Development of a Crystal Digital RT-PCR for the Detection of Atypical Porcine Pestivirus
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Vaccine Strains and Clinical Samples
2.2. Primers and Probe
2.3. RNA Extraction and Reverse Transcription
2.4. Preparation of Standard Plasmid
2.5. Optimization of the Reaction Conditions
2.6. Generation of the Standard Curves
2.7. Specificity Analysis
2.8. Sensitivity Analysis
2.9. Repeatability Analysis
2.10. Evaluation of the Clinical Samples
3. Results
3.1. Acquirement of the Standard Plasmid
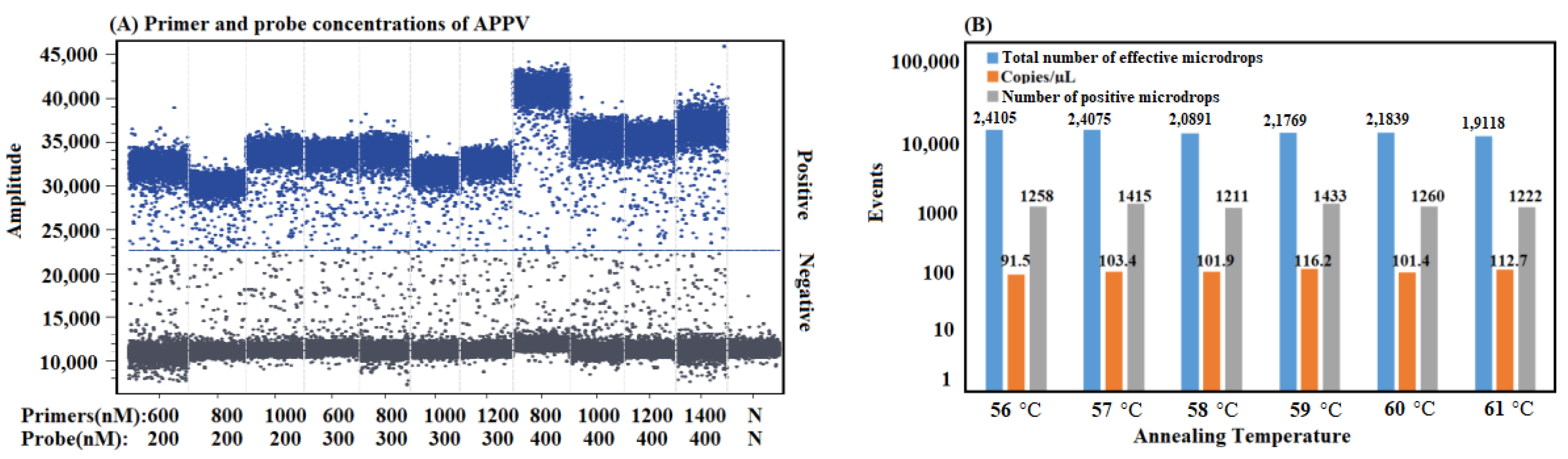
3.2. Optimization of the Reaction Conditions
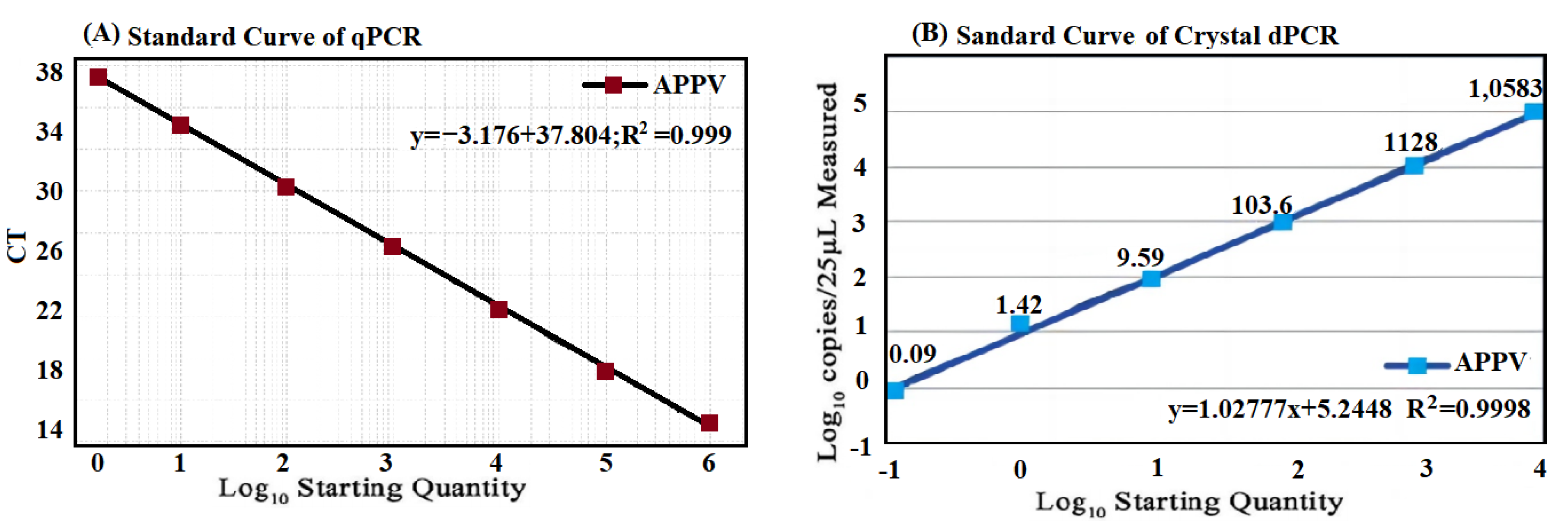
3.3. Generation of Standard Curve
3.4. Analysis on the Specificity
3.5. Analysis on the Sensitivity
3.6. Analysis on the Repeatability
3.7. Evaluation of the Field Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smith, D.B.; Meyers, G.; Bukh, J.; Gould, E.A.; Monath, T.; Scott Muerhoff, A.; Pletnev, A.; Rico-Hesse, R.; Stapleton, J.T.; Simmonds, P.; et al. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J. Gen. Virol. 2017, 98, 2106–2112. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Collin, E.A.; Peddireddi, L.; Yuan, F.; Chen, Z.; Hesse, R.A.; Gauger, P.C.; Clement, T.; Fang, Y.; Anderson, G. Discovery of a novel putative atypical porcine pestivirus in pigs in the USA. J. Gen. Virol. 2015, 96, 2994–2998. [Google Scholar] [CrossRef]
- Arruda, B.L.; Arruda, P.H.; Magstadt, D.R.; Schwartz, K.J.; Dohlman, T.; Schleining, J.A.; Patterson, A.R.; Visek, C.A.; Victoria, J.G. Identification of a divergent lineage porcine pestivirus in nursing piglets with congenital tremors and reproduction of disease following experimental inoculation. PLoS ONE 2016, 11, e0150104. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, K.; Liu, J.; Ge, S.; Xiao, Y.; Shang, Y.; Ning, Z. Identification of atypical porcine pestivirus infection in swine herds in China. Transbound. Emerg. Dis. 2017, 64, 1020–1023. [Google Scholar] [CrossRef]
- Postel, A.; Meyer, D.; Cagatay, G.N.; Feliziani, F.; De Mia, G.M.; Fischer, N.; Grundhoff, A.; Milićević, V.; Deng, M.C.; Chang, C.Y.; et al. High abundance and genetic variability of atypical porcine pestivirus in pigs from Europe and Asia. Emerg. Infect. Dis. 2017, 23, 2104–2107. [Google Scholar] [CrossRef] [PubMed]
- Beer, M.; Wernike, K.; Dräger, C.; Höper, D.; Pohlmann, A.; Bergermann, C.; Schröder, C.; Klinkhammer, S.; Blome, S.; Hoffmann, B. High prevalence of highly variable atypical porcine pestiviruses found in Germany. Transbound. Emerg. Dis. 2017, 64, e22–e26. [Google Scholar] [CrossRef]
- Muñoz-González, S.; Canturri, A.; Pérez-Simó, M.; Bohórquez, J.A.; Rosell, R.; Cabezón, O.; Segalés, J.; Domingo, M.; Ganges, L. First report of the novel atypical porcine pestivirus in Spain and a retrospective study. Transbound. Emerg. Dis. 2017, 64, 1645–1649. [Google Scholar] [CrossRef]
- Dénes, L.; Biksi, I.; Albert, M.; Szeredi, L.; Knapp, D.G.; Szilasi, A.; Bálint, Á.; Balka, G. Detection and phylogenetic characterization of atypical porcine pestivirus strains in Hungary. Transbound. Emerg. Dis. 2018, 65, 2039–2042. [Google Scholar] [CrossRef] [PubMed]
- Mósena, A.C.S.; Weber, M.N.; da Cruz, R.A.S.; Cibulski, S.P.; da Silva, M.S.; Puhl, D.E.; Hammerschmitt, M.E.; Takeuti, K.L.; Driemeier, D.; de Barcellos, D.E.S.N.; et al. Presence of atypical porcine pestivirus (APPV) in Brazilian pigs. Transbound. Emerg. Dis. 2018, 65, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.; Kristensen, C.S.; Strandbygaard, B.; Bøtner, A.; Rasmussen, T.B. Detection of atypical porcine pestivirus in piglets from Danish sow herds. Viruses 2021, 13, 717. [Google Scholar] [CrossRef]
- Kasahara-Kamiie, M.; Kagawa, M.; Shiokawa, M.; Sunaga, F.; Fukase, Y.; Aihara, N.; Shiga, T.; Kamiie, J.; Aoki, H.; Nagai, M. Detection and genetic analysis of a novel atypical porcine pestivirus from piglets with congenital tremor in Japan. Transbound. Emerg. Dis. 2022, 69, 1761–1769. [Google Scholar] [CrossRef]
- Pan, S.; Yan, Y.; Shi, K.; Wang, M.; Mou, C.; Chen, Z. Molecular characterization of two novel atypical porcine pestivirus (APPV) strains from piglets with congenital tremor in China. Transbound. Emerg. Dis. 2019, 66, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Liu, X.; Zhang, P.; Wang, L.; Liu, Y.; Zhang, L.; Liang, P.; Song, C. Identification and characterization of atypical porcine pestivirus genomes in newborn piglets with congenital tremor in China. J. Vet. Sci. 2018, 19, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.L.; Li, Y.Y.; He, L.L.; Wu, J.L.; Tang, X.Y.; Chen, G.H.; Mai, K.J.; Wu, R.T.; Li, Q.N.; Chen, Y.H.; et al. 12 novel atypical porcine pestivirus genomes from neonatal piglets with congenital tremors: A newly emerging branch and high prevalence in China. Virology 2019, 533, 50–58. [Google Scholar] [CrossRef]
- Zhou, K.; Yue, H.; Tang, C.; Ruan, W.; Zhou, Q.; Zhang, B. Prevalence and genome characteristics of atypical porcine pestivirus in southwest China. J. Gen. Virol. 2019, 100, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Xie, S.; Sun, W.; Liu, H.; Zhao, J.; Yin, Y.; Si, H.; Qu, S.; Lu, W. Evolution and genetic diversity of atypical porcine pestivirus (APPV) from piglets with congenital tremor in Guangxi Province, Southern China. Vet. Med. Sci. 2021, 7, 714–723. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, Y.; Song, C.; Cao, Z. Genetic characterization of atypical porcine pestivirus (APPV) in China and the successful isolation of a novel APPV strain within genotype 2. Microb. Pathog. 2021, 161, 105282. [Google Scholar] [CrossRef]
- Dall Agnol, A.M.; Alfieri, A.F.; Alfieri, A.A. Pestivirus K (atypical porcine pestivirus): Update on the virus, viral infection, and the association with congenital tremor in newborn piglets. Viruses 2020, 12, 903. [Google Scholar] [CrossRef]
- Stenberg, H.; Hellman, S.; Lindström, L.; Jacobson, M.; Fossum, C.; Hayer, J.; Malmberg, M. Congenital tremor and splay leg in piglets—Insights into the virome, local cytokine response, and histology. BMC Vet. Res. 2022, 18, 348. [Google Scholar] [CrossRef]
- Schwarz, L.; Riedel, C.; Högler, S.; Sinn, L.J.; Voglmayr, T.; Wöchtl, B.; Dinhopl, N.; Rebel-Bauder, B.; Weissenböck, H.; Ladinig, A.; et al. Congenital infection with atypical porcine pestivirus (APPV) is associated with disease and viral persistence. Vet. Res. 2017, 48, 1. [Google Scholar] [CrossRef]
- Folgueiras-González, A.; van den Braak, R.; Simmelink, B.; Deijs, M.; van der Hoek, L.; de Groof, A. Atypical porcine pestivirus circulation and molecular evolution within an affected swine herd. Viruses 2020, 12, 1080. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.C.; Falkenberg, S.M.; Palmer, M.V.; Arruda, P.H.; Magstadt, D.R.; Schwartz, K.J.; Gatto, I.R.; Neill, J.D.; Arruda, B.L. Distribution and persistence of atypical porcine pestivirus in experimentally inoculated pigs. J. Vet. Diagn. Investig. 2021, 33, 952–955. [Google Scholar] [CrossRef] [PubMed]
- Cagatay, G.N.; Antos, A.; Meyer, D.; Maistrelli, C.; Keuling, O.; Becher, P.; Postel, A. Frequent infection of wild boar with atypical porcine pestivirus (APPV). Transbound. Emerg. Dis. 2018, 65, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, E.; Salogni, C.; Lelli, D.; Barbieri, I.; Moreno, A.; Alborali, G.L.; Lavazza, A. Molecular survey and phylogenetic analysis of atypical porcine pestivirus (APPV) identified in swine and wild boar from Northern Italy. Viruses 2019, 11, 1142. [Google Scholar] [CrossRef]
- Gatto, I.R.H.; Harmon, K.; Bradner, L.; Silva, P.; Linhares, D.C.L.; Arruda, P.H.; de Oliveira, L.G.; Arruda, B.L. Detection of atypical porcine pestivirus in Brazil in the central nervous system of suckling piglets with congenital tremor. Transbound. Emerg. Dis. 2018, 65, 375–380. [Google Scholar] [CrossRef]
- Liu, J.; Li, Z.; Ren, X.; Li, H.; Lu, R.; Zhang, Y.; Ning, Z. Viral load and histological distribution of atypical porcine pestivirus in different tissues of naturally infected piglets. Arch. Virol. 2019, 164, 2519–2523. [Google Scholar] [CrossRef]
- De Groof, A.; Deijs, M.; Guelen, L.; van Grinsven, L.; van Os-Galdos, L.; Vogels, W.; Derks, C.; Cruijsen, T.; Geurts, V.; Vrijenhoek, M.; et al. Atypical porcine pestivirus: A possible cause of congenital tremor type A-II in newborn piglets. Viruses 2016, 8, 271. [Google Scholar] [CrossRef]
- Sutton, K.M.; Lahmers, K.K.; Harris, S.P.; Wijesena, H.R.; Mote, B.E.; Kachman, S.D.; Borza, T.; Ciobanu, D.C. Detection of atypical porcine pestivirus genome in newborn piglets affected by congenital tremor and high preweaning mortality. J. Anim. Sci. 2019, 97, 4093–4100. [Google Scholar] [CrossRef]
- Ganges, L.; Crooke, H.R.; Bohórquez, J.A.; Postel, A.; Sakoda, Y.; Becher, P.; Ruggli, N. Classical swine fever virus: The past, present and future. Virus Res. 2020, 289, 198151. [Google Scholar] [CrossRef]
- Zheng, H.H.; Fu, P.F.; Chen, H.Y.; Wang, Z.Y. Pseudorabies virus: From pathogenesis to prevention strategies. Viruses 2022, 14, 1638. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, X.; Su, D.; Feng, J.; Wei, L.; Cai, W.; Li, J.; Lin, S.; Yan, H.; He, D. Detection and genetic characterization of atypical porcine pestivirus in piglets with congenital tremors in Southern China. Front. Microbiol. 2019, 10, 1406. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Fu, J.; Liu, X.; Bai, J.; Peddireddi, L. Development of a quantitative real time RT-PCR assay for sensitive and rapid detection of emerging atypical porcine pestivirus associated with congenital tremor in pigs. J. Virol. Methods 2021, 296, 114220. [Google Scholar] [CrossRef]
- Liu, H.; Shi, K.; Sun, W.; Zhao, J.; Yin, Y.; Si, H.; Qu, S.; Lu, W. Development a multiplex RT-PCR assay for simultaneous detection of African swine fever virus, classical swine fever virus and atypical porcine pestivirus. J. Virol. Methods 2021, 287, 114006. [Google Scholar] [CrossRef]
- Liu, H.; Shi, K.; Zhao, J.; Yin, Y.; Chen, Y.; Si, H.; Qu, S.; Long, F.; Lu, W. Development of a one-step multiplex qRT-PCR assay for the detection of African swine fever virus, classical swine fever virus and atypical porcine pestivirus. BMC Vet. Res. 2022, 18, 43. [Google Scholar] [CrossRef] [PubMed]
- Kojabad, A.A.; Farzanehpour, M.; Galeh, H.E.G.; Dorostkar, R.; Jafarpour, A.; Bolandian, M.; Nodooshan, M.M. Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J. Med. Virol. 2021, 93, 4182–4197. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.L.; Loganathan, N.; Agarwalla, S.; Yang, C.; Yuan, W.; Zeng, J.; Wu, R.; Wang, W.; Duraiswamy, S. Current commercial dPCR platforms: Technology and market review. Crit. Rev. Biotechnol. 2023, 43, 433–464. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Deng, L.; Yang, X.; Xu, Z.; Li, F.; Zhao, J.; Deng, H.; Jian, Z.; Sun, X.; Zhu, L. Development and use of a droplet digital PCR (ddPCR) assay to achieve sensitive and fast atypical porcine pestivirus detection. Braz. J. Microbiol. 2022, 53, 625–631. [Google Scholar] [CrossRef]
- Choe, S.; Park, G.N.; Cha, R.M.; Hyun, B.H.; Park, B.K.; An, D.J. Prevalence and genetic diversity of atypical porcine pestivirus (APPV) detected in South Korean wild boars. Viruses 2020, 12, 680. [Google Scholar] [CrossRef]
- Possatti, F.; Headley, S.A.; Leme, R.A.; Dall Agnol, A.M.; Zotti, E.; de Oliveira, T.E.S.; Alfieri, A.F.; Alfieri, A.A. Viruses associated with congenital tremor and high lethality in piglets. Transbound. Emerg. Dis. 2018, 65, 331–337. [Google Scholar] [CrossRef]
- Espy, M.J.; Uhl, J.R.; Sloan, L.M.; Buckwalter, S.P.; Jones, M.F.; Vetter, E.A.; Yao, J.D.; Wengenack, N.L.; Rosenblatt, J.E.; Cockerill, F.R.; et al. Real-time PCR in clinical microbiology: Applications for routine laboratory testing. Clin. Microbiol. Rev. 2006, 19, 165–256. [Google Scholar] [CrossRef] [PubMed]
- Salipante, S.J.; Jerome, K.R. Digital PCR-An emerging technology with broad applications in microbiology. Clin. Chem. 2020, 66, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Postel, A.; Hansmann, F.; Baechlein, C.; Fischer, N.; Alawi, M.; Grundhoff, A.; Derking, S.; Tenhündfeld, J.; Pfankuche, V.M.; Herder, V.; et al. Presence of atypical porcine pestivirus (APPV) genomes in newborn piglets correlates with congenital tremor. Sci. Rep. 2016, 6, 27735. [Google Scholar] [CrossRef]
- Dessureault, F.G.; Choinière, M.; Provost, C.; Gagnon, C.A. First report of atypical porcine pestivirus in piglets with congenital tremor in Canada. Can. Vet. J. 2018, 59, 429–432. [Google Scholar]
- Kaufmann, C.; Stalder, H.; Sidler, X.; Renzullo, S.; Gurtner, C.; Grahofer, A.; Schweizer, M. Long-term circulation of atypical porcine pestivirus (APPV) within Switzerland. Viruses 2019, 11, 653. [Google Scholar] [CrossRef]
- Yuan, F.; Feng, Y.; Bai, J.; Liu, X.; Arruda, B.; Anbalagan, S.; Peddireddi, L. Genetic diversity and prevalence of atypical porcine pestivirus in the midwest of US swine herds during 2016-2018. Transbound. Emerg. Dis. 2022, 69, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Han, Z.; Li, J.; Huang, Y.; Yang, J.; Ding, H.; Zhang, J.; Zhu, M.; Zhang, Y.; Liao, J.; et al. Atypical porcine pestivirus as a novel type of pestivirus in pigs in China. Front. Microbiol. 2017, 8, 862. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Li, W.; Zhang, M.; Yang, Z.; Lin, L.; Ghonaim, A.H.; He, Q. The diversity and spatiotemporally evolutionary dynamic of atypical porcine pestivirus in China. Front. Microbiol. 2022, 13, 937918. [Google Scholar] [CrossRef]


| Primer/Probe | Sequence (5′→3′) | Product/bp |
|---|---|---|
| APPV-F | GGCGTGCCCAAAGAGAAAT | 90 |
| APPV-R | GGCACTCTATCAAGCAGTAAGGTCTA | |
| APPV-P | FAM-TCGGGTCCACCATGCCCCTTT-BHQ1 |
| cdRT-PCR | qRT-PCR | |||
|---|---|---|---|---|
| Volume (μL) | Final Concentration (nM) | Volume (μL) | Final Concentration (nM) | |
| qScript XLT One-Step RT-qPCR ToughMix (2×) (Quanta Bio., Gaithersburg, MD, USA) | 12.5 | 1× | / | / |
| Fluorescein sodium salt (1000 nM) (Apexbio Bio., Beijing, China), | 2.5 | 100 | / | / |
| One Step RT-PCR Buffer (2×) (TaKaRa, Dalian, China) | / | / | 12.5 | 1× |
| Ex Taq HS (5000 nM) (TaKaRa, Dalian, China) | / | / | 0.5 | 100 |
| Primer Script RT Enzyme Mix (5000 nM) (TaKaRa, Dalian, China) | / | / | 0.5 | 100 |
| APPV-F (25,000 nM) | 0.8 | 800 | 0.4 | 400 |
| APPV-R (25,000 nM) | 0.8 | 800 | 0.4 | 400 |
| APPV-P (25,000 nM) | 0.4 | 400 | 0.3 | 300 |
| Total nucleic acids | 2.5 | / | 2.5 | / |
| Distilled water | Up to 25 | / | Up to 25 | / |
| Concentration | cdRT-PCR | qRT-PCR | ||||||
|---|---|---|---|---|---|---|---|---|
| Intra-Assay | Inter-Assay | Intra-Assay | Inter-Assay | |||||
| CV% | CV% | CV% | CV% | |||||
| 103 | 1114.67 ± 22.23 | 1.99 | 1121.22 ± 9.14 | 0.82 | 26.70 ± 0.24 | 0.90 | 26.64 ± 0.09 | 0.34 |
| 102 | 109.07 ± 4.78 | 4.38 | 108.84 ± 0.63 | 0.58 | 30.20 ± 0.18 | 0.60 | 30.23 ± 0.24 | 0.79 |
| 101 | 9.68 ± 0.51 | 5.27 | 9.50 ± 0.20 | 2.11 | 34.57 ± 0.25 | 0.72 | 34.42 ± 0.15 | 0.44 |
| qRT-PCR | Coincidence Rate (%) | Kappa Value | ||||
|---|---|---|---|---|---|---|
| Positive | Negative | Total | ||||
| Positive | 14 | 1 | 15 | 98.33% | 0.955 | |
| cdRT-PCR | Negative | 0 | 45 | 45 | ||
| Total | 14 | 46 | 60 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Shi, K.; Feng, S.; Yin, Y.; Long, F.; Si, H. Development of a Crystal Digital RT-PCR for the Detection of Atypical Porcine Pestivirus. Vet. Sci. 2023, 10, 330. https://doi.org/10.3390/vetsci10050330
Liu H, Shi K, Feng S, Yin Y, Long F, Si H. Development of a Crystal Digital RT-PCR for the Detection of Atypical Porcine Pestivirus. Veterinary Sciences. 2023; 10(5):330. https://doi.org/10.3390/vetsci10050330
Chicago/Turabian StyleLiu, Huixin, Kaichuang Shi, Shuping Feng, Yanwen Yin, Feng Long, and Hongbin Si. 2023. "Development of a Crystal Digital RT-PCR for the Detection of Atypical Porcine Pestivirus" Veterinary Sciences 10, no. 5: 330. https://doi.org/10.3390/vetsci10050330
APA StyleLiu, H., Shi, K., Feng, S., Yin, Y., Long, F., & Si, H. (2023). Development of a Crystal Digital RT-PCR for the Detection of Atypical Porcine Pestivirus. Veterinary Sciences, 10(5), 330. https://doi.org/10.3390/vetsci10050330







