A Single-Cell Atlas of an Early Mongolian Sheep Embryo
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Experiment Animals
2.2. Embryo Sample Collection and Preparation of Single-Cell Embryo Cell Suspension
2.3. Embryos Single Cell RNA-Seq Performances and Sequencing
2.4. Mapping and Clustering Analysis
2.5. High-Density Plots
2.6. Pseudo-Time Analysis
2.7. RNA Velocity
2.8. qRT-PCR Analysis
2.9. Statistical Analysis
3. Results
3.1. Cell Type Identification and Cluster Analysis at E16 in Ujumqin and Hulunbuir Sheep
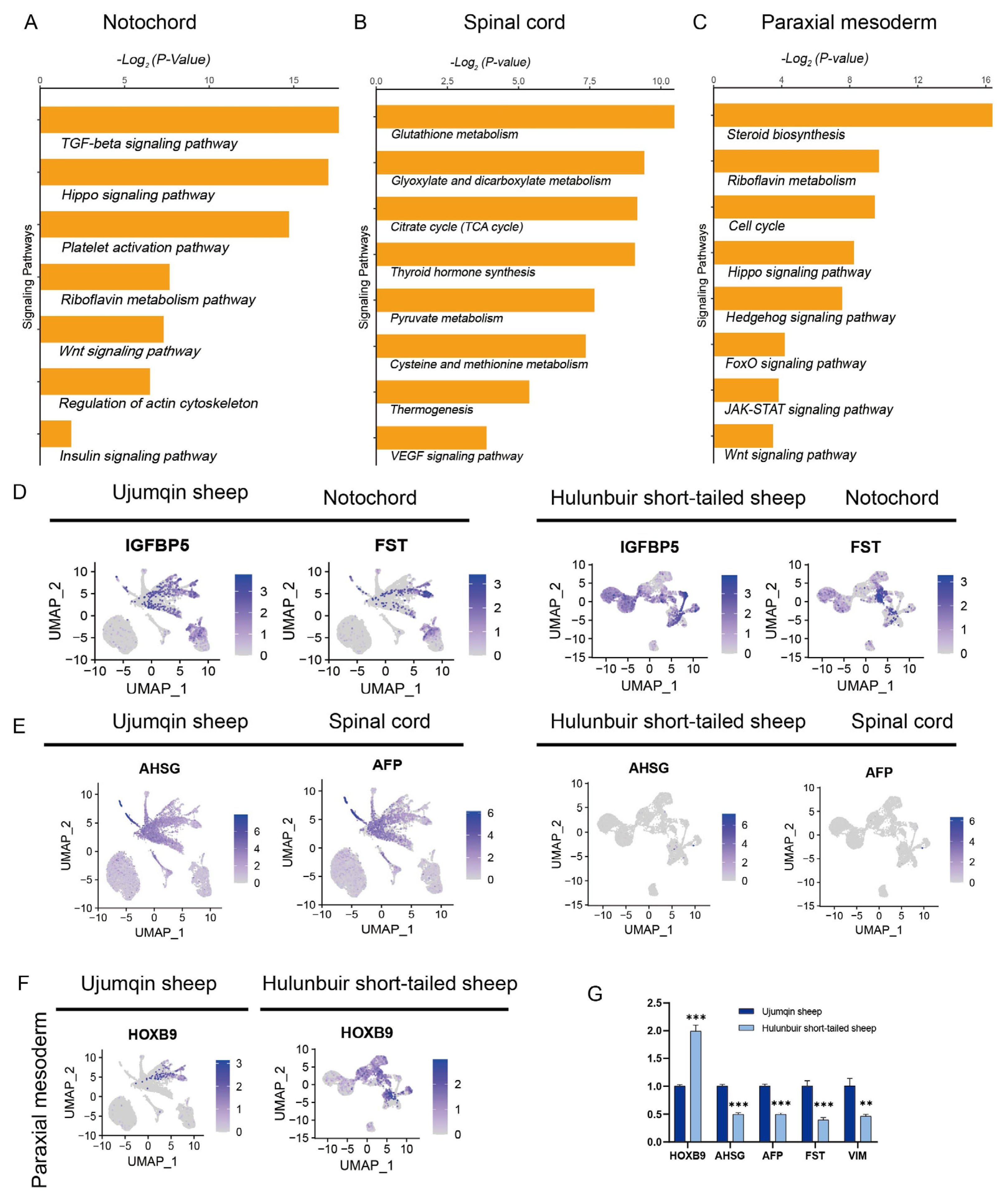
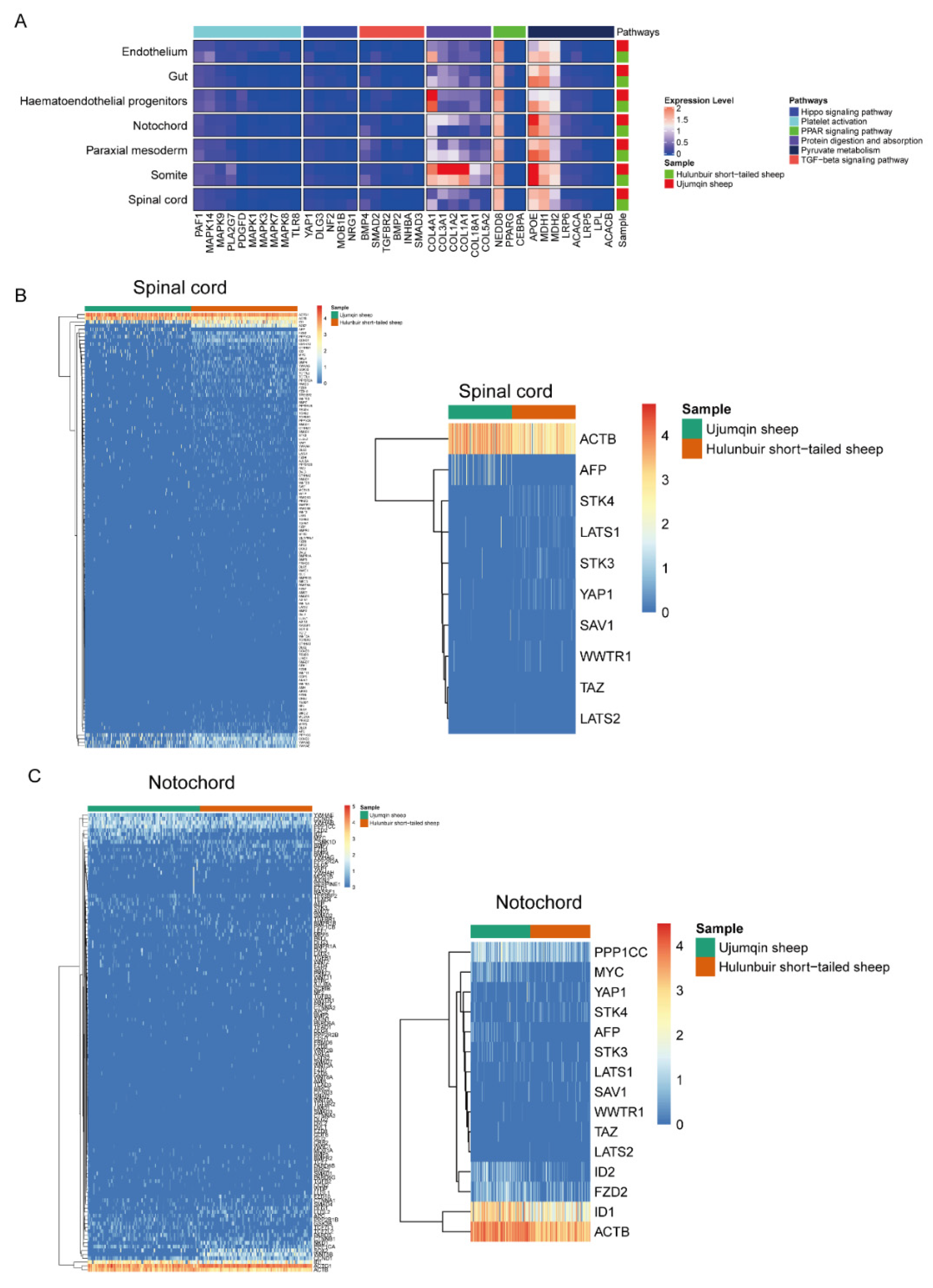
3.2. Functional Enrichment Analysis of E16 in Ujumqin and Hulunbuir Sheep
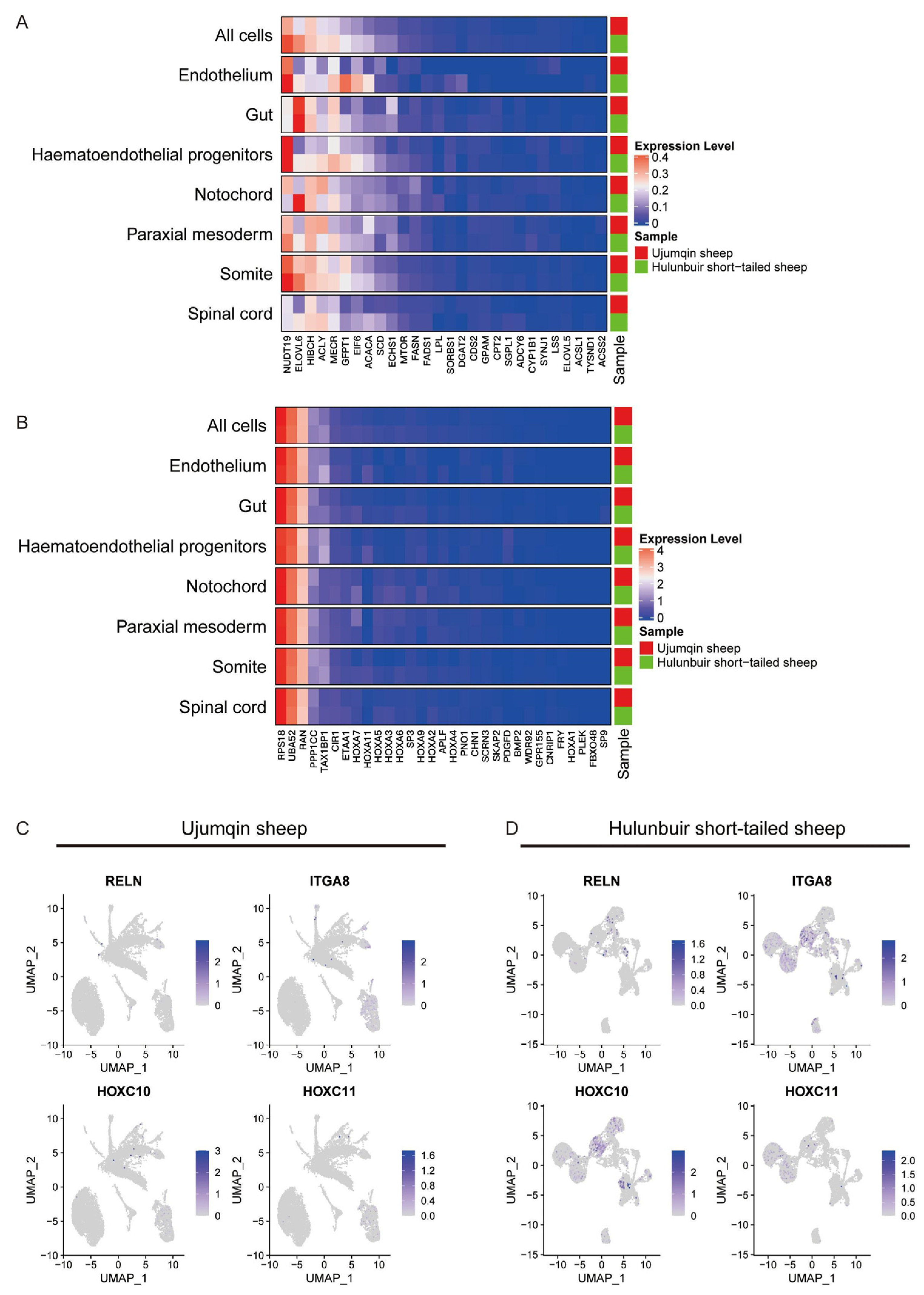
3.3. Different Expression Gene Analysis in Every Cell Cluster at E16 in Ujumqin and Hulunbuir Sheep
3.4. Sc-RNAseq Analysis of Notochord, Paraxial Mesoderm, Somite, and Spinal Cord Cell Characteristics in Hulunbuir and Ujumqin Sheep
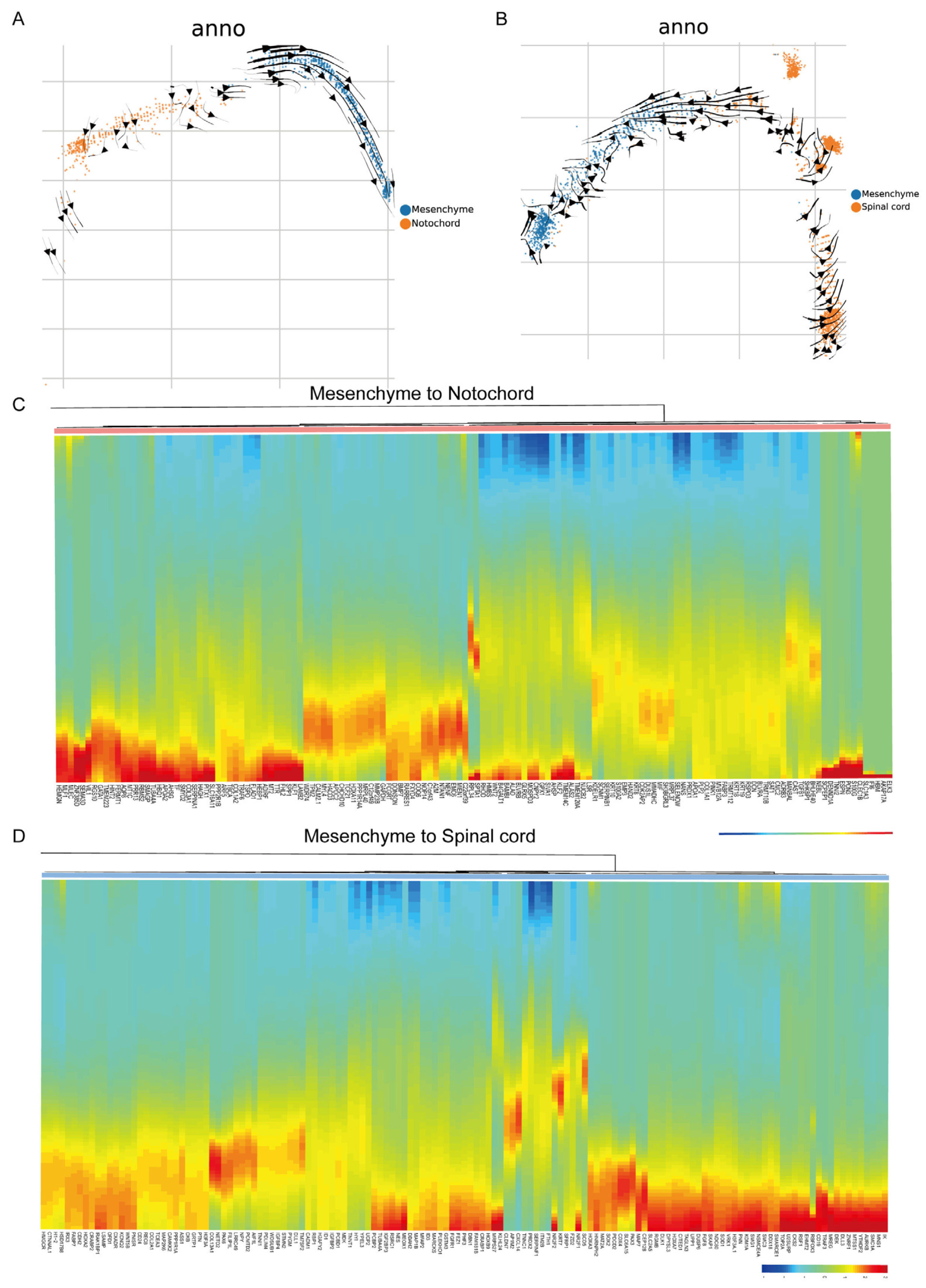
3.5. RNA Velocity and Gene Expression Analyses Support the Mesenchyme–Notochord Cell Transformation Theory in Ujumqin Sheep
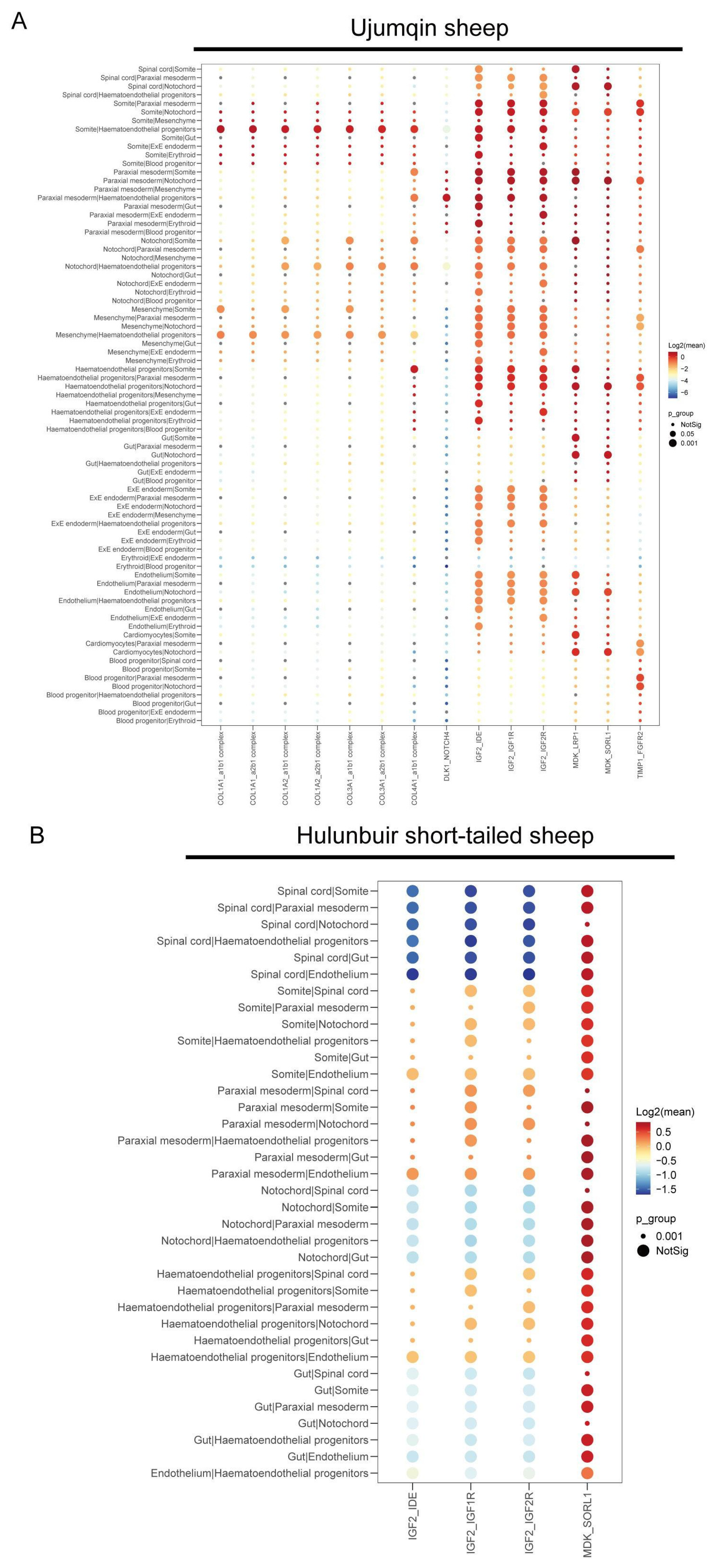
3.6. Cell–Cell Communication Analysis in Ujumqin and Hulunbuir Sheep
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, S.; Luo, R.; Lai, D.; Ma, M.; Hao, F.; Qi, X.; Liu, X.; Liu, D. Whole-genome resequencing of Ujumqin sheep to investigate the determinants of the multi-vertebral trait. Genome 2018, 61, 653–661. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, M.; Jin, Y.; Erdenee, S.; Hu, L.; Chen, H.; Cai, Y.; Lan, X. Comparative Transcriptome Profiling of mRNA and lncRNA Related to Tail Adipose Tissues of Sheep. Front. Genet. 2018, 9, 365. [Google Scholar] [CrossRef]
- Zhi, D.; Da, L.; Liu, M.; Cheng, C.; Zhang, Y.; Wang, X.; Li, X.; Tian, Z.; Yang, Y.; He, T.; et al. Whole Genome Sequencing of Hulunbuir Short-Tailed Sheep for Identifying Candidate Genes Related to the Short-Tail Phenotype. G3 Genes Genomes Genet. 2018, 8, 377–383. [Google Scholar] [CrossRef]
- Han, J.; Yang, M.; Guo, T.; Niu, C.; Liu, J.; Yue, Y.; Yuan, C.; Yang, B. Two linked TBXT (brachyury) gene polymorphisms are associated with the tailless phenotype in fat-rumped sheep. Anim. Genet. 2019, 50, 772–777. [Google Scholar] [CrossRef]
- Lolas, M.; Valenzuela, P.D.T.; Tjian, R.; Liu, Z. Charting Brachyury-mediated developmental pathways during early mouse embryogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 4478–4483. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Sun, X.; Hu, X.-S.; Zhuang, Y.; Liu, Y.-C.; Meng, H.; Miao, L.; Yu, H.; Luo, S.-J. Whole Genome Sequencing Identifies a Missense Mutation in HES7 Associated with Short Tails in Asian Domestic Cats. Sci. Rep. 2016, 6, 31583. [Google Scholar] [CrossRef]
- Choi, Y.B.; Kim, G.A.; Oh, H.J.; Kim, M.J.; Jo, Y.K.; Setyawan, E.M.; Lee, S.H.; Lee, B.C. Cloning of the short-tailed Gyeongju Donggyeong dog via SCNT: Conserving phenotypic inheritance. J. Vet. Med. Sci. 2016, 78, 329–331. [Google Scholar] [CrossRef][Green Version]
- Hytönen, M.K.; Grall, A.; Hédan, B.; Dréano, S.; Seguin, S.J.; Delattre, D.; Thomas, A.; Galibert, F.; Paulin, L.; Lohi, H.; et al. Ancestral T-Box Mutation Is Present in Many, but Not All, Short-Tailed Dog Breeds. J. Hered. 2008, 100, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Yoo, D.; Kim, K.; Kim, H.; Cho, S.; Kim, J.N.; Lim, D.; Choi, S.-G.; Choi, B.-H.; Kim, H. The Genetic Origin of Short Tail in Endangered Korean Dog, DongGyeongi. Sci. Rep. 2017, 7, 10048. [Google Scholar] [CrossRef]
- Fan, X.; Tang, D.; Liao, Y.; Li, P.; Zhang, Y.; Wang, M.; Liang, F.; Wang, X.; Gao, Y.; Wen, L.; et al. Single-cell RNA-seq analysis of mouse preimplantation embryos by third-generation sequencing. PLoS Biol. 2020, 18, e3001017. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Tang, F. Human Germline Cell Development: From the Perspective of Single-Cell Sequencing. Mol. Cell 2019, 76, 320–328. [Google Scholar] [CrossRef]
- Wen, L.; Tang, F. Single-cell sequencing in stem cell biology. Genome Biol. 2016, 17, 71. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, K.; Watt, F.M. Lineage tracing. Cell 2012, 148, 33–45. [Google Scholar] [CrossRef]
- Seita, J.; Weissman, I.L. Hematopoietic stem cell: Self-renewal versus differentiation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 640–653. [Google Scholar] [CrossRef]
- Sulston, J.E.; Schierenberg, E.; White, J.G.; Thomson, J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983, 100, 64–119. [Google Scholar] [CrossRef] [PubMed]
- Gulati, G.S.; Sikandar, S.S.; Wesche, D.J.; Manjunath, A.; Bharadwaj, A.; Berger, M.J.; Ilagan, F.; Kuo, A.H.; Hsieh, R.W.; Cai, S.; et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 2020, 367, 405–411. [Google Scholar] [CrossRef]
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Chal, J.; Pourquié, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef]
- Dequéant, M.L.; Glynn, E.; Gaudenz, K.; Wahl, M.; Chen, J.; Mushegian, A.; Pourquié, O. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science 2006, 314, 1595–1598. [Google Scholar] [CrossRef]
- Brink, S.C.v.D.; Alemany, A.; van Batenburg, V.; Moris, N.; Blotenburg, M.; Vivié, J.; Baillie-Johnson, P.; Nichols, J.; Sonnen, K.F.; Arias, A.M.; et al. Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature 2020, 582, 405–409. [Google Scholar] [CrossRef]
- Zhang, S.; Cui, Y.; Ma, X.; Yong, J.; Yan, L.; Yang, M.; Ren, J.; Tang, F.; Wen, L.; Qiao, J. Single-cell transcriptomics identifies divergent developmental lineage trajectories during human pituitary development. Nat. Commun. 2020, 11, 5275. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.-A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018, 37, 38–44. [Google Scholar] [CrossRef]
- McDavid, A.; Finak, G.; Chattopadyay, P.K.; Dominguez, M.; Lamoreaux, L.; Ma, S.S.; Roederer, M.; Gottardo, R. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 2013, 29, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, V.Y.; Kirschner, K.; Schaub, M.T.; Andrews, T.; Yiu, A.; Chandra, T.; Natarajan, K.N.; Reik, W.; Barahona, M.; Green, A.R.; et al. SC3: Consensus clustering of single-cell RNA-seq data. Nat. Methods 2017, 14, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, M.S.; Tirosh, I.; Heckl, D.; Rao, T.N.; Dixit, A.; Haas, B.J.; Schneider, R.K.; Wagers, A.J.; Ebert, B.L.; Regev, A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hema-topoietic stem cells. Genome Res. 2015, 25, 1860–1872. [Google Scholar] [CrossRef]
- Song, Y.; Guerrero-Juarez, C.F.; Chen, Z.; Tang, Y.; Ma, X.; Lv, C.; Bi, X.; Deng, M.; Bu, L.; Tian, Y.; et al. The Msi1-mTOR pathway drives the pathogenesis of mammary and extramammary Paget’s disease. Cell Res. 2020, 30, 854–872. [Google Scholar] [CrossRef]
- Guerrero-Juarez, C.F.; Dedhia, P.H.; Jin, S.; Ruiz-Vega, R.; Ma, D.; Liu, Y.; Yamaga, K.; Shestova, O.; Gay, D.L.; Yang, Z.; et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun. 2019, 10, 650. [Google Scholar] [CrossRef]
- Qiu, X.; Hill, A.; Packer, J.; Lin, D.; Ma, Y.-A.; Trapnell, C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 2017, 14, 309–315. [Google Scholar] [CrossRef]
- Sheng, X.; Lin, Z.; Lv, C.; Shao, C.; Bi, X.; Deng, M.; Xu, J.; Guerrero-Juarez, C.F.; Li, M.; Wu, X.; et al. Cycling Stem Cells Are Radioresistant and Regenerate the Intestine. Cell Rep. 2020, 32, 107952. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; MacLean, A.L.; Peng, T.; Nie, Q. scEpath: Energy landscape-based inference of transition probabilities and cellular trajectories from single-cell transcriptomic data. Bioinformatics 2018, 34, 2077–2086. [Google Scholar] [CrossRef]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A.; et al. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, J.; Shen, M.; Xie, X.-L.; Liu, G.-J.; Xu, Y.-X.; Lv, F.-H.; Yang, H.; Yang, Y.-L.; Liu, C.-B.; et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and ag-ronomic traits. Nat. Commun. 2020, 11, 2815. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Kim, K.Y.; Wang, Z.; Park, J.-H.; Bae, S.M.; Kim, S.-Y.; Song, H.-Y.; Jeon, J.Y. EW-7197, a Transforming Growth Factor-Beta Type I Receptor Kinase Inhibitor, Ameliorates Acquired Lymphedema in a Mouse Tail Model. Lymphat. Res. Biol. 2020, 18, 433–438. [Google Scholar] [CrossRef]
- Sauvegarde, C.; Paul, D.; Bridoux, L.; Jouneau, A.; Degrelle, S.; Hue, I.; Rezsohazy, R.; Donnay, I. Dynamic Pattern of HOXB9 Protein Localization during Oocyte Maturation and Early Embryonic Development in Mammals. PLoS ONE 2016, 11, e0165898. [Google Scholar] [CrossRef]
- Finckenstein, F.G.; Davicioni, E.; Osborn, K.G.; Cavenee, W.K.; Arden, K.C.; Anderson, M.J. Transgenic mice expressing PAX3-FKHR have multiple defects in muscle development, including ectopic skeletal myogenesis in the developing neural tube. Transgenic Res. 2006, 15, 595–614. [Google Scholar] [CrossRef]
- Yasuoka, Y.; Shinzato, C.; Satoh, N. The Mesoderm-Forming Gene brachyury Regulates Ectoderm-Endoderm Demarcation in the Coral Acropora digitifera. Curr. Biol. 2016, 26, 2885–2892. [Google Scholar] [CrossRef]
- Wu, B.; Shao, Y.; Chen, B.; Liu, C.; Xue, Z.; Wu, P.; Li, H. Identification of a Novel Mouse Brachyury (T) Allele Causing a Short Tail Mutation in Mice. Cell Biochem. Biophys. 2010, 58, 129–135. [Google Scholar] [CrossRef]
- Gourain, V.; Duluc, I.; Domon-Dell, C.; Freund, J.-N. A Core Response to the CDX2 Homeoprotein during Development and in Pathologies. Front. Genet. 2021, 12, 744165. [Google Scholar] [CrossRef]
- Xu, J.; Tang, Y.; Sheng, X.; Tian, Y.; Deng, M.; Du, S.; Lv, C.; Li, G.; Pan, Y.; Song, Y.; et al. Secreted stromal protein ISLR promotes intestinal regeneration by suppressing epithelial Hippo signaling. EMBO J. 2020, 39, e103255. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Lee, Y.; Lee, K.; Lee, H.; Yoo, J.E.; Ahn, S.; Park, Y.N.; Kim, H. The Clinicopathological Significance of YAP/TAZ Expression in Hepatocellular Carcinoma with Relation to Hypoxia and Stemness. Pathol. Oncol. Res. 2021, 27, 604600. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Barry, J.; Anthony, R. The pregnant sheep as a model for human pregnancy. Theriogenology 2008, 69, 55–67. [Google Scholar] [CrossRef]
- Sanchez, D.J.D.; Vasconcelos, F.R.; Teles-Filho, A.C.A.; Viana, A.G.A.; Martins, A.M.A.; Sousa, M.V.; Castro, M.S.; Ricart, C.A.; Fontes, W.; Bertolini, M.; et al. Proteomic profile of pre-implantational ovine embryos produced in vivo. Reprod. Domest. Anim. 2021, 56, 586–603. [Google Scholar] [CrossRef] [PubMed]
- Ptak, G.E.; D’Agostino, A.; Toschi, P.; Fidanza, A.; Zacchini, F.; Czernik, M.; Monaco, F.; Loi, P. Post-implantation mortality of in vitro produced embryos is associated with DNA methyltransferase 1 dys-function in sheep placenta. Hum. Reprod. 2013, 28, 298–305. [Google Scholar] [CrossRef]
- Mor, A.; Mondal, S.; Reddy, I.; Nandi, S.; Gupta, P. Molecular cloning and expression of FGF2 gene in pre-implantation developmental stages of in vitro-produced sheep embryos. Reprod. Domest. Anim. 2018, 53, 895–903. [Google Scholar] [CrossRef]
- Clark, E.L.; Bush, S.J.; McCulloch, M.E.B.; Farquhar, I.L.; Young, R.; Lefevre, L.; Pridans, C.; Tsang, H.G.; Wu, C.; Afrasiabi, C.; et al. A high resolution atlas of gene expression in the domestic sheep (Ovis aries). PLoS Genet. 2017, 13, e1006997. [Google Scholar] [CrossRef] [PubMed]
- Guillomot, M.; Turbe, A.; Hue, I.; Renard, J.P. Staging of ovine embryos and expression of the T-box genes Brachyury and Eomesodermin around gas-trulation. Reproduction 2004, 127, 491–501. [Google Scholar] [CrossRef] [PubMed][Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, T.; Guo, W.; Yang, G.; Su, H.; Dou, A.; Chen, L.; Ma, T.; Su, J.; Liu, M.; Su, B.; et al. A Single-Cell Atlas of an Early Mongolian Sheep Embryo. Vet. Sci. 2023, 10, 543. https://doi.org/10.3390/vetsci10090543
He T, Guo W, Yang G, Su H, Dou A, Chen L, Ma T, Su J, Liu M, Su B, et al. A Single-Cell Atlas of an Early Mongolian Sheep Embryo. Veterinary Sciences. 2023; 10(9):543. https://doi.org/10.3390/vetsci10090543
Chicago/Turabian StyleHe, Tingyi, Wenrui Guo, Guang Yang, Hong Su, Aolei Dou, Lu Chen, Teng Ma, Jie Su, Moning Liu, Budeng Su, and et al. 2023. "A Single-Cell Atlas of an Early Mongolian Sheep Embryo" Veterinary Sciences 10, no. 9: 543. https://doi.org/10.3390/vetsci10090543
APA StyleHe, T., Guo, W., Yang, G., Su, H., Dou, A., Chen, L., Ma, T., Su, J., Liu, M., Su, B., Qi, W., Li, H., Mao, W., Wang, X., Li, X., Yang, Y., Song, Y., & Cao, G. (2023). A Single-Cell Atlas of an Early Mongolian Sheep Embryo. Veterinary Sciences, 10(9), 543. https://doi.org/10.3390/vetsci10090543






