blasco, F.B.; Yanez, L.; Changoluisa, D.; Echeverría, G.; et al. High Prevalence of Prototheca bovis Infection in Dairy Cattle with Chronic Mastitis in Ecuador. Vet. Sci. 2022, 9, 659. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, X.; Ni, F.; Xia, W. Cutaneous Protothecosis with Meningitis Due to Prototheca wickerhamii in an Immunocompetent Teenager: Case Report and Literature Review. Infect. Drug Resist. 2021, 14, 2787–2794. [Google Scholar] [CrossRef] [PubMed]
- Falcaro, C.; Furlanello, T.; Binanti, D.; Fondati, A.; Bonfanti, U.; Krockenberger, M.; Malik, R.; Danesi, P. Molecular characterization of Prototheca in 11 symptomatic dogs. J. Vet. Diagn. Investig. 2021, 33, 156–161. [Google Scholar] [CrossRef]
- Ely, V.L.; Felipetto Cargnelutti, J.; Segabinazzi Ries, A.; Gressler, L.T.; Costa, S.; Braz, P.H.; Pötter, L.; da Costa, M.M.; Júnior, F.G.d.S.; de Oliveira, H.P.; et al. Prototheca bovis in goats: Experimental mastitis and treatment with polypyrrole. Microb. Pathog. 2023, 174, 105950. [Google Scholar] [CrossRef]
- Corbellini, L.G.; Driemeier, D.; Cruz, C.; Dias, M.M.; Ferreiro, L. Bovine mastitis due to Prototheca zopfii: Clinical, epidemiological and pathological aspects in a Brazilian dairy herd. Trop. Anim. Health Prod. 2001, 33, 463–470. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, H.; Hu, C.; Jin, E.; Li, D.; Hu, X.; Chen, J. Isolation, identification and drug sensitivity analysis of main pathogenic microorganisms of mastitis in a dairy farm in Hubei Province. Chin. J. Vet. Med. 2022, 58, 1–8. [Google Scholar]
- Tashakkori, N.; Rahmani, H.K.; Khoramian, B. Genotypic and phenotypic diversity of Prototheca spp. recovered from bovine mastitis in terms of antimicrobial resistance and biofilm formation ability. BMC Vet. Res. 2022, 18, 452. [Google Scholar] [CrossRef]
- Lerche, M. Eine durch Algen (Prototheca) hervorgerufene Mastitis der Kuh. Berl. Munch. Tierarztl. Wochenschr. 1954, 4, 64–69. [Google Scholar]
- Möller, A.; Truyen, U.; Roesler, U. Prototheca zopfii genotype 2: The causative agent of bovine protothecal mastitis? Vet. Microbiol. 2007, 120, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Buzzini, P.; Turchetti, B.; Facelli, R.; Baudino, R.; Cavarero, F.; Mattalia, L.; Mosso, P.; Martini, A. First large-scale isolation of Prototheca zopfii from milk produced by dairy herds in Italy. Mycopathologia 2004, 158, 427–430. [Google Scholar] [CrossRef]
- Aouay, A.; Coppée, F.; Cloet, S.; Cuvelier, P.; Belayew, A.; Lagneau, P.E.; Mullender, C. Molecular characterization of Prototheca strains isolated from bovine mastitis. J. De Mycol. Médicale 2008, 18, 224–227. [Google Scholar] [CrossRef]
- Jagielski, T.; Roeske, K.; Bakuła, Z.; Piech, T.; Wlazło, Ł.; Bochniarz, M.; Woch, P.; Krukowski, H. A survey on the incidence of Prototheca mastitis in dairy herds in Lublin province, Poland. J. Dairy Sci. 2019, 102, 619–628. [Google Scholar] [CrossRef]
- Osumi, T.; Kishimoto, Y.; Kano, R.; Maruyama, H.; Onozaki, M.; Makimura, K.; Ito, T.; Matsubara, K.; Hasegawa, A. Prototheca zopfii genotypes isolated from cow barns and bovine mastitis in Japan. Vet. Microbiol. 2008, 131, 419–423. [Google Scholar] [CrossRef]
- Park, H.S.; Moon, D.C.; Hyun, B.H.; Lim, S.K. Short communication: Occurrence and persistence of Prototheca zopfii in dairy herds of Korea. J. Dairy Sci. 2019, 102, 2539–2543. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, H.Q.; He, J.Z.; He, Y.H.; Li, S.M.; Hou, R.G.; Wu, Q.-X.; Gao, Y.; Han, B. Characterization of Prototheca zopfii associated with outbreak of bovine clinical mastitis in herd of Beijing, China. Mycopathologia 2012, 173, 275–281. [Google Scholar] [CrossRef]
- Li, J.; Chen, X.; Jin, E.; Wang, G.; Wu, L.; Shao, Z.; Wan, P.; Hu, C.; Li, J.; Chen, J.; et al. A survey of Prototheca bovis infection in dairy farms of the Hubei province, China. J. Vet. Med. Sci. 2021, 83, 1248–1255. [Google Scholar] [CrossRef]
- Hsieh, J.C.; Hsieh, Y.F.; Chuang, S.T. Prototheca from bovine milk and associated minimal algaecide concentration of chlorhexidine and povidone-iodine in Taiwan. Tierarztl. Prax. Ausg. G Grosstiere Nutztiere 2020, 48, 380–385. [Google Scholar] [CrossRef]
- Pieper, L.; Godkin, A.; Roesler, U.; Polleichtner, A.; Slavic, D.; Leslie, K.E.; Kelton, D. Herd characteristics and cow-level factors associated with Prototheca mastitis on dairy farms in Ontario, Canada. J. Dairy Sci. 2012, 95, 5635–5644. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Ali, T.; Zhang, L.; Hou, R.; Zhang, S.; Ding, L.; Han, D.; Deng, Z.; Rahman, A.; Han, B. Characterization of Prototheca zopfii Genotypes Isolated from Cases of Bovine Mastitis and Cow Barns in China. Mycopathologia 2016, 181, 185–195. [Google Scholar] [CrossRef] [PubMed]
- National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals, 8th ed.; National Academies Press: Washington, DC, USA, 2011; pp. 12–35.
- Liu, J.; Li, F.; Guo, J. Application of Dairy Herd Improvement Data in Dairy Farm. China Dairy 2023, 2, 44–48. [Google Scholar]
- Zhang, H. Establishment of the Fluorescence Quantitative PCR and Loop-Mediated Isothermal Amplification Methods of Prototheca bovis and Application. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2022. [Google Scholar]
- McDonald, J.S.; Richard, J.L.; Cheville, N.F. Natural and experimental bovine intramammary infection with Prototheca zopfii. Am. J. Vet. Res. 1984, 45, 592–595. [Google Scholar] [PubMed]
- Ricchi, M.; Goretti, M.; Branda, E.; Cammi, G.; Garbarino, C.A.; Turchetti, B.; Moroni, P.; Arrigoni, N.; Buzzini, P. Molecular characterization of Prototheca strains isolated from Italian dairy herds. J. Dairy Sci. 2010, 93, 4625–4631. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, J.; Wu, Q.; Gao, Y.; Han, B. Bovine mastitis caused by Prototheca zopfii. Chin. J. Vet. Med. 2011, 47, 13–15. [Google Scholar]
- Reneau, J.K. Effective use of dairy herd improvement somatic cell counts in mastitis control. J. Dairy Sci. 1986, 69, 1708–1720. [Google Scholar] [CrossRef]
- Masoud, W.; Vogensen, F.K.; Lillevang, S.; Abu Al-Soud, W.; Sørensen, S.J.; Jakobsen, M. The fate of indigenous microbiota, starter cultures, Escherichia coli, Listeria innocua and Staphylococcus aureus in Danish raw milk and cheeses determined by pyrosequencing and quantitative real time (qRT)-PCR. Int. J. Food Microbiol. 2012, 153, 192–202. [Google Scholar] [CrossRef]
- Hornik, B.; Czarny, J.; Staninska-Pięta, J.; Wolko, Ł.; Cyplik, P.; Piotrowska-Cyplik, A. The Raw Milk Microbiota from Semi-Subsistence Farms Characteristics by NGS Analysis Method. Molecules 2021, 26, 5029. [Google Scholar] [CrossRef]
- Marques, S.; Huss, V.A.; Pfisterer, K.; Grosse, C.; Thompson, G. Internal transcribed spacer sequence-based rapid molecular identification of Prototheca zopfii and Prototheca blaschkeae directly from milk of infected cows. J. Dairy Sci. 2015, 98, 3001–3009. [Google Scholar] [CrossRef]
- Bacova, R.; Kralik, P.; Kucharovicova, I.; Seydlova, R.; Moravkova, M. A novel TaqMan qPCR assay for rapid detection and quantification of pro-inflammatory microalgae Prototheca spp. in milk samples. Med. Mycol. 2021, 59, 784–792. [Google Scholar] [CrossRef]
- Wu, H.; Yao, S.; Wang, T.; Wang, J.; Ren, K.; Yang, H.; Ma, W.; Ji, P.; Lu, Y.; Ma, H.; et al. Effects of Melatonin on Dairy Herd Improvement (DHI) of Holstein Cow with High SCS. Molecules 2021, 26, 834. [Google Scholar] [CrossRef] [PubMed]
- Bueno, V.F.; de Mesquita, A.J.; Neves, R.B.; de Souza, M.A.; Ribeiro, A.R.; Nicolau, E.S.; de Oliveira, A.N. Epidemiological and clinical aspects of the first outbreak of bovine mastitis caused by Prototheca zopfii in Goiás State, Brazil. Mycopathologia 2006, 161, 141–145. [Google Scholar] [CrossRef]
- Costa, E.O.; Carciofi, A.C.; Melville, P.A.; Prada, M.S.; Schalch, U. Prototheca sp. outbreak of bovine mastitis. Zentralbl. Vet. B 1996, 43, 321–324. [Google Scholar] [CrossRef]
- Shahid, M.; Cavalcante, P.A.; Knight, C.G.; Barkema, H.W.; Han, B.; Gao, J.; Cobo, E.R. Murine and Human Cathelicidins Contribute Differently to Hallmarks of Mastitis Induced by Pathogenic Prototheca bovis Algae. Front. Cell. Infect. Microbiol. 2020, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; He, F.; Barkema, H.W.; Xu, S.; Gao, J.; Liu, G.; Deng, Z.; Shahid, M.; Shi, Y.; Kastelic, J.P.; et al. Prototheca spp. induce an inflammatory response via mtROS-mediated activation of NF-κB and NLRP3 inflammasome pathways in bovine mammary epithelial cell cultures. Vet. Res. 2021, 52, 144. [Google Scholar] [CrossRef]
- Hu, X. Research on Follow-up Monitoring and Disinfection Measures for Prototheca bovis Mastitis in Dairy Cows in Hubei Region. Master’s Thesis, Yangtze University, Jingzhou, China, 2023. [Google Scholar]
- Sobukawa, H.; Watanabe, M.; Kano, R.; Ito, T.; Onozaki, M.; Hasegawa, A.; Kamata, H. In vitro algaecide effect of disinfectants on Prototheca zopfii genotypes 1 and 2. J. Vet. Med. Sci. 2011, 73, 1527–1529. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, J.L.; Lee, S.H.; de Paula Arruda, E.; Pedroso Galles, D.; Camargo Caetano, V.; Fernandes de Oliveira, C.A.; Fernandes, A.M.; dos Santos, M.V. Biofilm-producing ability and efficiency of sanitizing agents against Prototheca zopfii isolates from bovine subclinical mastitis. J. Dairy Sci. 2015, 98, 3613–3621. [Google Scholar] [CrossRef]
- Libisch, B.; Picot, C.; Ceballos-Garzon, A.; Moravkova, M.; Klimesová, M.; Telkes, G.; Chuang, S.-T.; Le Pape, P. Prototheca Infections and Ecology from a One Health Perspective. Microorganisms 2022, 10, 938. [Google Scholar] [CrossRef] [PubMed]
| Primer | Sequence (5′-3′) | Target Fragment Size |
|---|---|---|
| 16s rDNA 27-R | AGAGTTTGATCCTGGCTCAG | 1500 bp |
| 16s rDNA 1492-F | TACGGCTACCTTGTTACGACT | |
| cob-5-F | CTAGTTATTCAAGTCCTCG | 131 bp |
| cob-5-R | AATTACTGTAGCACCCC |
| Month of Year in 2022 | Number of Samples | P. bovis Positive Samples | P. bovis Negative Samples | P. bovis Positivity Rate (%) |
|---|---|---|---|---|
| August | 71 | 38 | 33 | 53.5 |
| September | 65 | 38 | 27 | 58.5 |
| October | 76 | 46 | 30 | 60.5 |
| November | 65 | 33 | 32 | 50.8 |
| December | 71 | 55 | 16 | 77.5 |
| Total | 348 | 210 | 138 | 60.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Hu, X.; Li, G.; Wan, P.; Shao, Z.; Jin, E.; Liu, X.; Yang, Q.; Long, A.; Qian, Y. Investigation of Prototheca bovis Infection and Its Correlation with Dairy Herd Improvement Data from a Dairy Farm in Central China. Vet. Sci. 2024, 11, 37. https://doi.org/10.3390/vetsci11010037
Chen J, Hu X, Li G, Wan P, Shao Z, Jin E, Liu X, Yang Q, Long A, Qian Y. Investigation of Prototheca bovis Infection and Its Correlation with Dairy Herd Improvement Data from a Dairy Farm in Central China. Veterinary Sciences. 2024; 11(1):37. https://doi.org/10.3390/vetsci11010037
Chicago/Turabian StyleChen, Jie, Xiuxiu Hu, Guohong Li, Pingmin Wan, Zhiyong Shao, Erguang Jin, Xiaohua Liu, Qian Yang, Anying Long, and Yunguo Qian. 2024. "Investigation of Prototheca bovis Infection and Its Correlation with Dairy Herd Improvement Data from a Dairy Farm in Central China" Veterinary Sciences 11, no. 1: 37. https://doi.org/10.3390/vetsci11010037
APA StyleChen, J., Hu, X., Li, G., Wan, P., Shao, Z., Jin, E., Liu, X., Yang, Q., Long, A., & Qian, Y. (2024). Investigation of Prototheca bovis Infection and Its Correlation with Dairy Herd Improvement Data from a Dairy Farm in Central China. Veterinary Sciences, 11(1), 37. https://doi.org/10.3390/vetsci11010037





