Alternative Samples for Porcine Reproductive and Respiratory Syndrome Surveillance in an Endemic PRRSV-1-Infected Breeding Herd: A Descriptive Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Collection
- -
- Blood from one piglet per litter, targeting the weakest piglet within the litter
- -
- FOF
- -
- UW
2.3. Diagnostic Testing
2.4. Pooling
2.5. Data Analysis
2.5.1. Comparison of the Rate of Detection between Sample Types and Agreement between Them
2.5.2. Evaluation of Pooling Ability to Detect PRRSV-1
3. Results
3.1. Tests Abilities to Detect PRRSV
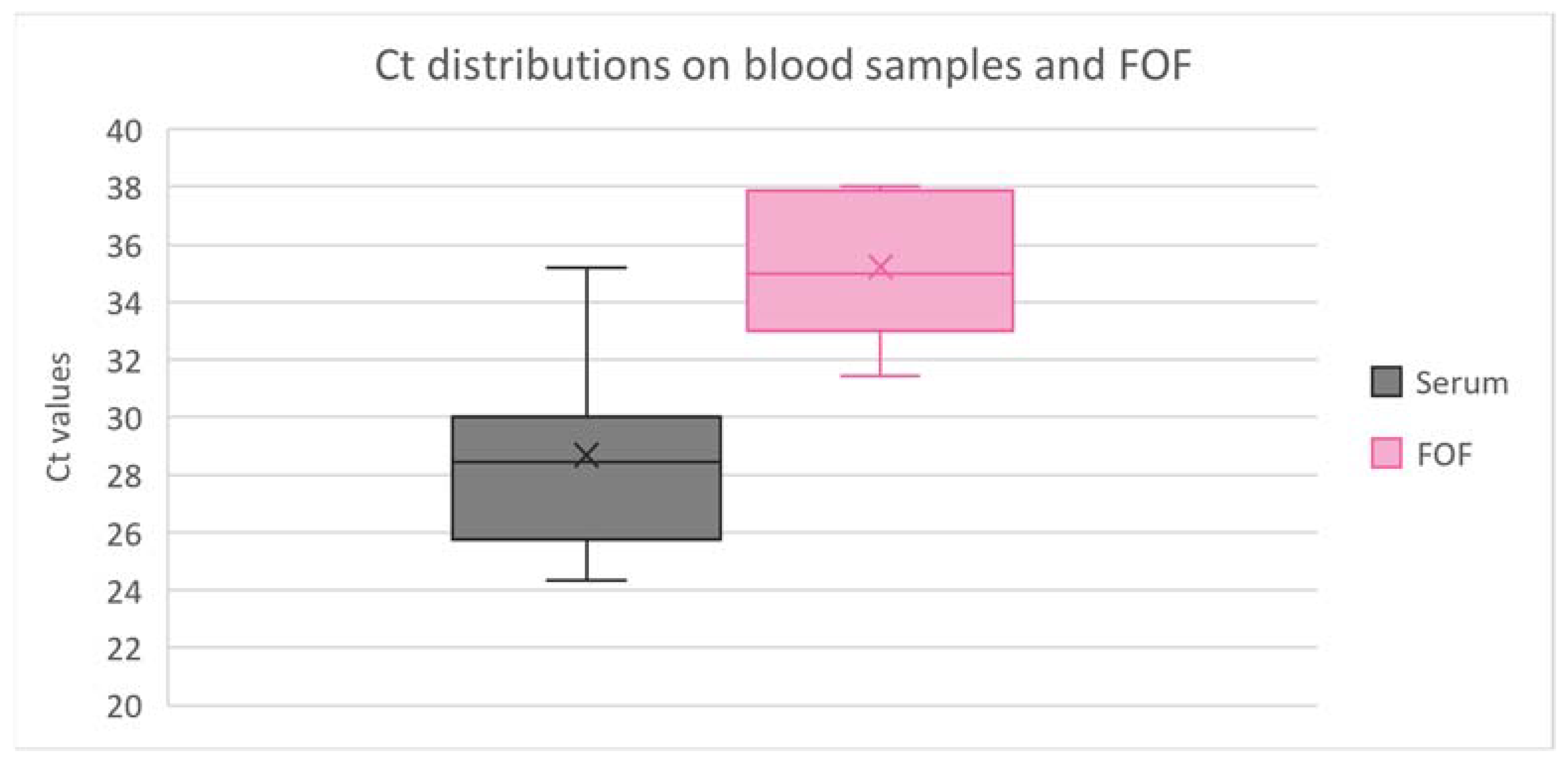
3.2. Comparative Ct in Samples Analyzed Individually
3.3. Evaluation of Pooling
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holtkamp, D.; Kliebenstein, J.; Neumann, E.; Zimmerman, J.; Rotto, H.; Yoder, T.; Wang, C.; Yeske, P.; Mowrer, C.; Haley, C. Assessment of the Economic Impact of Porcine Reproductive and Respiratory Syndrome Virus on U.S. Pork Producers. J. Swine Health Prod. 2013, 21, 72–84. [Google Scholar]
- Nathues, H.; Alarcon, P.; Rushton, J.; Jolie, R.; Fiebig, K.; Jimenez, M.; Geurts, V.; Nathues, C. Cost of Porcine Reproductive and Respiratory Syndrome Virus at Individual Farm Level—An Economic Disease Model. Prev. Vet. Med. 2017, 142, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Renken, C.; Nathues, C.; Swam, H.; Fiebig, K.; Weiss, C.; Eddicks, M.; Ritzmann, M.; Nathues, H. Application of an Economic Calculator to Determine the Cost of Porcine Reproductive and Respiratory Syndrome at Farm-Level in 21 Pig Herds in Germany. Porc. Health Manag. 2021, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Pileri, E.; Mateu, E. Review on the Transmission Porcine Reproductive and Respiratory Syndrome Virus between Pigs and Farms and Impact on Vaccination. Vet. Res. 2016, 47, 108. [Google Scholar] [CrossRef] [PubMed]
- Martín-Valls, G.E.; Cortey, M.; Allepuz, A.; Illas, F.; Tello, M.; Mateu, E. Introduction of a PRRSV-1 Strain of Increased Virulence in a Pig Production Structure in Spain: Virus Evolution and Impact on Production. Porc. Health Manag. 2023, 9, 1. [Google Scholar] [CrossRef]
- Holtkamp, D.J.; Morrison, B.; Rowland, R.R.; Snelson, H. Terminology for Classifying Swine Herds by Porcine Reproductive and Respiratory Syndrome Virus Status. J. Swine Health Prod. 2011, 19, 13. [Google Scholar]
- Kittawornrat, A.; Panyasing, Y.; Goodell, C.; Wang, C.; Gauger, P.; Harmon, K.; Rauh, R.; Desfresne, L.; Levis, I.; Zimmerman, J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Surveillance Using Pre-Weaning Oral Fluid Samples Detects Circulation of Wild-Type PRRSV. Vet. Microbiol. 2014, 168, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Lebret, A.; Boulbria, G.; Berton, P.; Moalic, P.-Y.; Le Guennec, J.; Bouchet, F.; Auvigne, V.; Normand, V. Monitoring PRRSV-1 in Suckling Piglets in an Endemic Herd Using Reverse Transcriptase Quantitative Real Time Polymerase Chain Reaction: Comparison of the Rate of Detection in Serum and Oral Fluid Samples and Evaluation of Pooling. Porc. Health Manag. 2019, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.N.; Rotto, H.; Schneider, P.; Robb, C.; Zimmerman, J.J.; Holtkamp, D.J.; Rademacher, C.J.; Linhares, D.C.L. Collecting Oral Fluid Samples from Due-to-Wean Litters. Prev. Vet. Med. 2020, 174, 104810. [Google Scholar] [CrossRef]
- Osemeke, O.H.; de Freitas Costa, E.; Almeida, M.N.; Trevisan, G.; Ghosh, A.P.; Silva, G.S.; Linhares, D.C.L. Effect of Pooling Family Oral Fluids on the Probability of PRRSV RNA Detection by RT-RtPCR. Prev. Vet. Med. 2022, 206, 105701. [Google Scholar] [CrossRef]
- Vilalta, C.; Sanhueza, J.M.; Schwartz, M.; Kikuti, M.; Torremorell, M.; Corzo, C.A. Assessing the Litter Level Agreement of RT-PCR Results for Porcine Reproductive and Respiratory Syndrome Virus in Testicles, Tails and Udder Wipes Diagnostic Samples Relative to Serum from Piglets. Prev. Vet. Med. 2021, 186, 105211. [Google Scholar] [CrossRef] [PubMed]
- López, W.A.; Zimmerman, J.J.; Gauger, P.C.; Harmon, K.M.; Bradner, L.; Zhang, M.; Giménez-Lirola, L.; Ramirez, A.; Cano, J.P.; Linhares, D.C.L. Practical Aspects of PRRSV RNA Detection in Processing Fluids Collected in Commercial Swine Farms. Prev. Vet. Med. 2020, 180, 105021. [Google Scholar] [CrossRef] [PubMed]
- Vilalta, C.; Sanhueza, J.; Alvarez, J.; Murray, D.; Torremorell, M.; Corzo, C.; Morrison, R. Use of Processing Fluids and Serum Samples to Characterize Porcine Reproductive and Respiratory Syndrome Virus Dynamics in 3 Day-Old Pigs. Vet. Microbiol. 2018, 225, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Martín-Valls, G.E.; Hidalgo, M.; Cano, E.; Mateu, E. Testing of Umbilical Cords by Real Time PCR Is Suitable for Assessing Vertical Transmission of Porcine Reproductive and Respiratory Syndrome Virus under Field Conditions. Vet. J. 2018, 234, 27–29. [Google Scholar] [CrossRef]
- Baliellas, J.; Novell, E.; Enric-Tarancón, V.; Vilalta, C.; Fraile, L. Porcine Reproductive and Respiratory Syndrome Surveillance in Breeding Herds and Nurseries Using Tongue Tips from Dead Animals. Vet. Sci. 2021, 8, 259. [Google Scholar] [CrossRef]
- Machado, I.F.; Magalhães, E.S.; Silva, A.P.S.P.; Moraes, D.C.A.; Cezar, G.; Mil-Homens, M.P.; Osemeke, O.H.; Paiva, R.; Moura, C.A.A.; Gauger, P.; et al. Porcine Reproductive and Respiratory Syndrome Virus RNA Detection in Tongue Tips from Dead Animals. Front. Vet. Sci. 2022, 9, 993442. [Google Scholar] [CrossRef]
- Holtkamp, D.; Torremorell, M.; Corzo, C.; Linhares, D.; Nunes de Almeida, M.; Polson, D.; Snelson, H.; Silva, G.; Sanhueza, J.; Vilalta, C.; et al. Proposed Modifications to Porcine Reproductive and Respiratory Syndrome Virus Herd Classification. J. Swine Health Prod. 2021, 29, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Gibert, E.; Martín-Valls, G.; Mateu, E. Comparison of Protocols for the Analysis of Type 1 Porcine Reproductive and Respiratory Syndrome Virus by RT-PCR Using Oral Fluids. J. Virol. Methods 2017, 243, 190–195. [Google Scholar] [CrossRef]
- Almeida, M.N.; Zhang, M.; Zimmerman, J.J.; Holtkamp, D.J.; Linhares, D.C.L. Finding PRRSV in Sow Herds: Family Oral Fluids vs. Serum Samples from Due-to-Wean Pigs. Prev. Vet. Med. 2021, 193, 105397. [Google Scholar] [CrossRef]
- Boulbria, G.; Normand, V.; Leblanc-Maridor, M.; Belloc, C.; Berton, P.; Bouchet, F.; Lebret, A. Feasibility of Pooled Oral Fluid Collection from Pre-Weaning Piglets Using Cotton Ropes. Vet. Anim. Sci. 2020, 9, 100099. [Google Scholar] [CrossRef]
- Gerber, P.F.; O’Neill, K.; Owolodun, O.; Wang, C.; Harmon, K.; Zhang, J.; Halbur, P.G.; Zhou, L.; Meng, X.-J.; Opriessnig, T. Comparison of Commercial Real-Time Reverse Transcription-PCR Assays for Reliable, Early, and Rapid Detection of Heterologous Strains of Porcine Reproductive and Respiratory Syndrome Virus in Experimentally Infected or Noninfected Boars by Use of Different Sample Types. J. Clin. Microbiol. 2013, 51, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Vilalta, C.; Sanhueza, J.; Garrido, J.; Murray, D.; Morrison, R.; Corzo, C.A.; Torremorell, M. Indirect Assessment of Porcine Reproductive and Respiratory Syndrome Virus Status in Pigs Prior to Weaning by Sampling Sows and the Environment. Vet. Microbiol. 2019, 237, 108406. [Google Scholar] [CrossRef] [PubMed]
- Lopez, W.A.; Gauger, P.C.; Harmon, K.; Bradner, L.; Cano, J.P.; Silva, G.; Macedo, N.; Angulo, J.; Linhares, D.C.L. Modeling the Dilution Effect of PRRSV RNA in Processing Fluid Field Samples on the Probability of Virus Detection by QRT-PCR. Annu. Meet. Am. Assoc. Swine Vet. 2019, 1, 48–49. [Google Scholar]
- Rovira, A.; Clement, T.; Christopher-Hennings, J.; Thompson, B.; Engle, M.; Reicks, D.; Muñoz-Zanzi, C. Evaluation of the Sensitivity of Reverse-Transcription Polymerase Chain Reaction to Detect Porcine Reproductive and Respiratory Syndrome Virus on Individual and Pooled Samples from Boars. J. Vet. Diagn. Investig. 2007, 19, 502–509. [Google Scholar] [CrossRef]
- Vilalta, C.; Baker, J.; Sanhueza, J.; Murray, D.; Sponheim, A.; Alvarez, J.; Sylvia, F.; Polson, D.; Torremorell, M.; Corzo, C.; et al. Effect of Litter Aggregation and Pooling on Detection of Porcine Reproductive and Respiratory Virus in Piglet Processing Fluids. J. Vet. Diagn. Investig. 2019, 31, 625–628. [Google Scholar] [CrossRef]
| No. of Litters | RT-qPCR + | |||
|---|---|---|---|---|
| Serum | FOF | UW | ||
| Batch 1 | 30 | 3 | 1 | 0 |
| Batch 2 | 29 | 7 | 5 | 0 |
| Batch 3 | 30 | 2 | 2 | 1 |
| Batch 4 | 30 | 1 | 1 | 0 |
| 119 | 13 | 9 | 1 | |
| Serum | ||||
|---|---|---|---|---|
| NEG | POS | Total | ||
| FOF | NEG | 103 | 7 | 110 |
| POS | 3 | 6 | 9 | |
| Total | 106 | 13 | 119 | |
| SERA | FOF | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample Identification | Batch | Ct Individual | Ct Pool 1:3 | Ct Pool 1:5 | Sample Identification | Batch | Ct Individual | Ct Pool 1:3 | Ct Pool 1:5 |
| Serum-1 | 1 | 24.9 | 28 | 28.9 | FOF-1 | 1 | 31.4 | 32.5 | 34.8 |
| Serum-2 | 1 | 24.3 | 28 | 28.9 | FOF-2 | 2 | 33 | >40 | >40 |
| Serum-3 | 1 | 25.5 | 28.3 | 29.4 | FOF-3 | 2 | 38 | >40 | >40 |
| Serum-4 | 2 | 26 | 28.4 | 29.2 | FOF-4 | 2 | 38 | >40 | >40 |
| Serum-5 | 2 | 33 | 34.4 | 37 | FOF-5 | 2 | 35 | 35.4 | >40 |
| Serum-6 | 2 | 28 | 31.2 | 32 | FOF-6 | 2 | 33 | >40 | >40 |
| Serum-7 | 2 | 30 | 33.8 | 34.9 | FOF-7 | 3 | 36.8 | >40 | >40 |
| Serum-8 | 2 | 30 | 32.8 | 34.3 | FOF-8 | 3 | 37.8 | >40 | >40 |
| Serum-9 | 2 | 30 | >40 | >40 | FOF-9 | 4 | 34.2 | >40 | >40 |
| Serum-10 | 2 | 29 | ND | ND | |||||
| Serum-11 | 3 | 35.2 | >40 | >40 | |||||
| Serum-12 | 3 | 28.4 | 30.7 | 31.6 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebret, A.; Normand, V.; Berton, P.; Nicolazo, T.; Teixeira Costa, C.; Chevance, C.; Brissonnier, M.; Boulbria, G. Alternative Samples for Porcine Reproductive and Respiratory Syndrome Surveillance in an Endemic PRRSV-1-Infected Breeding Herd: A Descriptive Study. Vet. Sci. 2023, 10, 558. https://doi.org/10.3390/vetsci10090558
Lebret A, Normand V, Berton P, Nicolazo T, Teixeira Costa C, Chevance C, Brissonnier M, Boulbria G. Alternative Samples for Porcine Reproductive and Respiratory Syndrome Surveillance in an Endemic PRRSV-1-Infected Breeding Herd: A Descriptive Study. Veterinary Sciences. 2023; 10(9):558. https://doi.org/10.3390/vetsci10090558
Chicago/Turabian StyleLebret, Arnaud, Valérie Normand, Pauline Berton, Théo Nicolazo, Charlotte Teixeira Costa, Céline Chevance, Mathieu Brissonnier, and Gwenaël Boulbria. 2023. "Alternative Samples for Porcine Reproductive and Respiratory Syndrome Surveillance in an Endemic PRRSV-1-Infected Breeding Herd: A Descriptive Study" Veterinary Sciences 10, no. 9: 558. https://doi.org/10.3390/vetsci10090558
APA StyleLebret, A., Normand, V., Berton, P., Nicolazo, T., Teixeira Costa, C., Chevance, C., Brissonnier, M., & Boulbria, G. (2023). Alternative Samples for Porcine Reproductive and Respiratory Syndrome Surveillance in an Endemic PRRSV-1-Infected Breeding Herd: A Descriptive Study. Veterinary Sciences, 10(9), 558. https://doi.org/10.3390/vetsci10090558






