Isolation, Identification, and Whole Genome Analysis of Chicken Infectious Anemia Virus in an Outbreak of Disease in Adult Layer Hens
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Reagents and Materials
2.3. Primer Design and Synthesis
2.4. Sample Handling
2.4.1. PCR Detection
2.4.2. Virus Isolation and Identification
2.4.3. MDCC-MSB1 Cells Inoculation of CIAV Isolate
2.5. Whole Genome Amplification of CIAV Isolates
2.6. Experimental Infection of 1-Day-Old SPF Chicks with CIAV
2.7. Genome-Wide Bioinformatics Analysis
Genome-Wide Genetic Evolutionary Analysis
3. Results
3.1. Pathological Changes
3.2. PCR Test Results of the Samples
3.3. Virus Isolation Results
3.4. MDCC-MSB1 Culture Results
3.5. Whole Genome Amplification Results
3.6. CIAV GX21121 Pathogenicity to One-Day-Old SPF Chicks
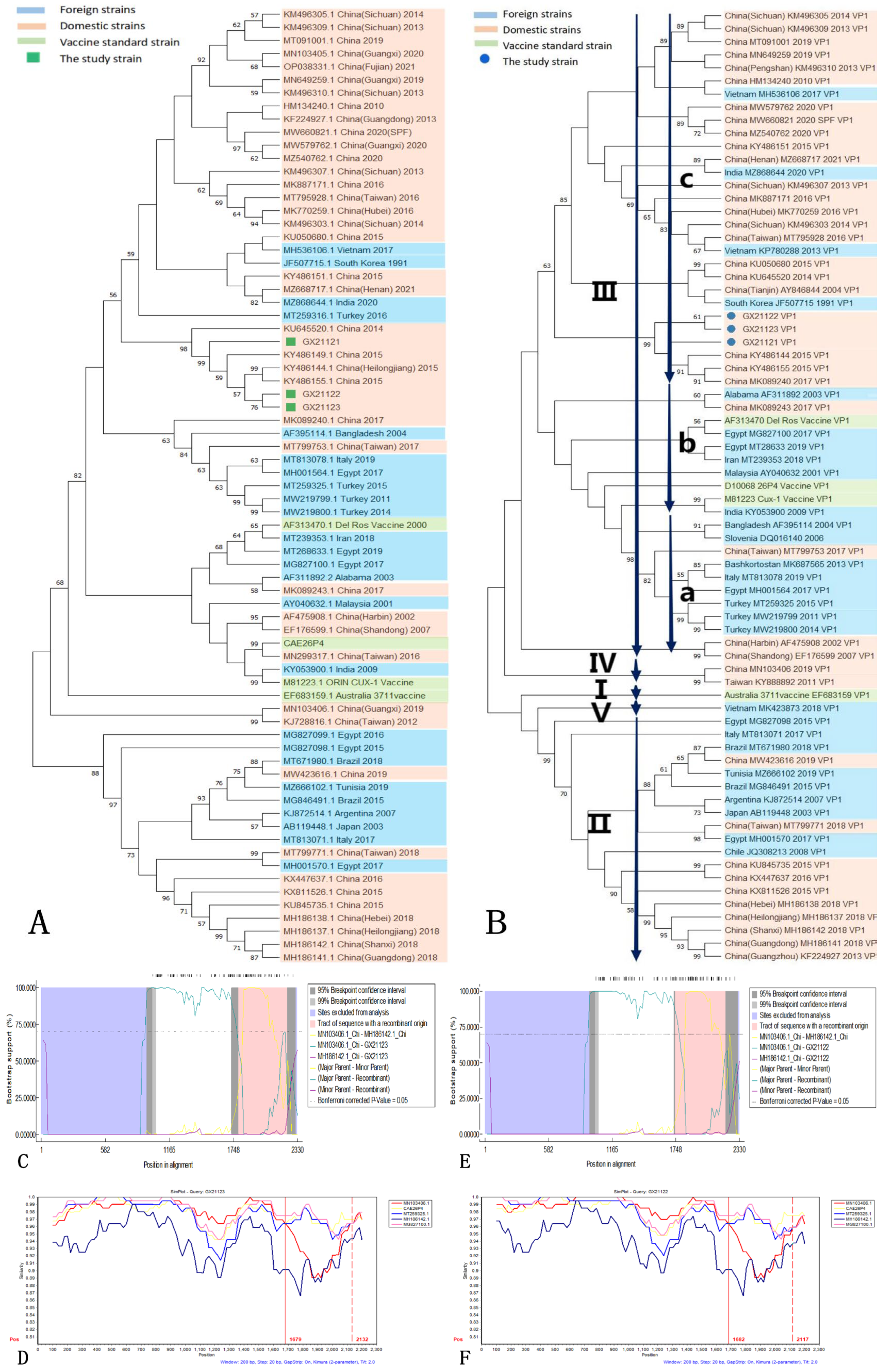
3.7. Genome-Wide Bioinformatics Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| No. | Accession No. | Country | Isolate | Year | Nucleotides (bp) |
| 1 | ON596886 | China | GX2112 | 2021 | 2298 |
| 2 | MH186142 | China | CIAV-Shanxi7 | 2018 | 2291 |
| 3 | HM134240 | China | \ | 2010 | 2298 |
| 4 | KU050680 | China | GD-101 | 2015 | 2298 |
| 5 | KU645520 | China | HN1405 | 2014 | 2298 |
| 6 | KU845735 | China | CIAV-F10 | 2015 | 2298 |
| 7 | KX447637 | China | LY-2 | 2016 | 2298 |
| 8 | KX811526 | China | SD15 | 2015 | 2298 |
| 9 | KY486144 | China | HLJ15170 | 2015 | 2298 |
| 10 | KY486151 | China | NX15140 | 2015 | 2298 |
| 11 | KY486155 | China | LN15170 | 2015 | 2298 |
| 12 | MK089240 | China | 17CC0509 | 2017 | 2298 |
| 13 | MK089243 | China | 17SY0902 | 2017 | 2298 |
| 14 | MK887171 | China | N1 | 2016 | 2298 |
| 15 | MN103406 | China | GX1904B | 2019 | 2298 |
| 16 | MN649259 | China | GX1907B | 2019 | 2298 |
| 17 | MT091001 | China | TBC19 | 2019 | 2298 |
| 18 | MW423616 | China | TZC1910 | 2019 | 2298 |
| 19 | MW579762 | China | GX2020-D6 | 2020 | 2298 |
| 20 | MW660821 | China | SDSPF2020 | 2020 | 2298 |
| 21 | MZ540762 | China | YN04 | 2020 | 2298 |
| 22 | MH186141 | China | CIAV-Guangdong11 | 2018 | 2291 |
| 23 | KF224927 | China | GD-C-12 | 2013 | 2298 |
| 24 | AF475908 | China | \ | 2002 | 2298 |
| 25 | MH186138 | China | CIAV-Hebei12 | 2018 | 2293 |
| 26 | MH186137 | China | CIAV-Heilongjiang16 | 2018 | 2295 |
| 27 | MZ668717 | China | HN2021-1415 | 2021 | 2298 |
| 28 | MK770259 | China | XH16 | 2016 | 2298 |
| 29 | KM496310 | China | SC-PS | 2013 | 2298 |
| 30 | EF176599 | China | C14 | 2007 | 2298 |
| 31 | KM496303 | China | SC-HY | 2014 | 2298 |
| 32 | KM496305 | China | SC-MS | 2014 | 2298 |
| 33 | KM496307 | China | SC-MZ42A | 2013 | 2298 |
| 34 | KM496309 | China | SC-NC2 | 2013 | 2298 |
| 35 | KJ728816 | China | 4 | 2012 | 2298 |
| 36 | MT795928 | China | 1512TW | 2016 | 2298 |
| 37 | MT799753 | China | 1635TW | 2017 | 2298 |
| 38 | MT799771 | China | 1874TW | 2018 | 2298 |
| 39 | AY846844 | China | TJBD40 | 2004 | 2298 |
| 40 | AF311892 | Alabama | 98D02152 | 2003 | 2298 |
| 41 | KJ872514 | Argentina | CAV-18 | 2007 | 2298 |
| 42 | AF395114 | Bangladesh | BD-3 | 2004 | 2298 |
| 44 | MG846491 | Brazil | RS/BR/15/1R | 2015 | 2298 |
| 45 | MT671980 | Brazil | CAV | 2018 | 2298 |
| 46 | MG827098 | Egypt | CAV-CA1-2015 | 2015 | 2298 |
| 47 | MG827099 | Egypt | CAV-GZ2-2016 | 2016 | 2298 |
| 48 | MG827100 | Egypt | CAV-SK4-2017 | 2017 | 2298 |
| 49 | MH001570 | Egypt | CAV-EG-28 | 2017 | 2298 |
| 50 | MT268633 | Egypt | EG-ElBeheira-2019 | 2019 | 2298 |
| 51 | KY053900 | India | CAV/NAM/TANUVAS/09 | 2009 | 2263 |
| 52 | MZ868644 | India | NIAB-DPR | 2020 | 2298 |
| 53 | MT239353 | Iran | UT-Zahraee | 2018 | 2298 |
| 54 | MT813071 | Italy | CIAV/IT/CK/855/17 | 2017 | 2189 |
| 55 | MT813078 | Italy | CIAV/IT/CK/1188/19 | 2019 | 2186 |
| 56 | AY040632 | Malaysia | 3-1P60 | 2001 | 2298 |
| 57 | MZ666102 | Tunisia | CIAV_TN_7-15 | 2019 | 2181 |
| 58 | MT259325 | Turkey | GDA6K | 2015 | 2296 |
| 59 | MW219799 | Turkey | CIAV/2011/Akcdg/TUR | 2011 | 2298 |
| 60 | MW219800 | Turkey | CIAV/2014/Elzg-66/TUR | 2014 | 2298 |
| 61 | MH536106 | Vietnam | G17.3.1 | 2017 | 2298 |
| 62 | D10068 | \ | 26P4 (vaccine) | 2007 | 2298 |
| 63 | AF313470 | USA | Del Ros (vaccine) | 2000 | 2294 |
| 64 | M81223 | \ | CUX-1 (vaccine) | 1993 | 2298 |
| 65 | EF683159 | Australia | 3711 (vaccine) | 2007 | 2279 |
| 66 | MH001564 | Egypt | CAV-EG-26 | 2017 | 2298 |
| 67 | JF507715 | South Korea | CIAV89-69 | 1991 | 2298 |
| 68 | AB119448 | Japan | G6 | 2009 | 2298 |
| 69 | DQ016140 | Slovenia | 69 | 2006 | 1350 |
| 70 | JQ308213 | Chile | CL37 | 2008 | 1831 |
| 71 | KY888892 | China | 1102PT01 | 2011 | 1823 |
| 72 | MK423873 | Vietnam | Vietnam/PT1/17 | 2019 | 1823 |
| 73 | KP780288 | Vietnam | CAV/Vietnam/HN1/13 | 2018 | 1823 |
| 74 | DQ991394 | USA | 01-4201 | 2007 | 2298 |
| 75 | KY888907 | China | 1310CH05 | 2013 | 1823 |
References
- Shao, H.; Li, J.; Yuan, H.; Ji, L.; Zhang, J.; Jin, W.; Qian, K.; Ye, J.; Qin, A. Isolation and Molecular Characteristics of a CIAV Isolate From Pigeons, China. Front. Vet. Sci. 2021, 8, 669154. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Chen, L.; Wang, Q.; Zhou, M.; Zhao, H.; Chi, Z.; Wang, Y.; Chang, S.; Zhao, P. Genomic Characterization of CIAV Detected in Contaminated Attenuated NDV Vaccine: Epidemiological Evidence of Source and Vertical Transmission From SPF Chicken Embryos in China. Front. Vet. Sci. 2022, 9, 930887. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Breitbart, M.; Harrach, B.; Segalés, J.; Delwart, E.; Biagini, P.; Varsani, A. Revisiting the Taxonomy of the Family Circoviridae: Establishment of the Genus Cyclovirus and Removal of the Genus Gyrovirus. Arch. Virol. 2017, 162, 1447–1463. [Google Scholar] [CrossRef]
- Miller, M.M.; Ealey, K.A.; Oswald, W.B.; Schat, K.A. Detection of Chicken Anemia Virus DNA in Embryonal Tissues and Eggshell Membranes. Avian Dis. 2003, 47, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Noteborn, M.H.; de Boer, G.F.; van Roozelaar, D.J.; Karreman, C.; Kranenburg, O.; Vos, J.G.; Jeurissen, S.H.; Hoeben, R.C.; Zantema, A.; Koch, G. Characterization of Cloned Chicken Anemia Virus DNA That Contains All Elements for the Infectious Replication Cycle. J. Virol. 1991, 65, 3131–3139. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, N.; Taniguchi, T.; Yoshida, I. Isolation and Some Characteristics of an Agent Inducing Anemia in Chicks. Avian Dis. 1979, 23, 366. [Google Scholar] [CrossRef]
- Chu, D.K.W.; Poon, L.L.M.; Chiu, S.S.S.; Chan, K.H.; Ng, E.M.; Bauer, I.; Cheung, T.K.; Ng, I.H.Y.; Guan, Y.; Wang, D.; et al. Characterization of a Novel Gyrovirus in Human Stool and Chicken Meat. J. Clin. Virol. 2012, 55, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Fatoba, A.J.; Adeleke, M.A. Chicken Anemia Virus: A Deadly Pathogen of Poultry. Acta Virol. 2019, 63, 19–25. [Google Scholar] [CrossRef]
- Fang, L.; Li, Y.; Wang, Y.; Fu, J.; Cui, S.; Li, X.; Chang, S.; Zhao, P. Genetic Analysis of Two Chicken Infectious Anemia Virus Variants-Related Gyrovirus in Stray Mice and Dogs: The First Report in China, 2015. BioMed Res. Int. 2017, 2017, 6707868. [Google Scholar] [CrossRef]
- Kim, H.-R.; Kwon, Y.-K.; Bae, Y.-C.; Oem, J.-K.; Lee, O.-S. Molecular Characterization of Chicken Infectious Anemia Viruses Detected from Breeder and Broiler Chickens in South Korea. Poult. Sci. 2010, 89, 2426–2431. [Google Scholar] [CrossRef]
- Miller, M.M.; Schat, K.A. Chicken Infectious Anemia Virus: An Example of the Ultimate Host–Parasite Relationship. Avian Dis. 2004, 48, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Todd, D. Avian Circovirus Diseases: Lessons for the Study of PMWS. Vet. Microbiol. 2004, 98, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, Y.; Cui, S.; Fu, J.; Wang, Y.; Cui, Z.; Fang, L.; Chang, S.; Zhao, P. Molecular Characterization of Chicken Infectious Anemia Virus from Contaminated Live-Virus Vaccines. Poult. Sci. 2017, 96, 1045–1051. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Wu, B.; Sun, B.; Chen, F.; Ji, J.; Ma, J.; Xie, Q. Phylogenetic and Molecular Characterization of Chicken Anemia Virus in Southern China from 2011 to 2012. Sci. Rep. 2013, 3, 3519. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, N. Propagation and Infectivity Titration of the Gifu-1 Strain of Chicken Anemia Agent in a Cell Line (MDCC-MSB1) Derived from Marek’s Disease Lymphoma. Natl. Inst. Anim. Health Q. 1983, 23, 13–20. [Google Scholar]
- Akiyama, Y.; Kato, S. Two Cell Lines from Lymphomas of Marek’s Disease. Biken J. 1974, 17, 105–116. [Google Scholar]
- Renshaw, R.W.; Soiné, C.; Weinkle, T.; O’Connell, P.H.; Ohashi, K.; Watson, S.; Lucio, B.; Harrington, S.; Schat, K.A. A Hypervariable Region in VP1 of Chicken Infectious Anemia Virus Mediates Rate of Spread and Cell Tropism in Tissue Culture. J. Virol. 1996, 70, 8872–8878. [Google Scholar] [CrossRef]
- Lacorte, C.; Lohuis, H.; Goldbach, R.; Prins, M. Assessing the Expression of Chicken Anemia Virus Proteins in Plants. Virus Res. 2007, 129, 80–86. [Google Scholar] [CrossRef]
- Koch, G.; van Roozelaar, D.J.; Verschueren, C.A.; van der Eb, A.J.; Noteborn, M.H. Immunogenic and Protective Properties of Chicken Anaemia Virus Proteins Expressed by Baculovirus. Vaccine 1995, 13, 763–770. [Google Scholar] [CrossRef]
- Douglas, A.J.; Phenix, K.; Mawhinney, K.A.; Todd, D.; Mackie, D.P.; Curran, W.L. Identification of a 24 KDa Protein Expressed by Chicken Anaemia Virus. J. Gen. Virol. 1995, 76 Pt 7, 1557–1562. [Google Scholar] [CrossRef]
- Trinh, D.Q.; Ogawa, H.; Bui, V.N.; Baatartsogt, T.; Kizito, M.K.; Yamaguchi, S.; Imai, K. Characterization of MAbs to Chicken Anemia Virus and Epitope Mapping on Its Viral Protein, VP1. J. Gen. Virol. 2015, 96, 1086–1097. [Google Scholar] [CrossRef] [PubMed]
- Noteborn, M.H.; Kranenburg, O.; Zantema, A.; Koch, G.; de Boer, G.F.; van der Eb, A.J. Transcription of the Chicken Anemia Virus (CAV) Genome and Synthesis of Its 52-KDa Protein. Gene 1992, 118, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Peters, M.A.; Jackson, D.C.; Crabb, B.S.; Browning, G.F. Chicken Anemia Virus VP2 Is a Novel Dual Specificity Protein Phosphatase. J. Biol. Chem. 2002, 277, 39566–39573. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Zhang, Y.; Chen, L.; Fang, L.; Chang, S.; Wang, Y.; Zhao, P. Construction of Chicken Infectious Anemia Virus Infectious Clone and Study on Its Pathogenicity. Front. Microbiol. 2022, 13, 1016784. [Google Scholar] [CrossRef]
- Simultaneous Expression of Recombinant Baculovirus-Encoded Chicken Anaemia Virus (CAV) Proteins VP1 and VP2 Is Required for Formation of the CAV-Specific Neutralizing Epitope—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/9880024/ (accessed on 18 February 2023).
- Chowdhury, S.M.Z.H.; Omar, A.R.; Aini, I.; Hair-Bejo, M.; Jamaluddin, A.A.; Md-Zain, B.M.; Kono, Y. Pathogenicity, Sequence and Phylogenetic Analysis of Malaysian Chicken Anaemia Virus Obtained after Low and High Passages in MSB-1 Cells. Arch. Virol. 2003, 148, 2437–2448. [Google Scholar] [CrossRef] [PubMed]
- Hailemariam, Z.; Omar, A.R.; Hair-Bejo, M.; Giap, T.C. Detection and Characterization of Chicken Anemia Virus from Commercial Broiler Breeder Chickens. Virol. J. 2008, 5, 128. [Google Scholar] [CrossRef] [PubMed]
- Farkas, T.; Tanaka, A.; Kai, K.; Kanoe, M. Cloning and Sequencing of the Genome of Chicken Anaemia Virus (CAV) TK-5803 Strain and Comparison with Other CAV Strains. J. Vet. Med. Sci. 1996, 58, 681–684. [Google Scholar] [CrossRef]
- Ducatez, M.F.; Owoade, A.A.; Abiola, J.O.; Muller, C.P. Molecular Epidemiology of Chicken Anemia Virus in Nigeria. Arch. Virol. 2006, 151, 97–111. [Google Scholar] [CrossRef]
- Ou, S.-C.; Lin, H.-L.; Liu, P.-C.; Huang, H.-J.; Lee, M.-S.; Lien, Y.-Y.; Tsai, Y.-L. Epidemiology and Molecular Characterization of Chicken Anaemia Virus from Commercial and Native Chickens in Taiwan. Transbound. Emerg. Dis. 2018, 65, 1493–1501. [Google Scholar] [CrossRef]
- Van Dong, H.; Tran, G.T.H.; Van Nguyen, G.; Dao, T.D.; Bui, V.N.; Huynh, L.T.M.; Takeda, Y.; Ogawa, H.; Imai, K. Chicken Anemia Virus in Northern Vietnam: Molecular Characterization Reveals Multiple Genotypes and Evidence of Recombination. Virus Genes 2019, 55, 643–653. [Google Scholar] [CrossRef]
- Tekelemariam, T.H.; Walkden-Brown, S.; Atire, F.A.; Tefera, D.A.; Alemayehu, D.H.; Gerber, P.F. Detection of Chicken Respiratory Pathogens in Live Markets of Addis Ababa, Ethiopia, and Epidemiological Implications. Vet. Sci. 2022, 9, 503. [Google Scholar] [CrossRef] [PubMed]
- Fadly, A.; Garcia, M.C. Detection of Reticuloendotheliosis Virus in Live Virus Vaccines of Poultry. Dev. Biol. 2006, 126, 301–305. [Google Scholar]
- Gopal, S.; Manoharan, P.; Kathaperumal, K.; Chidambaram, B.; Divya, K.C. Differential Detection of Avian Oncogenic Viruses in Poultry Layer Farms and Turkeys by Use of Multiplex PCR. J. Clin. Microbiol. 2012, 50, 2668–2673. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.; Wang, Z.; Xu, Z.; Pang, Y.; Leng, M.; Tang, S.; Zhang, X.; Qin, J.; Chen, F.; Lin, W. Pathogenicity and Molecular Characterization of Infectious Bursal Disease Virus in China. Poult. Sci. 2021, 101, 101502. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-Length Human Immunodeficiency Virus Type 1 Genomes from Subtype C-Infected Seroconverters in India, with Evidence of Intersubtype Recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef]
- Li, Y.; Fang, L.; Cui, S.; Fu, J.; Li, X.; Zhang, H.; Cui, Z.; Chang, S.; Shi, W.; Zhao, P. Genomic Characterization of Recent Chicken Anemia Virus Isolates in China. Front. Microbiol. 2017, 8, 401. [Google Scholar] [CrossRef]
- Miles, A.M.; Reddy, S.M.; Morgan, R.W. Coinfection of Specific-Pathogen-Free Chickens with Marek’s Disease Virus (MDV) and Chicken Infectious Anemia Virus: Effect of MDV Pathotype. Avian Dis. 2001, 45, 9–18. [Google Scholar] [CrossRef]
- Gimeno, I.M.; Schat, K.A. Virus-Induced Immunosuppression in Chickens. Avian Dis. 2018, 62, 272–285. [Google Scholar] [CrossRef]
- Li, X.; Zhang, K.; Pei, Y.; Xue, J.; Ruan, S.; Zhang, G. Development and Application of an MRT-QPCR Assay for Detecting Coinfection of Six Vertically Transmitted or Immunosuppressive Avian Viruses. Front. Microbiol. 2020, 11, 1581. [Google Scholar] [CrossRef]
- Lucio, B.; Schat, K.A.; Shivaprasad, H.L. Identification of the Chicken Anemia Agent, Reproduction of the Disease, and Serological Survey in the United States. Avian Dis. 1990, 34, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.L.; Priyanka, E.; Kannaki, T.R.; Subbiah, M. Whole Genome Analysis and Molecular Characterization of Chicken Infectious Anemia Virus from an Outbreak in a Layer Flock Reveals Circulation of Genogroup IIIb in South India. Virus Res. 2022, 308, 198649. [Google Scholar] [CrossRef] [PubMed]

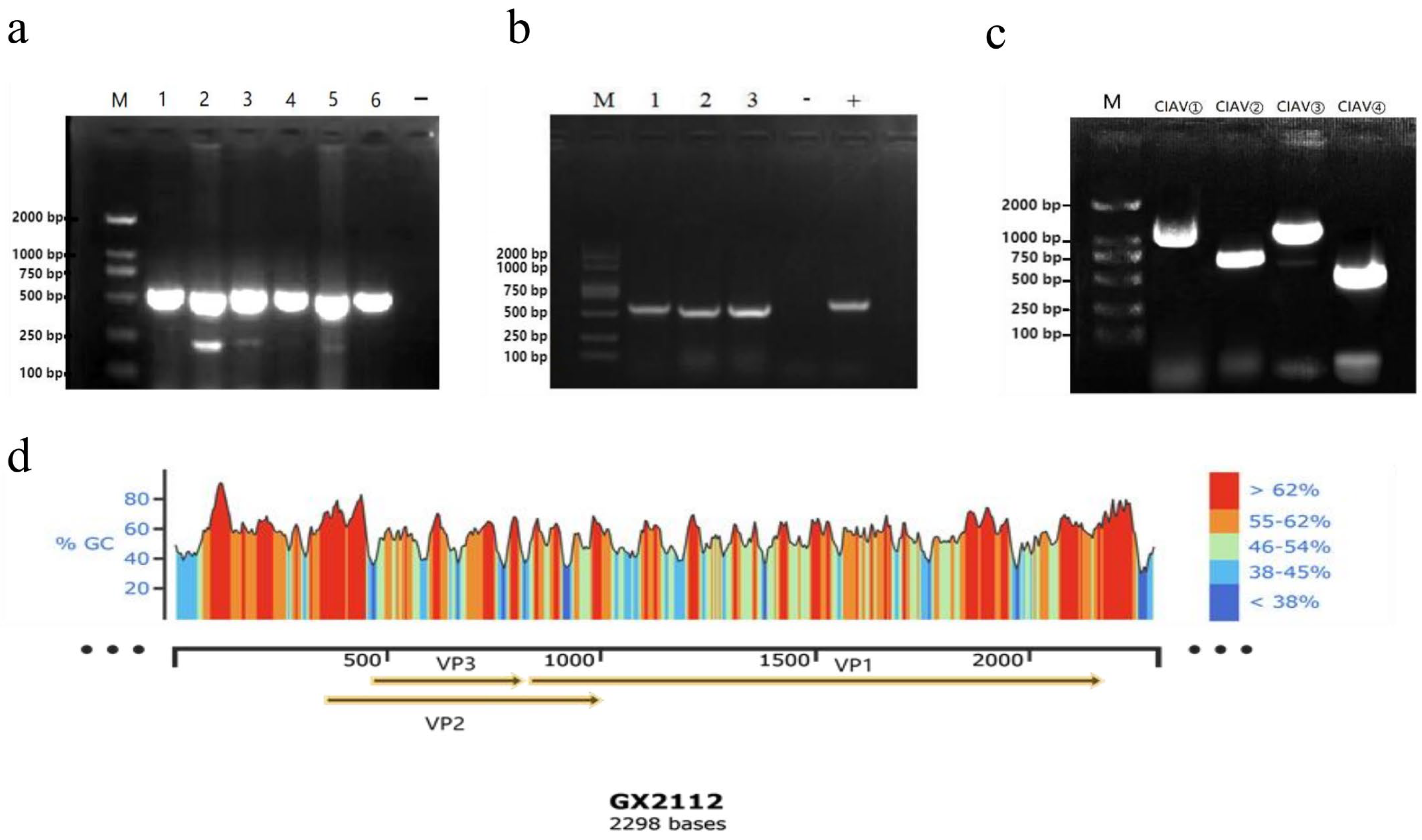

| Primers | (Primer Sequence 5′ to 3′) | Length |
|---|---|---|
| CIAV ①-F | GACCGATCAACCCAAGCCTCC | 1010 bp |
| CIAV ①-R | ATCTTCCCGGTCGCATAAGCA | |
| CIAV ②-F | CTTGCCGGTTCTTTAATCACCCT | 751 bp |
| CIAV ②-R | CTCTTACCCAGCTGCCACACC | |
| CIAV ③-F | CTACATGGCAGCACCCGCATC | 1055 bp |
| CIAV ③-R | TCCGGCACATTCTTA(G)AAACCAG | |
| CIAV ④-F | AATGAACGCTCTCCAAGAAG | 540 bp |
| CIAV ④-R | AGCGGATAGTCATAGTAGAT |
| Primers | Pre-Denatured | Denatured × 35 | Annealing × 35 | Extension × 35 | Post Elongation |
|---|---|---|---|---|---|
| CIAV ① | 95 °C, 5 min | 94 °C, 30 s | 61 °C, 30 s | 72 °C, 70 s | 72 °C, 7 min |
| CIAV ② | \ | \ | 62 °C, 30 s | 72 °C, 45 s | 72 °C, 7 min |
| CIAV ③ | \ | \ | 58.7 °C, 30 s | 72 °C, 70 s | 72 °C, 7 min |
| CIAV ④ | \ | \ | 50.5 °C, 30 s | 72 °C, 30 s | 72 °C, 8 min |
| Method | Recombination p Value |
|---|---|
| RDP | 1.403 × 10−2 |
| GENECONV | 2.624 × 10−2 |
| BootScan | 3.598 × 10−2 |
| MaxChi | 4.106 × 10−3 |
| Chimaera | 1.071 × 10−2 |
| SiScan | 2.675 × 10−4 |
| 3Seq | 6.864 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, Y.; Zhang, H.; Zhang, H. Isolation, Identification, and Whole Genome Analysis of Chicken Infectious Anemia Virus in an Outbreak of Disease in Adult Layer Hens. Vet. Sci. 2023, 10, 481. https://doi.org/10.3390/vetsci10070481
Zeng Y, Zhang H, Zhang H. Isolation, Identification, and Whole Genome Analysis of Chicken Infectious Anemia Virus in an Outbreak of Disease in Adult Layer Hens. Veterinary Sciences. 2023; 10(7):481. https://doi.org/10.3390/vetsci10070481
Chicago/Turabian StyleZeng, Yueyan, Hui Zhang, and Huanrong Zhang. 2023. "Isolation, Identification, and Whole Genome Analysis of Chicken Infectious Anemia Virus in an Outbreak of Disease in Adult Layer Hens" Veterinary Sciences 10, no. 7: 481. https://doi.org/10.3390/vetsci10070481
APA StyleZeng, Y., Zhang, H., & Zhang, H. (2023). Isolation, Identification, and Whole Genome Analysis of Chicken Infectious Anemia Virus in an Outbreak of Disease in Adult Layer Hens. Veterinary Sciences, 10(7), 481. https://doi.org/10.3390/vetsci10070481






