Genomic Characteristics of Feline Anelloviruses Isolated from Domestic Cats in Shanghai, China
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Viral Metagenomics of the Fecal Samples
2.2. Sample Pretreatment
2.3. Library Generation and Sequencing
2.4. Data Analysis
2.5. Validation of FcTTV Infection
2.6. Full-Length Sequence of the Shanghai FcTTV Isolates
2.7. Phylogenetic Analysis of the FcTTV Shanghai Isolates
3. Results
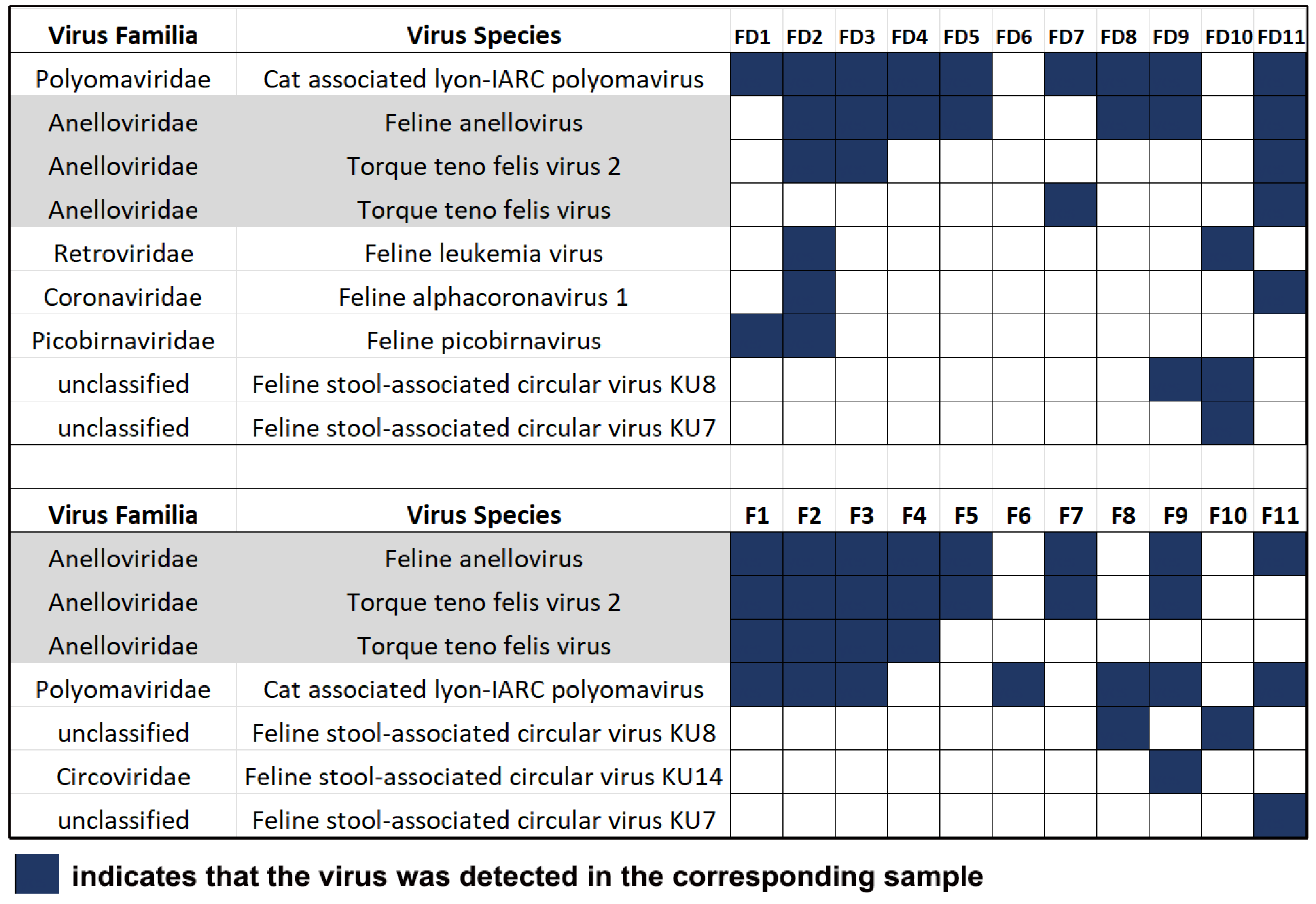
3.1. Cat-Associated Viruses Detected by Viral Metagenomics
3.2. Validation of FcTTV Infection
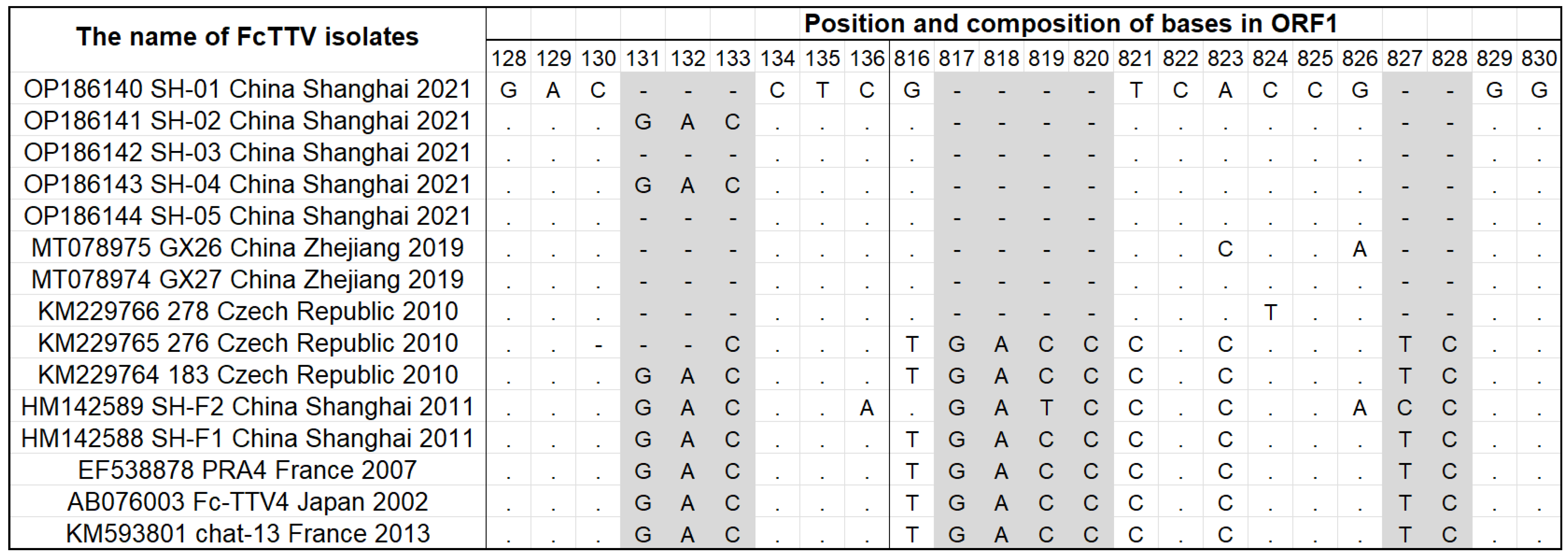
3.3. Full-Length Sequence of the Shanghai FcTTV Isolates
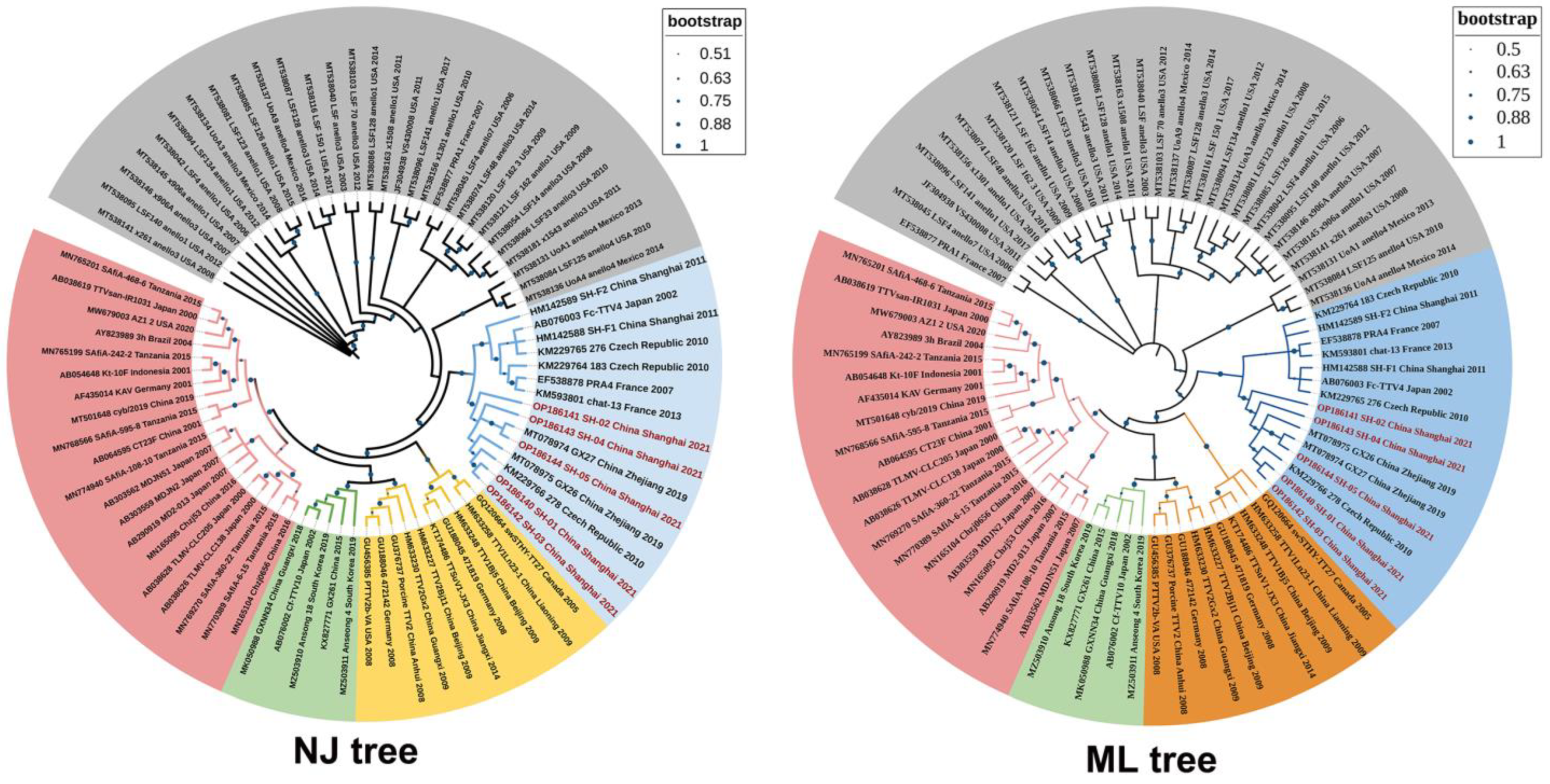
3.4. The Results of Phylogenetic Analysis
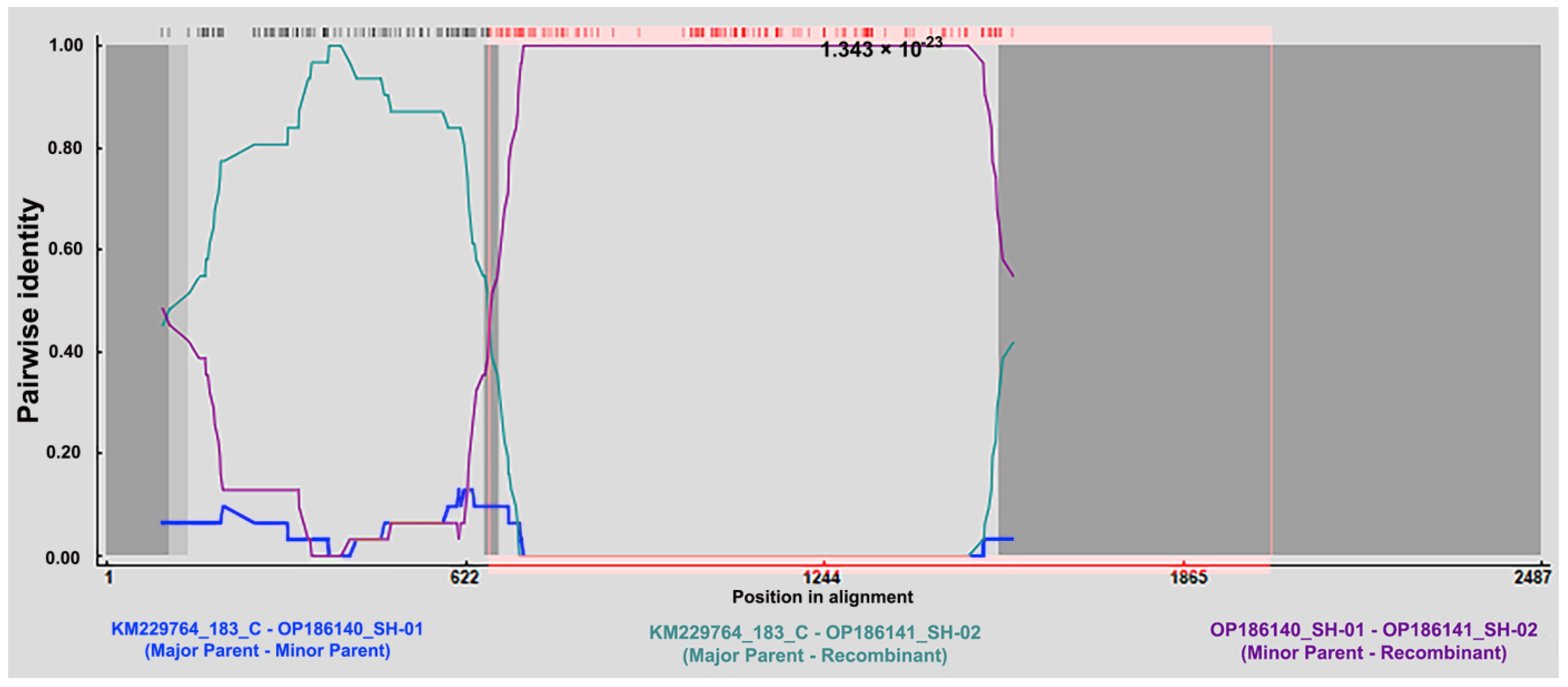
3.5. The Recombination Event Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mihalov-Kovács, E.; Fehér, E.; Martella, V.; Bányai, K.; Farkas, S.L. The fecal virome of domesticated animals. Virusdisease 2014, 25, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Miyata, H.; Tsunoda, H.; Kazi, A.; Yamada, A.; Khan, M.A.; Murakami, J.; Kamahora, T.; Shiraki, K.; Hino, S. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J. Virol. 1999, 73, 3582–3586. [Google Scholar] [CrossRef] [PubMed]
- Kaczorowska, J.; van der Hoek, L. Human anelloviruses: Diverse, omnipresent and commensal members of the virome. FEMS Microbiol. Rev. 2020, 44, 305–313. [Google Scholar] [CrossRef]
- Ning, S.-Y.; Zhou, M.-M.; Yang, J.; Zeng, J.; Wang, J.-P. Viral metagenomics reveals two novel anelloviruses in feces of experimental rats. Virol. J. 2021, 18, 252. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, T.; Okamoto, H.; Konishi, K.; Yoshizawa, H.; Miyakawa, Y.; Mayumi, M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 1997, 241, 92–97. [Google Scholar] [CrossRef]
- Fauquet, C.M.; Mayo, M.A.; Maniloff, J.; Desselberger, U.; Ball, L.A. Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses; Academic Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Okamoto, H.; Takahashi, M.; Nishizawa, T.; Tawara, A.; Fukai, K.; Muramatsu, U.; Naito, Y.; Yoshikawa, A. Genomic characterization of TT viruses (TTVs) in pigs, cats and dogs and their relatedness with species-specific TTVs in primates and tupaias. J. Gen. Virol. 2002, 83, 1291–1297. [Google Scholar] [CrossRef]
- Leary, T.P.; Erker, J.C.; Chalmers, M.L.; Desai, S.M.; Mushahwar, I.K. Improved detection systems for TT virus reveal high prevalence in humans, non-human primates and farm animals. J. Gen. Virol. 1999, 80, 2115–2120. [Google Scholar] [CrossRef]
- Okamoto, H.; Fukuda, M.; Tawara, A.; Nishizawa, T.; Itoh, Y.; Hayasaka, I.; Tsuda, F.; Tanaka, T.; Miyakawa, Y.; Mayumi, M. Species-specific TT viruses and cross-species infection in nonhuman primates. Biochem. J. Virol. 2000, 74, 1132–1139. [Google Scholar] [CrossRef]
- Verschoor, E.J.; Langenhuijzen, S.; Heeney, J.L. TT viruses (TTV) of non-human primates and their relationship to the human TTV genotypes. J. Gen. Virol. 1999, 80, 2491–2499. [Google Scholar] [CrossRef]
- Bigarré, L.; Beven, V.; De Boisséson, C.; Grasland, B.; Rose, N.; Biagini, P.; Jestin, A. Pig anelloviruses are highly prevalent in swine herds in France. J. Gen. Virol. 2005, 86, 631–635. [Google Scholar] [CrossRef]
- Nishiyama, S.; Dutia, B.M.; Stewart, J.P.; Meredith, A.L.; Shaw, D.J.; Simmonds, P.; Sharp, C.P. Identification of novel anelloviruses with broad diversity in UK rodents. J. Gen. Virol. 2014, 95, 1544. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Li, Y.; Lu, L.; Zheng, D.; Liu, B.; Yang, L.; Su, H.; Dong, J.; Sun, L.; Zhu, Y. Biodiversity of rodent anelloviruses in China. Emerg. Microbes Infect. 2018, 7, 1–3. [Google Scholar] [CrossRef]
- Cibulski, S.P.; Teixeira, T.F.; de Sales Lima, F.E.; do Santos, H.F.; Franco, A.C.; Roehe, P.M. A novel Anelloviridae species detected in Tadarida brasiliensis bats: First sequence of a chiropteran Anellovirus. Genome Announc. 2014, 2, e01028-14. [Google Scholar] [CrossRef]
- Ng, T.F.F.; Suedmeyer, W.K.; Wheeler, E.; Gulland, F.; Breitbart, M. Novel anellovirus discovered from a mortality event of captive California sea lions. J. Gen. Virol. 2009, 90, 1256–1261. [Google Scholar] [CrossRef]
- Gergely Jr, P.; Perl, A.; Poór, G. Possible pathogenic nature of the recently discovered TT virus: Does it play a role in autoimmune rheumatic diseases? Autoimmun. Rev. 2006, 6, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Maggi, F.; Andreoli, E.; Riente, L.; Meschi, S.; Rocchi, J.; Delle Sedie, A.; Vatteroni, M.; Ceccherini-Nelli, L.; Specter, S.; Bendinelli, M. Torquetenovirus in patients with arthritis. Rheumatology 2007, 46, 885–886. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Okamoto, H.; Luengrojanakul, P.; Chainuvati, T.; Tsuda, F.; Tanaka, T.; Miyakawa, Y.; Mayumi, M. Infection with an unenveloped DNA virus (TTV) associated with posttransfusion non-A to G hepatitis in hepatitis patients and healthy blood donors in Thailand. J. Med. Virol. 1998, 56, 234–238. [Google Scholar] [CrossRef]
- Naoumov, N.V.; Petrova, E.P.; Thomas, M.G.; Williams, R. Presence of a newly described human DNA virus (TTV) in patients with liver disease. Lancet 1998, 352, 195–197. [Google Scholar] [CrossRef]
- Mancuso, R.; Saresella, M.; Hernis, A.; Agostini, S.; Piancone, F.; Caputo, D.; Maggi, F.; Clerici, M. Torque teno virus (TTV) in multiple sclerosis patients with different patterns of disease. J. Med. Virol. 2013, 85, 2176–2183. [Google Scholar] [CrossRef]
- Komijani, M.; Bouzari, M.; Etemadifar, M.; Zarkesh-Esfahani, H.; Shaykh-Baygloo, N.; Ghazimorad, A.; Mostajeran, M.; Nasr-Azadani, A.; Maghzi, A.-H. Torque teno mini virus infection and multiple sclerosis. Int. J. Neurosci. 2011, 121, 437–441. [Google Scholar] [CrossRef]
- Bernardin, F.; Operskalski, E.; Busch, M.; Delwart, E. Transfusion transmission of highly prevalent commensal human viruses. Transfusion 2010, 50, 2474–2483. [Google Scholar] [CrossRef] [PubMed]
- Kraberger, S.; Serieys, L.E.; Richet, C.; Fountain-Jones, N.M.; Baele, G.; Bishop, J.M.; Nehring, M.; Ivan, J.S.; Newkirk, E.S.; Squires, J.R. Complex evolutionary history of felid anelloviruses. Virology 2021, 562, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Jarošová, V.; Hrazdilová, K.; Filipejová, Z.; Schánilec, P.; Celer, V. Whole genome sequencing and phylogenetic analysis of feline anelloviruses. Infect. Genet. Evol. 2015, 32, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; evolution. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef]
- Zhu, C.; Shan, T.; Cui, L.; Luo, X.; Liu, Z.; Tang, S.; Liu, Z.; Yuan, C.; Lan, D.; Zhao, W. Molecular detection and sequence analysis of feline Torque teno virus (TTV) in China. Virus Res. 2011, 156, 13–16. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Okamoto, H. TT Viruses in Animals. TT Viruses: The Still Elusive Human Pathogens; Springer: Berlin/Heidelberg, Germany, 2009; pp. 35–52. [Google Scholar]
- Biagini, P.; Uch, R.; Belhouchet, M.; Attoui, H.; Cantaloube, J.-F.; Brisbarre, N.; de Micco, P. Circular genomes related to anelloviruses identified in human and animal samples by using a combined rolling-circle amplification/sequence-independent single primer amplification approach. J. Gen. Virol. 2007, 88, 2696–2701. [Google Scholar] [CrossRef]
- Arze, C.A.; Springer, S.; Dudas, G.; Patel, S.; Bhattacharyya, A.; Swaminathan, H.; Brugnara, C.; Delagrave, S.; Ong, T.; Kahvejian, A.; et al. Global genome analysis reveals a vast and dynamic anellovirus landscape within the human virome. Cell Host Microbe 2021, 29, 1305–1315.e1306. [Google Scholar] [CrossRef]
- Fahsbender, E.; Burns, J.M.; Kim, S.; Kraberger, S.; Frankfurter, G.; Eilers, A.A.; Shero, M.R.; Beltran, R.; Kirkham, A.; McCorkell, R. Diverse and highly recombinant anelloviruses associated with Weddell seals in Antarctica. Virus Evol. 2017, 3, vex017. [Google Scholar] [CrossRef]
- Cebriá-Mendoza, M.; Beamud, B.; Andreu-Moreno, I.; Arbona, C.; Larrea, L.; Díaz, W.; Sanjuán, R.; Cuevas, J.M. Human Anelloviruses: Influence of Demographic Factors, Recombination, and Worldwide Diversity. Microbiol. Spectr. 2023, 11, e04928-22. [Google Scholar] [CrossRef]
- Kim, D.-Y.; Ahn, H.-S.; Han, S.-H.; Go, H.-J.; Kim, D.-H.; Kim, J.-H.; Lee, J.-B.; Park, S.-Y.; Song, C.-S.; Lee, S.-W. Genetic Analysis of Torque Teno Canis Virus Identified in Republic of Korea. Vet. Sci. 2022, 9, 693. [Google Scholar] [CrossRef]
- Sun, W.; Xie, C.; Zheng, M.; Zhao, G.; Zhang, P.; Han, J.; Jing, J.; Wen, S.; Xiao, P.; Cui, Z. Molecular detection and genomic characterization of Torque teno canis virus in domestic dogs in Guangxi Province, China. J. Biotechnol. 2017, 252, 50–54. [Google Scholar] [CrossRef]
- Hamer, S.A.; Ghai, R.R.; Zecca, I.B.; Auckland, L.D.; Roundy, C.M.; Davila, E.; Busselman, R.E.; Tang, W.; Pauvolid-Corrêa, A.; Killian, M.L. SARS-CoV-2 B. 1.1. 7 variant of concern detected in a pet dog and cat after exposure to a person with COVID-19, USA. Transbound. Emerg. Dis. 2022, 69, 1656–1658. [Google Scholar] [CrossRef]
- Lenz, O.C.; Marques, A.D.; Kelly, B.J.; Rodino, K.G.; Cole, S.D.; Perera, R.A.; Weiss, S.R.; Bushman, F.D.; Lennon, E.M. SARS-CoV-2 Delta variant (AY. 3) in the feces of a domestic cat. Viruses 2022, 14, 421. [Google Scholar] [CrossRef]
| Primer Name | Sequence (5′-3′) | Product Size (bp) | Position (nt) a |
|---|---|---|---|
| FcTTV-P1 | GTAAGTACACTGACGAATGGCT | 8-585 | |
| FcTTV-P2 | CAGTTACCACAGTTCGAGGTCGT | ||
| FcTTV-P3 | ACTGGTGACAGGACGTGCGA | 314 | 50-363 |
| FcTTV-P4 | TGCGGAGACAAGTTGCTTCC | ||
| FcTTV-P1Q | TCGCACGTCCTGTCACCAGT | 344-69 | |
| FcTTV-P2Q | GGAAGCAACTTGTCTCCGCA | ||
| FcTTV-P3Q | TCAGCCATTCGTCAGTGTACTTACT | 1528 | 567-31 |
| FcTTV-P4Q | CGACCTCGAACTGTGGTAACTG | ||
| FcTTV-P5Q | GTTAGTGTTGCTTTACGGCAG | 1969-753 | |
| FcTTV-P6Q | TGAACCAGTGGTGTCCCCAG | ||
| FcTTV-P1 | GTAAGTACACTGACGAATGGCT | 579 | 7-585 |
| FcTTV-P2 | CAGTTACCACAGTTCGAGGTCGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Liu, C.; Yi, J.; Shi, Y.; Li, H.; Liu, H. Genomic Characteristics of Feline Anelloviruses Isolated from Domestic Cats in Shanghai, China. Vet. Sci. 2023, 10, 444. https://doi.org/10.3390/vetsci10070444
Gao J, Liu C, Yi J, Shi Y, Li H, Liu H. Genomic Characteristics of Feline Anelloviruses Isolated from Domestic Cats in Shanghai, China. Veterinary Sciences. 2023; 10(7):444. https://doi.org/10.3390/vetsci10070444
Chicago/Turabian StyleGao, Jun, Chengqian Liu, Jianzhong Yi, Ying Shi, Hong Li, and Huili Liu. 2023. "Genomic Characteristics of Feline Anelloviruses Isolated from Domestic Cats in Shanghai, China" Veterinary Sciences 10, no. 7: 444. https://doi.org/10.3390/vetsci10070444
APA StyleGao, J., Liu, C., Yi, J., Shi, Y., Li, H., & Liu, H. (2023). Genomic Characteristics of Feline Anelloviruses Isolated from Domestic Cats in Shanghai, China. Veterinary Sciences, 10(7), 444. https://doi.org/10.3390/vetsci10070444






