Using Machine Learning in Veterinary Medical Education: An Introduction for Veterinary Medicine Educators
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Introduction to Educational Data Mining and Machine Learning
1.2. Comparison of Classical Statistical Analysis and Machine Learning Models
1.3. Overview of the Main Types of Machine Learning Algorithms and Random Forest Machine Learning Models
1.4. Programming Languages and Tools
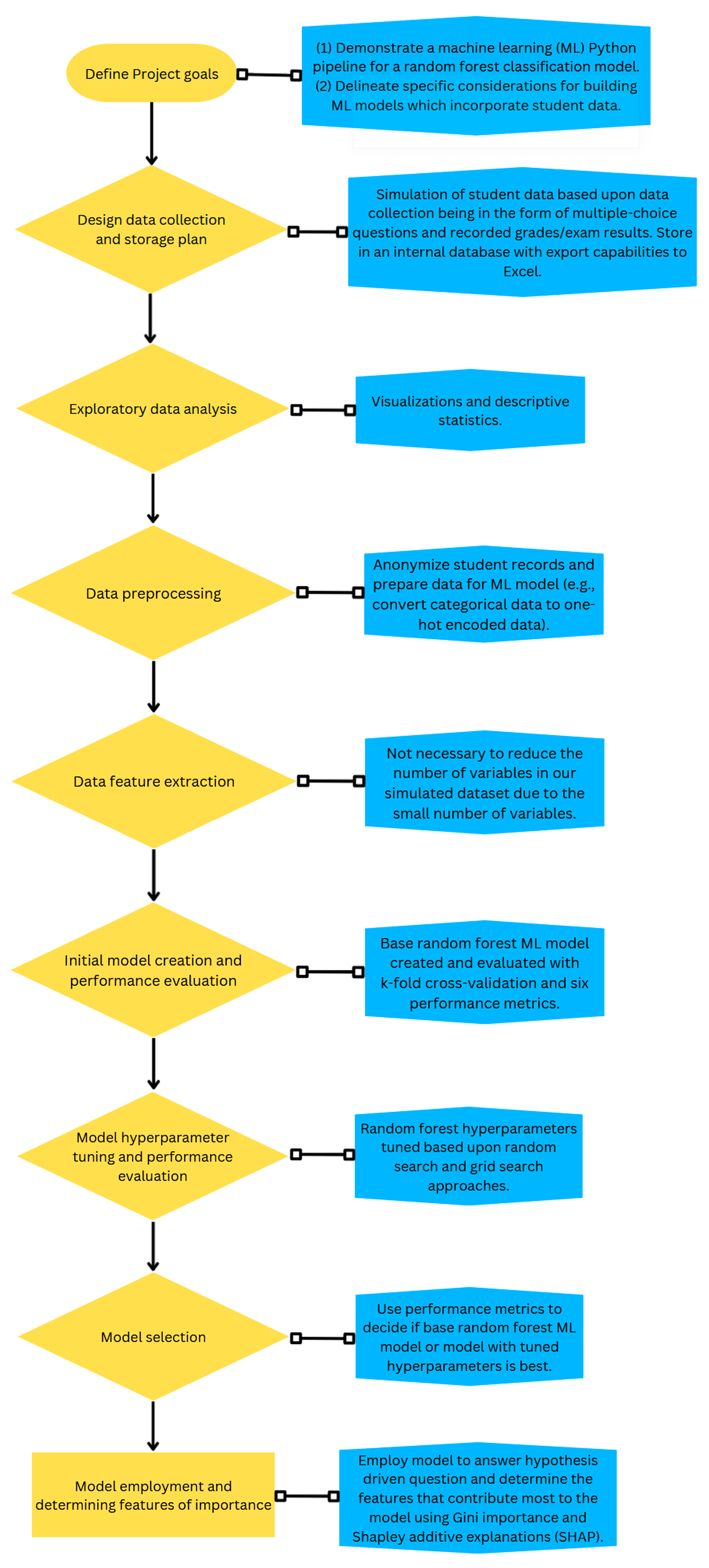
2. Simulation of Dataset and Creation of a Random Forest Machine Learning Model
2.1. Defining the Project Goals
2.2. Data Collection and Storage Plan
2.2.1. Simulated Data Collection and Storage
2.2.2. Importing the Dataset
- #Import required packages:
- Import pandas as pd
- #Import the dataset using the function pd.read_excel().
- Dataset = pd.read_excel(r’C:\location_of_data\name_of_excell_datafile.xlsx’,
- sheet_name = ‘name’)
- #To view the first 10 rows of the dataset with the column names:
- Dataset.head(10)
2.3. Exploratory Data Analysis
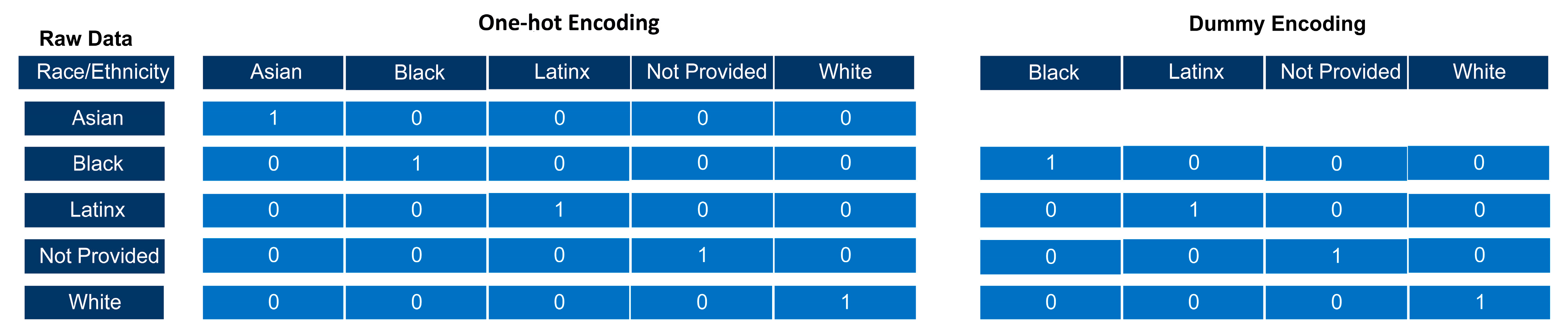
2.4. Data Preprocessing
- #To one-hot encode for race column, use the get_dummies() from the pandas package
- #We assign this transformed data to a new variable called dataset_OneHot
- #We also need the argument drop_first to not be true in order to perform one-hot
- #encoding.
- Dataset_OneHot = pd.get_dummies(dataset, columns = [“Race”], drop_first = False)
- dataset_OneHot.head() #To view the first several rows and column names
- #To dummy encode the gender column
- dataset_OneHot = pd.get_dummies(dataset_OneHot, columns = [“Gender”],
- drop_first = True)
- print(dataset_OneHot.head()) #To view the first several rows and column names
- #Import the first dataset with missing GRE values using the function pd.read_excel().
- Biased_dataset = pd.read_excel(r’C:\location_of_data/name_of_excell_datafile.xlsx’,
- sheet_name = ‘name’)
- #Code to drop delete each student record that does not have a GRE score reported
- #The “empty” GRE value will be noted as an “na” in Python, therefore we use the
- #dropna()
- #The argument axis = 0 means the row with the “na” will be dropped.
- #The argument how = ’any’ means that any “na” will result in the row being deleted
- #The argument inplace = True means that a new dataframe will not be created
- biased_dataset_OneHot.dropna(axis = 0, how = ‘any’, inplace = True)
- #Code to replace each missing GRE score with the mean of the GRE value
- biased_dataset_OneHot.fillna((biased_dataset_OneHot[‘GRE’].mean()), inplace = True)
2.5. Data Feature Extraction
2.6. Model Creation and Performance Evaluation
2.6.1. Generation of Base Random Forest Model
- #X is our variable dataframe and y is our target dataframe
- #create dataframe without target, [rows, columns], the : indicates to select all rows
- X = dataset_OneHot.loc[: , dataset_OneHot.columns != ‘Fail’]
- y = dataset_OneHot[‘Fail’] #target variable for prediction
- #Import required python function of SMOTE from Python package imbalanced-learn
- #version 0.10.1from imblearn.over_sampling import SMOTE,
- #Oversampling to allow 0 and 1 target to be equal
- #Assigning a value to the random state argument ensures that anyone can generate
- #the same set of random numbers again
- X_resampled, y_resampled = SMOTE(random_state = 23).fit_resample(X, y)
- #Import required functions:
- from sklearn.model_selection import train_test_split
- #Use the balanced data to create testing and training datasets with 70% of the data
- #being training and 30% of the data being testing.
- X_trainSMOTE, X_testSMOTE, y_trainSMOTE, y_testSMOTE =
- train_test_split(X_resampled, y_resampled, stratify = y_resampled, test_size = 0.3,
- random_state = 50)
- #Check sizes of arrays to make sure it they match each other
- print(‘Training Variables Shape:’, X_trainSMOTE.shape)
- print(‘Training Target Shape:’, y_trainSMOTE.shape)
- print(‘Testing Variables Shape:’, X_testSMOTE.shape)
- print(‘Testing Target Shape:’, y_testSMOTE.shape)
- #Import required functions:
- from sklearn.ensemble import RandomForestClassifier
- #Build base model without any changes to default settings
- forest_base = RandomForestClassifier(random_state = 23)
- #Train the model via fit()
- forest_base.fit(X_trainSMOTE, y_trainSMOTE) #using training data
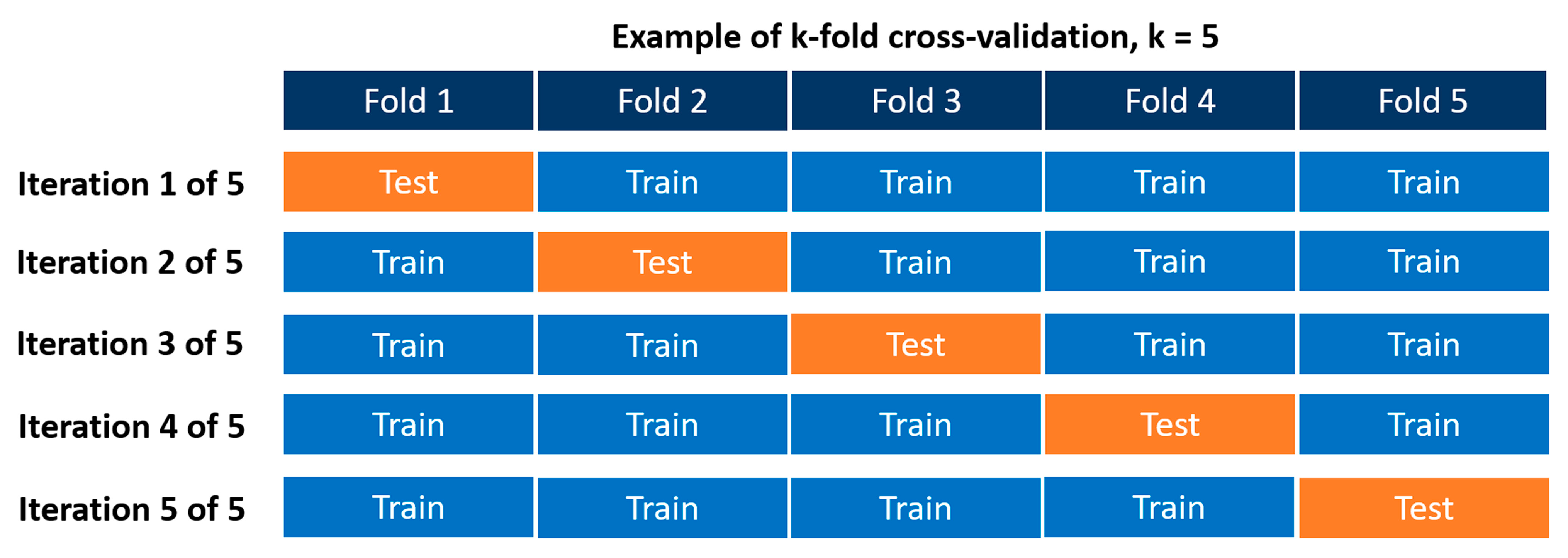
2.6.2. Evaluation of the Base Random Forest Model
- #Make predictions using testing data set
- y_predictions = forest_base.predict(X_testSMOTE)
- y_trueSMOTE = y_testSMOTE #Rename the test target dataframe
- #Import required function
- from sklearn.model_selection import KFold
- #Defining the cross-validation to be able to compute the performance metrics using
- #the k-fold CV
- kf = KFold(shuffle = True, n_splits = 5)
- #Import required function
- from sklearn.model_selection import cross_val_score
- #To calculate the accuracy of the model using k-fold cross validation
- score_accuracy_mean = cross_val_score(forest_base, X_testSMOTE, y_trueSMOTE,
- cv = kf, scoring = ‘accuracy’).mean()
- print(score_accuracy_mean) #View the mean of the CV validation results for
- #accuracy of the model.
- #To calculate the recall of the model using k-fold cross validation
- recall = cross_val_score(best_grid_model, X_testSMOTE, y_testSMOTE, cv = kf,
- scoring = ‘recall’).mean()
- print(recall) #View the mean of the CV validation results for recall of the model
- #Import required function
- from sklearn.metrics import make_scorer
- #Define specificity
- scoring = make_scorer(recall_score, pos_label = 0)
- #Use our defined specificity as the type of score that is calculated
- score_specificity_mean = cross_val_score(forest_base, X_testSMOTE, y_trueSMOTE,
- cv = kf, scoring = scoring).mean()
- cross_val_score(forest_base, X_testSMOTE, y_trueSMOTE, cv = kf, scoring = scoring)
- print(score_specificity_mean) #View the mean of the CV validation results for
- #specificity of the model
- # To calculate the precision of the model using k-fold cross validation
- score_precision_mean = cross_val_score(forest_base, X_testSMOTE, y_trueSMOTE,
- cv = kf, scoring = ‘precision’).mean()
- print(score_precision_mean) #View the mean of the CV validation results for
- #precision of the model
- #To calculate the F-score of the model using k-fold cross validation
- score_f1_mean = cross_val_score(forest_base, X_testSMOTE, y_trueSMOTE, cv = kf,
- scoring = ‘f1’).mean()
- print(score_f1_mean) #View the mean of the CV validation results for precision of
- #the model
- #To calculate the ROC curve AUC of the model using k-fold cross validation
- #score_auc_mean = cross_val_score(forest_base, X_testSMOTE, y_trueSMOTE, cv =
- kf, scoring = ‘roc_auc’).mean()
- print(score_auc_mean) ) #View the mean of the CV validation results for ROC curve
- #AUC of the model
2.6.3. Tuning of the Random Forest Model
- ##Assess hyperparameters to try to improve upon base model:
- #Import required functions:
- from sklearn.model_selection import RandomizedSearchCV
- from sklearn.model_selection import GridSearchCV
- # Create the hyperparameter grid for first the random search function
- hyper_grid = {# Number of trees to be included in random forest
- ‘n_estimators’: [150, 200, 250, 300, 350, 400],
- # Number of features to consider at every split
- ‘max_features’: [‘sqrt’],
- #Maximum number of levels in a tree
- ‘max_depth’: [10, 20, 40, 60, 80, 100, 120, 140, 160, 180, 200],
- # Minimum number of samples required to split a node
- ‘min_samples_split’: [2, 4, 6, 8, 10],
- # Minimum number of samples required at each leaf node
- ‘min_samples_leaf’: [1, 2, 4, 6, 8, 10],
- # Method of selecting samples for training each tree
- ‘bootstrap’: [True, False]}
- #Initiate random forest base model to tune
- best_params = RandomForestClassifier(random_state = (23))
- #Use random grid search to find best hyperparameters, uses k-fold validation as cross
- #validation method
- #Search 200 different combinations
- best_params_results = RandomizedSearchCV(estimator = best_params,
- param_distributions = hyper_grid, n_iter = 200, cv = kf, verbose = 5, random_state = (23))
- #Fit the random search model
- best_params_results.fit(X_trainSMOTE, y_trainSMOTE)
- #Find the best parameters from the grid search results
- Print(best_params_results.best_params_)
- #Build another hyperparameter grid using narrowed down parameter guidelines
- #from above
- #Then use GridSearchCV method to search every combination of grid
- new_grid = {‘n_estimators’: [250, 275, 300, 325, 332, 350, 375],
- ‘max_features’: [‘sqrt’],
- ‘max_depth’: [160, 165, 170, 175, 180, 185, 190, 195],
- ‘min_samples_split’: [1, 2, 3, 4, 5, 6],
- ‘min_samples_leaf’: [1, 2, 3],
- ‘bootstrap’: [True]}
- #Initiate random forest base model to tune
- best_params = RandomForestClassifier(random_state = (23))
- #Use GridSearchCV method to search every combination of grid
- best_params_grid_search = GridSearchCV(estimator = best_params, param_grid =
- new_grid, cv = kf, n_jobs = −1, verbose = 10)
- #Fit the gridsearch model
- best_params_grid_search.fit(X_trainSMOTE, y_trainSMOTE)
- #Get the results of the search grid form the random forest model
- best_params_grid_search.best_params_
- #Using the results of the best parameters, we will create a new model and show the
- #specific arguments.
- best_grid_model = RandomForestClassifier(n_estimators = 375, max_features = ‘sqrt’,
- max_depth = (160), min_samples_split = 2, min_samples_leaf = 2, bootstrap = True)
- #Best model based upon grid
- best_grid_model.fit(X_trainSMOTE, y_trainSMOTE)
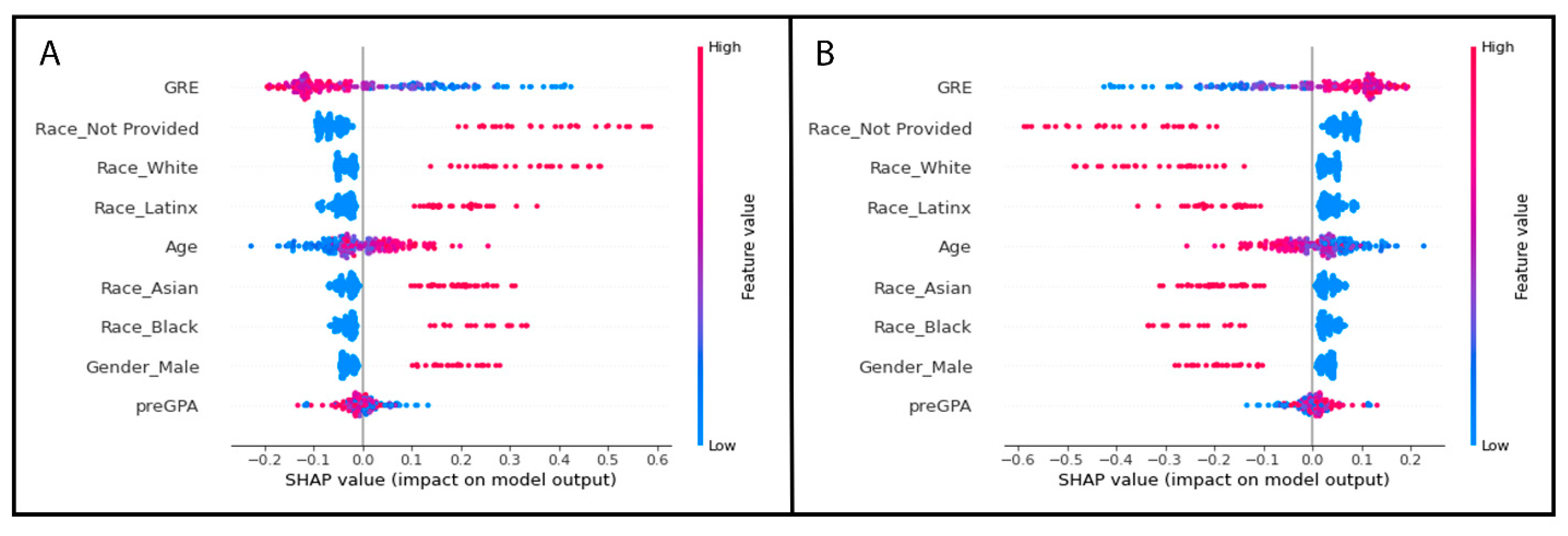
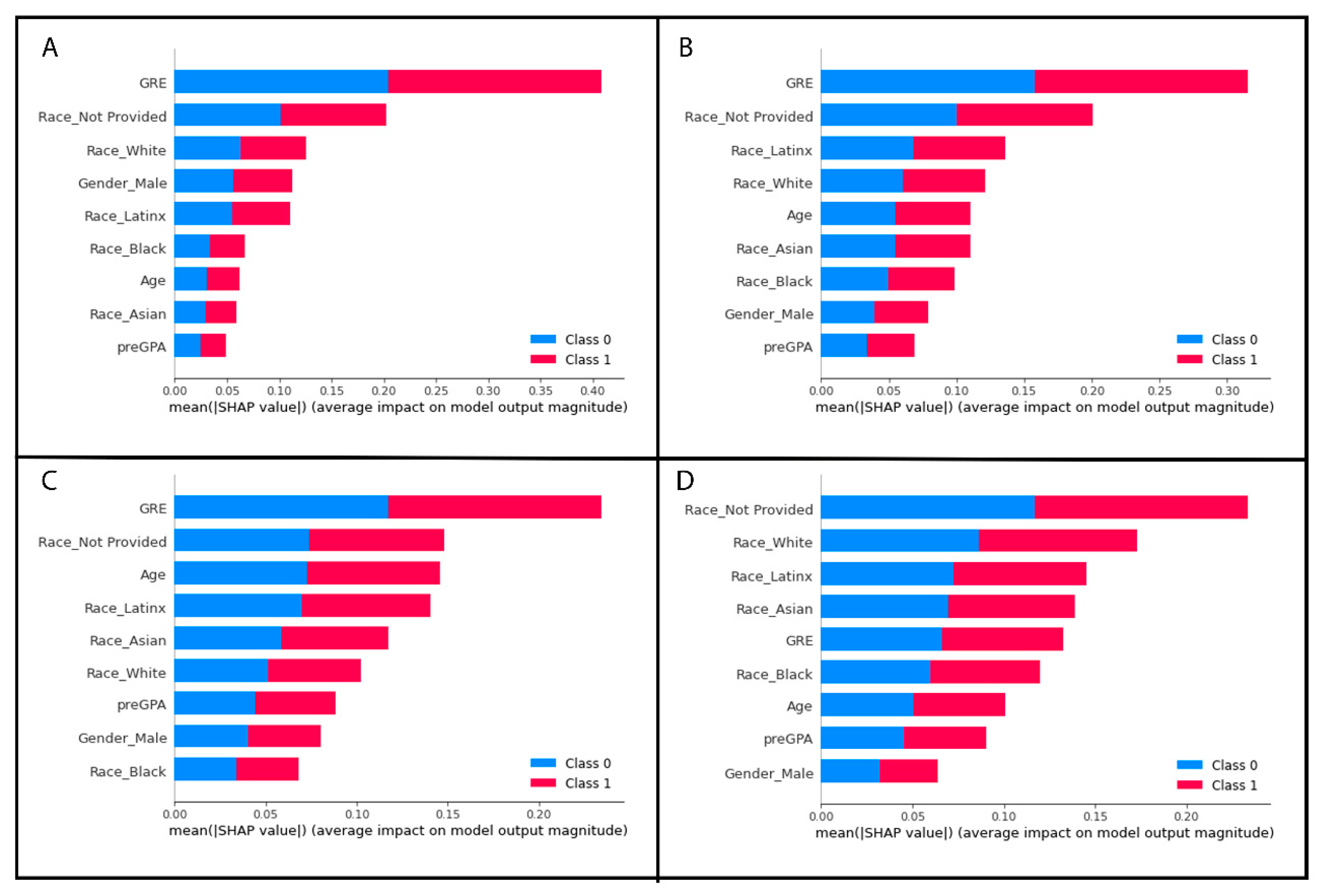
2.6.4. Determining the Most Important Features of the Random Forest Model
- #Most important features from best performing random forest model, Gini im
- #portance
- feature_imp = pd.Series(best_grid_model.feature_importances_, index = X.columns)
- feature_imp = feature_imp.sort_values(ascending = False)
- print(feature_imp)
- #Import required package
- import shap
- #Most important features from best performing random forest model, SHAP values
- shap_feature_imp = shap.TreeExplainer(best_grid_model)
- shap_values = shap_feature_imp.shap_values(X_testSMOTE)
- shap.summary_plot(shap_values, X_testSMOTE) #Shows results in a plot
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Basran, P.S.; Appleby, R.B. The unmet potential of artificial intelligence in veterinary medicine. Am. J. Vet. Res. 2022, 83, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Hennessey, E.; DiFazio, M.; Hennessey, R.; Cassel, N. Artificial intelligence in veterinary diagnostic imaging: A literature review. Vet. Radiol. Ultrasound 2022, 63, 851–870. [Google Scholar] [CrossRef] [PubMed]
- Katznelson, G.; Gerke, S. The need for health AI ethics in medical school education. Adv. Health Sci. Educ. 2021, 26, 1447. [Google Scholar] [CrossRef]
- Calvet Liñán, L.; Juan Pérez, Á.A. Educational Data Mining and Learning Analytics: Differences, similarities, and time evolution. Int. J. Educ. Technol. High. Educ. 2015, 12, 98–112. [Google Scholar] [CrossRef]
- Algarni, A. Data mining in education. Int. J. Adv. Comput. Sci. Appl. 2016, 7, 456–461. [Google Scholar] [CrossRef]
- Alyahyan, E.; Düştegör, D. Predicting academic success in higher education: Literature review and best practices. Int. J. Educ. Technol. High. Educ. 2020, 17, 1–21. [Google Scholar] [CrossRef]
- Samuel, A.L. Some studies in machine learning using the game of checkers. IBM J. Res. Dev. 1959, 3, 210–229. [Google Scholar] [CrossRef]
- Bi, Q.; Goodman, K.E.; Kaminsky, J.; Lessler, J. What is Machine Learning? A Primer for the Epidemiologist. Am. J. Epidemiol. 2019, 188, 2222–2239. [Google Scholar] [CrossRef]
- Von Davier, A.A.; Mislevy, R.J.; Hao, J. Introduction to Computational Psychometrics: Towards a Principled Integration of Data Science and Machine Learning Techniques into Psychometrics. In Computational Psychometrics: New Methodologies for a New Generation of Digital Learning and Assessment: With Examples in R and Python; von Davier, A.A., Mislevy, R.J., Hao, J., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 1–6. [Google Scholar]
- Khamisy-Farah, R.; Gilbey, P.; Furstenau, L.B.; Sott, M.K.; Farah, R.; Viviani, M.; Bisogni, M.; Kong, J.D.; Ciliberti, R.; Bragazzi, N.L. Big Data for Biomedical Education with a Focus on the COVID-19 Era: An Integrative Review of the Literature. Int J Env. Res Public Health 2021, 18, 8989. [Google Scholar] [CrossRef]
- Peers, I. Statistical Analysis for Education and Psychology Researchers: Tools for Researchers in Education and Psychology; Routledge: London, UK, 2006. [Google Scholar]
- Nie, R.; Guo, Q.; Morin, M. Machine Learning Literacy for Measurement Professionals: A Practical Tutorial. Educ. Meas. Issues Pract. 2023, 42, 9–23. [Google Scholar] [CrossRef]
- Van Vertloo, L.R.; Burzette, R.G.; Danielson, J.A. Predicting Academic Difficulty in Veterinary Medicine: A Case-Control Study. J. Vet. Med. Educ. 2022, 49, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, J.C. Logistic Regression: A Brief Primer. Acad. Emerg. Med. 2011, 18, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Mo, B.; Zhao, J. Theory-based residual neural networks: A synergy of discrete choice models and deep neural networks. Transp. Res. Part B Methodol. 2021, 146, 333–358. [Google Scholar] [CrossRef]
- Dass, S.; Gary, K.; Cunningham, J. Predicting Student Dropout in Self-Paced MOOC Course Using Random Forest Model. Information 2021, 12, 476. [Google Scholar] [CrossRef]
- He, L.; Levine, R.A.; Fan, J.; Beemer, J.; Stronach, J. Random forest as a predictive analytics alternative to regression in institutional research. Pract. Assess. Res. Eval. 2018, 23, 1. [Google Scholar]
- Sarker, I.H. Machine Learning: Algorithms, Real-World Applications and Research Directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef]
- Romero, C.; Ventura, S. Educational Data Mining: A Review of the State of the Art. IEEE Trans. Syst. Man Cybern. Part C (Appl. Rev.) 2010, 40, 601–618. [Google Scholar] [CrossRef]
- Louppe, G. Understanding random forests: From theory to practice. arXiv 2014, arXiv:1407.7502. [Google Scholar]
- Spoon, K.; Beemer, J.; Whitmer, J.C.; Fan, J.; Frazee, J.P.; Stronach, J.; Bohonak, A.J.; Levine, R.A. Random Forests for Evaluating Pedagogy and Informing Personalized Learning. J. Educ. Data Min. 2016, 8, 20–50. [Google Scholar] [CrossRef]
- Choudhary, R.; Gianey, H.K. Comprehensive Review On Supervised Machine Learning Algorithms. In Proceedings of the 2017 International Conference on Machine Learning and Data Science (MLDS), Noida, India, 14–15 December 2017; pp. 37–43. [Google Scholar]
- Kumar, N. Advantages and Disadvantages of Random Forest Algorithm in Machine Learning. Available online: http://theprofessionalspoint.blogspot.com/2019/02/advantages-and-disadvantages-of-random.html (accessed on 11 February 2022).
- Altman, N.; Krzywinski, M. Ensemble methods: Bagging and random forests. Nat. Methods 2017, 14, 933–935. [Google Scholar] [CrossRef]
- Dietterich, T.G. Ensemble Learning. In The Handbook of Brain Theory and Neural Networks, 2nd ed.; Arbib, M.A., Ed.; MIT Press: Cambridge, MA, USA, 2002. [Google Scholar]
- Anaconda Software Distribution. v22.9.0. October 2022. Available online: https://www.anaconda.com/download (accessed on 13 November 2022).
- Wang, Y.; Wen, M.; Liu, Y.; Wang, Y.; Li, Z.; Wang, C.; Yu, H.; Cheung, S.-C.; Xu, C.; Zhu, Z. Watchman: Monitoring dependency conflicts for python library ecosystem. In Proceedings of the ACM/IEEE 42nd International Conference on Software Engineering, Seoul, Republic of Korea, 27 June–19 July 2020; pp. 125–135. [Google Scholar]
- Gudivada, V.; Apon, A.; Ding, J. Data quality considerations for big data and machine learning: Going beyond data cleaning and transformations. Int. J. Adv. Softw. 2017, 10, 1–20. [Google Scholar]
- Hao, J.; Mislevy, R.J. A Data Science Perspective on Computational Psychometrics. In Computational Psychometrics: New Methodologies for a New Generation of Digital Learning and Assessment: With Examples in R and Python; von Davier, A.A., Mislevy, R.J., Hao, J., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 133–158. [Google Scholar]
- Adtalem Global Education. OutReach IQ; Adtalem Global Education: Chicago, IL ,USA, 2022. [Google Scholar]
- MicroBatVet. Rusvmcenter4/veterinary_education_ml_tutorial: Vet Ed ML Primer V1.1. Zenodo 2023. [Google Scholar] [CrossRef]
- McKinney, W. Data structures for statistical computing in python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June 28–3 July 2010; pp. 51–56. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Cutler, A.; Cutler, D.R.; Stevens, J.R. Random Forests. In Ensemble Machine Learning: Methods and Applications; Zhang, C., Ma, Y., Eds.; Springer: Boston, MA, USA, 2012; pp. 157–175. [Google Scholar]
- Horning, N. Random Forests: An algorithm for image classification and generation of continuous fields data sets. In Proceedings of the International Conference on Geoinformatics for Spatial Infrastructure Development in Earth and Allied Sciences, Osaka, Japan, 9–11 December 2010; pp. 1–6. [Google Scholar]
- Sullivan, L.M.; Velez, A.A.; Longe, N.; Larese, A.M.; Galea, S. Removing the Graduate Record Examination as an Admissions Requirement Does Not Impact Student Success. Public Health Rev. 2022, 43, 1605023. [Google Scholar] [CrossRef] [PubMed]
- Langin, K. A Wave of Graduate Programs Drops the GRE Application Requirement. Available online: https://www.science.org/content/article/wave-graduate-programs-drop-gre-application-requirement (accessed on 28 May 2023).
- Peng, J.; Harwell, M.; Liou, S.M.; Ehman, L.H. Advances in missing data methods and implications for educational research. Real Data Anal. 2006, 3178, 102. [Google Scholar]
- Pigott, T.D. A Review of Methods for Missing Data. Educ. Res. Eval. 2001, 7, 353–383. [Google Scholar] [CrossRef]
- Khalid, S.; Khalil, T.; Nasreen, S. A survey of feature selection and feature extraction techniques in machine learning. In Proceedings of the 2014 Science and Information Conference, London, UK, 27–29 August 2014; pp. 372–378. [Google Scholar]
- Chizi, B.; Maimon, O. Dimension Reduction and Feature Selection. In Data Mining and Knowledge Discovery Handbook; Maimon, O., Rokach, L., Eds.; Springer: Boston, MA, USA, 2010; pp. 83–100. [Google Scholar]
- Brownlee, J. Data Preparation for Machine Learning: Data Cleaning, Feature Selection, and Data Transforms in Python; Machine Learning Mastery: Vermont, Australia, 2020. [Google Scholar]
- Jabbar, H.; Khan, R.Z. Methods to avoid over-fitting and under-fitting in supervised machine learning (comparative study). Comput. Sci. Commun. Instrum. Devices 2015, 70, 163–172. [Google Scholar]
- Gu, J.; Oelke, D. Understanding bias in machine learning. arXiv 2019, arXiv:1909.01866. [Google Scholar]
- Ashfaq, U.; Poolan Marikannan, B.; Raheem, M. Managing Student Performance: A Predictive Analytics using Imbalanced Data. Int. J. Recent Technol. Eng. 2020, 8, 2277–2283. [Google Scholar] [CrossRef]
- Flores, V.; Heras, S.; Julian, V. Comparison of Predictive Models with Balanced Classes Using the SMOTE Method for the Forecast of Student Dropout in Higher Education. Electronics 2022, 11, 457. [Google Scholar] [CrossRef]
- Revathy, M.; Kamalakkannan, S.; Kavitha, P. Machine Learning based Prediction of Dropout Students from the Education University using SMOTE. In Proceedings of the 2022 4th International Conference on Smart Systems and Inventive Technology (ICSSIT), Tirunelveli, India, 20–22 January 2022; pp. 1750–1758. [Google Scholar]
- Berrar, D. Cross-Validation. In Encyclopedia of Bioinformatics and Computational Biology, Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C., Eds.; Academic Press: Oxford, 2019; pp. 542–545. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Ling, C.X. Using AUC and accuracy in evaluating learning algorithms. IEEE Trans. Knowl. Data Eng. 2005, 17, 299–310. [Google Scholar] [CrossRef]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Belete, D.M.; Huchaiah, M.D. Grid search in hyperparameter optimization of machine learning models for prediction of HIV/AIDS test results. Int. J. Comput. Appl. 2022, 44, 875–886. [Google Scholar] [CrossRef]
- Feature Importance Evaluation. Available online: https://scikit-learn.org/stable/modules/ensemble.html (accessed on 1 August 2022).
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Aljuaid, T.; Sasi, S. Proper imputation techniques for missing values in data sets. In Proceedings of the 2016 International Conference on Data Science and Engineering (ICDSE), Cochin, India, 23–25 August 2016; pp. 1–5. [Google Scholar]
- Newgard, C.D.; Lewis, R.J. Missing Data: How to Best Account for What Is Not Known. JAMA 2015, 314, 940–941. [Google Scholar] [CrossRef]
- Sawilowsky, S.S. Real Data Analysis; Information Age Pub.: Charlotte, NC, USA, 2007. [Google Scholar]
- Baudeu, R.; Wright, M.N.; Loecher, M. Are SHAP Values Biased Towards High-Entropy Features? Springer Nature: Cham, Switzerland, 2023; pp. 418–433. [Google Scholar]
- Seger, C. An Investigation of Categorical Variable Encoding Techniques in Machine Learning: Binary Versus One-Hot and Feature Hashing. Master’s Thesis, KTH Royal Institute of Technology School of Electrical Engineering and Computer Science, Stockholm, Sweden, 25 September 2018. [Google Scholar]
- Cerda, P.; Varoquaux, G.; Kégl, B. Similarity encoding for learning with dirty categorical variables. Mach. Learn. 2018, 107, 1477–1494. [Google Scholar] [CrossRef]
- Huang, J.; Galal, G.; Etemadi, M.; Vaidyanathan, M. Evaluation and Mitigation of Racial Bias in Clinical Machine Learning Models: Scoping Review. JMIR Med. Inf. 2022, 10, e36388. [Google Scholar] [CrossRef]
- Afrose, S.; Song, W.; Nemeroff, C.B.; Lu, C.; Yao, D. Subpopulation-specific machine learning prognosis for underrepresented patients with double prioritized bias correction. Commun. Med. 2022, 2, 111. [Google Scholar] [CrossRef]
- American Assocation of Veterinary Medical Colleges. Annual Data Report 2022–2023; American Assocation of Veterinary Medical Colleges: Washington, DC, USA, 2023; pp. 1–67. [Google Scholar]
- Boyajian, M.Y. Student Intervention System Using Machine Learning. Ph.D. Thesis, American University of Beirut, Beirut, Lebanon, 2019. [Google Scholar]
- Yakin, M.; Linden, K. Adaptive e-learning platforms can improve student performance and engagement in dental education. J. Dent. Educ. 2021, 85, 1309–1315. [Google Scholar] [CrossRef]
- Kuzminsky, J.; Phillips, H.; Sharif, H.; Moran, C.; Gleason, H.E.; Topulos, S.P.; Pitt, K.; McNeil, L.K.; McCoy, A.M.; Kesavadas, T. Reliability in performance assessment creates a potential application of artificial intelligence in veterinary education: Evaluation of suturing skills at a single institution. Am. J. Vet. Res. 2023, 84, 1–11. [Google Scholar] [CrossRef] [PubMed]


| Variable Name | Range of Values | Type of Data |
|---|---|---|
| Full Name | 400 randomly generated female and male names | Categorical |
| Gender | Male or Female | Categorical |
| Race/Ethnicity | Asian, Black, Latinx, Not Provided, White | Categorical |
| Age | 20–40 years | Numeric |
| Pre-Vet School GPA | 3.00–4.00 | Numeric |
| GRE | 260–330 | Numeric |
| Fail | 0–1 | Numeric |
| Actual Negative Class: 0, Student Who Did Not Fail | Actual Positive Class: 1, Student Who Did Fail | |
|---|---|---|
| Predicted negative Class: 0, student who did not fail | True negative (TN) | False negative (FN) |
| Predicted positive Class: 1, student who did fail | False positive (FP) | True positive (TP) |
| Performance Metric | Random Forest Base Model | Radom Forest Tuned Model |
|---|---|---|
| Accuracy | 87.07% | 86.61% |
| Recall/Sensitivity/TPR | 89.61% | 89.77% |
| Specificity/TNR | 87.15% | 88.11% |
| Precision | 86.46% | 86.21% |
| F1-Score | 86.24% | 88.40% |
| ROC curve AUC | 87.15% | 88.11% |
| All GRE Records | Feature Importance Score | Missing Low GRE Values Removed | Feature Importance Score | Missing Low GRE Values Replaced with Mean | Feature Importance Score | Random Missing GRE Values Removed | Feature Importance Score | Random Missing GRE Values Replaced with Mean | Feature Importance Score |
|---|---|---|---|---|---|---|---|---|---|
| GRE | 0.241850 | GRE | 0.370290 | GRE | 0.357720 | GRE | 0.291419 | preGPA | 0.218575 |
| Age | 0.150457 | Race_Not Provided | 0.131981 | preGPA | 0.152793 | preGPA | 0.199777 | GRE | 0.181400 |
| Race_Not Provided | 0.146913 | preGPA | 0.106498 | Age | 0.146683 | Age | 0.163021 | Age | 0.180350 |
| preGPA | 0.130455 | Age | 0.100696 | Race_Not Provided | 0.096973 | Race_Not Provided | 0.071514 | Race_Not Provided | 0.117902 |
| Race_White | 0.078924 | Race_White | 0.093832 | Race_Latinx | 0.062529 | Race_Asian | 0.062693 | Race_White | 0.073569 |
| Race_Latinx | 0.067359 | Gender_Male | 0.082514 | Race_White | 0.057276 | Race_White | 0.059140 | Race_Black | 0.071190 |
| Gender_Male | 0.063287 | Race_Latinx | 0.047706 | Race_Asian | 0.044694 | Race_Black | 0.051953 | Race_Asian | 0.062513 |
| Race_Black | 0.063043 | Race_Black | 0.041844 | Gender_Male | 0.042020 | Gender_Male | 0.050608 | Race_Latinx | 0.060454 |
| Race_Asian | 0.057713 | Race_Asian | 0.024639 | Race_Black | 0.039311 | Race_Latinx | 0.049876 | Gender_male | 0.034048 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hooper, S.E.; Hecker, K.G.; Artemiou, E. Using Machine Learning in Veterinary Medical Education: An Introduction for Veterinary Medicine Educators. Vet. Sci. 2023, 10, 537. https://doi.org/10.3390/vetsci10090537
Hooper SE, Hecker KG, Artemiou E. Using Machine Learning in Veterinary Medical Education: An Introduction for Veterinary Medicine Educators. Veterinary Sciences. 2023; 10(9):537. https://doi.org/10.3390/vetsci10090537
Chicago/Turabian StyleHooper, Sarah E., Kent G. Hecker, and Elpida Artemiou. 2023. "Using Machine Learning in Veterinary Medical Education: An Introduction for Veterinary Medicine Educators" Veterinary Sciences 10, no. 9: 537. https://doi.org/10.3390/vetsci10090537
APA StyleHooper, S. E., Hecker, K. G., & Artemiou, E. (2023). Using Machine Learning in Veterinary Medical Education: An Introduction for Veterinary Medicine Educators. Veterinary Sciences, 10(9), 537. https://doi.org/10.3390/vetsci10090537






