Influence of the At-Arrival Host Transcriptome on Bovine Respiratory Disease Incidence during Backgrounding
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Use and Study Enrollment
2.2. Sample Processing, Next-Generation RNA Sequencing, and Bioinformatic Processing
2.3. Differential Gene Expression and Functional Enrichment Analyses
2.4. Data Visualization and Model-Based Unsupervised Clustering Analyses
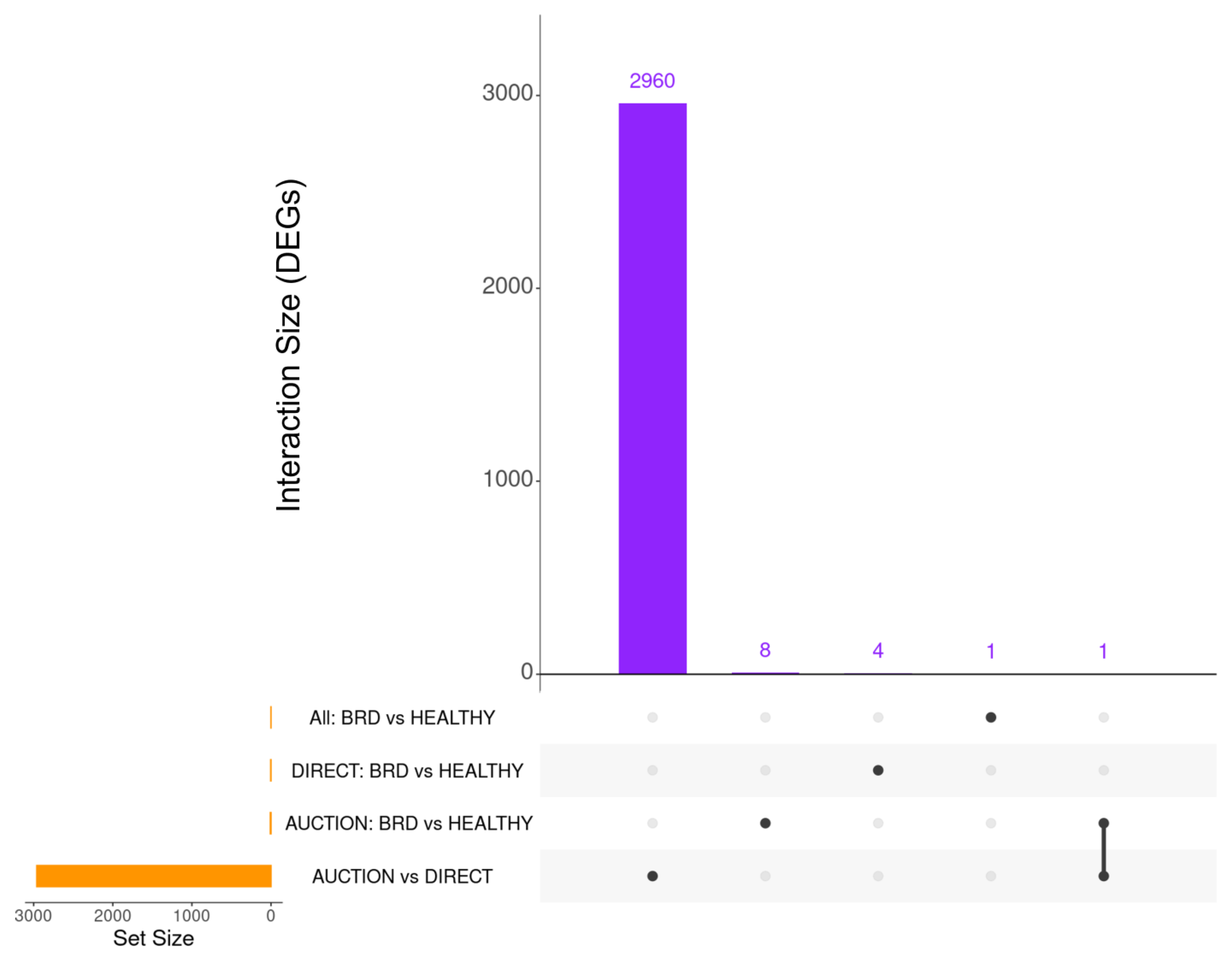
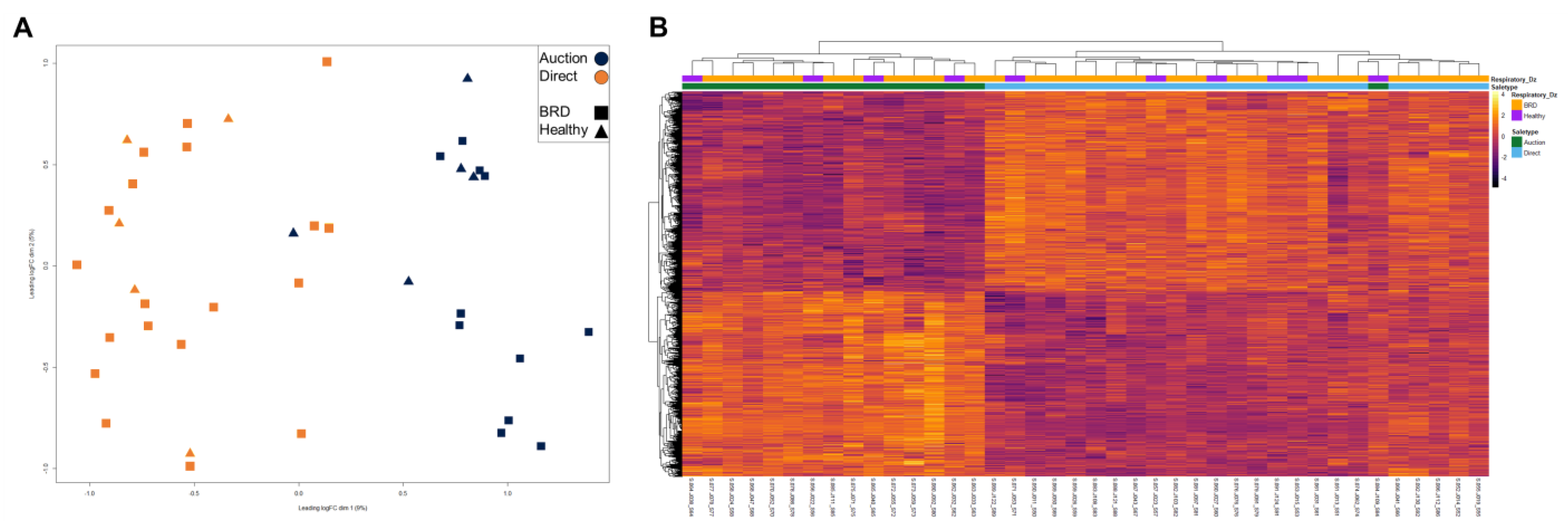
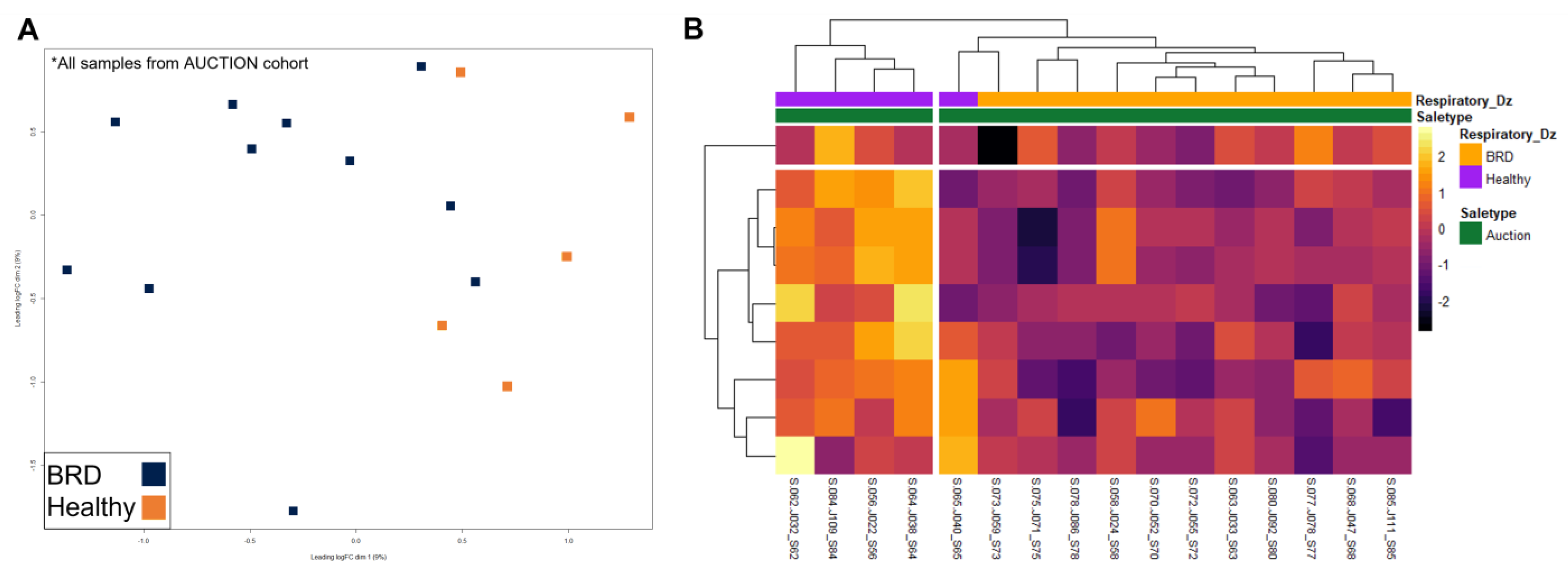
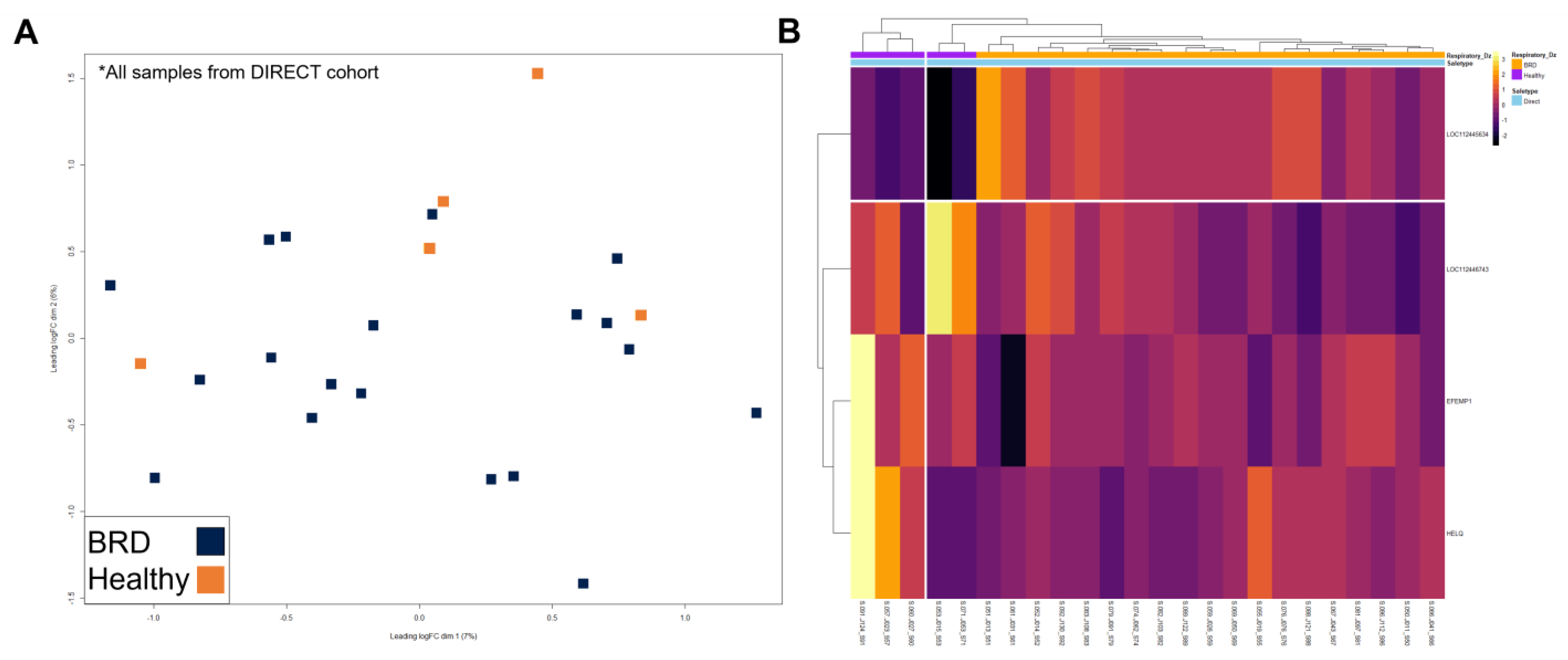
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- USDA. Part IV: Health and Health Management on U.S. Feedlots with a Capacity of 1000 or More Head; USDA-APHIS-VS-CEAH-NAHMS: Fort Collins, CO, USA, 2011. Available online: https://www.aphis.usda.gov/animal_health/nahms/feedlot/downloads/feedlot2011/Feed11_dr_PartIV_1.pdf (accessed on 14 November 2022).
- Dargatz, D.A.; Lombard, J.E. Summary of BRD data from the 2011 NAHMS feedlot and dairy heifer studies. Anim. Health Res. Rev. 2014, 15, 123–125. [Google Scholar] [CrossRef] [PubMed]
- Snowder, G.D.; Van Vleck, L.D.; Cundiff, L.V.; Bennett, G. Bovine respiratory disease in feedlot cattle: Environmental, genetic, and economic factors. J. Anim. Sci. 2006, 84, 1999–2008. [Google Scholar] [CrossRef] [PubMed]
- Blakebrough-Hall, C.; McMeniman, J.P.; González, L.A. An evaluation of the economic effects of bovine respiratory disease on animal performance, carcass traits, and economic outcomes in feedlot cattle defined using four BRD diagnosis methods. J. Anim. Sci. 2020, 98, skaa005. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.K.; Richards, C.J.; Step, D.L.; Krehbiel, C.R. Beef Species symposium: Best management practices for newly weaned calves for improved health and well-being1. J. Anim. Sci. 2017, 95, 2170–2182. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, M.W.; Dargatz, D.A.; Wagner, B.A. Risk factors for initial respiratory disease in United States’ feedlots based on producer-collected daily morbidity counts. Am. Jew. Hist. 2008, 49, 373–378. [Google Scholar]
- Woods, G.T.; E Mansfield, M.; Webb, R.J. A three year comparison of acute respiratory disease, shrink and weight gain in preconditioned and non-preconditioned Illinois beef calves sold at the same auction and mixed in a feedlot. Can. J. Comp. Med. 1973, 37, 249–255. [Google Scholar]
- Ribble, C.S.; Meek, A.H.; Shewen, P.E.; Guichon, P.T.; Jim, G.K. Effect of Pretransit Mixing on Fatal Fibrinous Pneumonia in Calves. J. Am. Vet. Med. Assoc. 1995, 207, 616–619. [Google Scholar]
- Step, D.L.; Krehbiel, C.R.; DePra, H.A.; Cranston, J.J.; Fulton, R.W.; Kirkpatrick, J.G.; Gill, D.R.; Payton, M.E.; Montelongo, M.A.; Confer, A.W. Effects of commingling beef calves from different sources and weaning protocols during a forty-two-day receiving period on performance and bovine respiratory disease1,2. J. Anim. Sci. 2008, 86, 3146–3158. [Google Scholar] [CrossRef]
- McGill, J.L.; Sacco, R.E. The Immunology of Bovine Respiratory Disease. Veter.-Clin. N. Am. Food Anim. Pract. 2020, 36, 333–348. [Google Scholar] [CrossRef]
- Smith, D.R. Risk factors for bovine respiratory disease in beef cattle. Anim. Health Res. Rev. 2020, 21, 149–152. [Google Scholar] [CrossRef]
- Mosier, D. Review of BRD pathogenesis: The old and the new. Anim. Health Res. Rev. 2014, 15, 166–168. [Google Scholar] [CrossRef] [PubMed]
- Gummow, B.; Mapham, P.H. A stochastic partial-budget analysis of an experimental Pasteurella haemolytica feedlot vaccine trial. Prev. Veter.-Med. 2000, 43, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.D.; Fulton, R.W.; Lehenbauer, T.W.; Step, D.L.; Confer, A.W. The Epidemiology of Bovine Respiratory Disease: What Is the Evidence for Predisposing Factors? Can. Vet. J. 2010, 51, 1095–1102. [Google Scholar] [PubMed]
- Schneider, M.J.; Tait, R.G.; Busby, W.D.; Reecy, J.M. An evaluation of bovine respiratory disease complex in feedlot cattle: Impact on performance and carcass traits using treatment records and lung lesion scores. J. Anim. Sci. 2009, 87, 1821–1827. [Google Scholar] [CrossRef]
- Du Sert, N.P.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. Br. J. Pharmacol. 2020, 177, 3617–3624. [Google Scholar] [CrossRef]
- Scott, M.A.; Woolums, A.R.; Karisch, B.B.; Harvey, K.M.; Capik, S.F. Impact of preweaning vaccination on host gene expression and antibody titers in healthy beef calves. Front. Veter.-Sci. 2022, 9, 1010039. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Khan, A.; Mathelier, A. Intervene: A tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinform. 2017, 18, 287. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Lee, H.; Han, B. FastRNA: An efficient solution for PCA of single-cell RNA-sequencing data based on a batch-accounting count model. Am. J. Hum. Genet. 2022, 109, 1974–1985. [Google Scholar] [CrossRef]
- Tipping, M.E.; Bishop, C.M. Probabilistic Principal Component Analysis. J. R. Stat. Soc. Ser. B Stat. Methodol. 1999, 61, 611–622. [Google Scholar] [CrossRef]
- Narasimhan, S.; Shah, S.L. Model identification and error covariance matrix estimation from noisy data using PCA. Control Eng. Pract. 2008, 16, 146–155. [Google Scholar] [CrossRef]
- Horn, J.L. A rationale and test for the number of factors in factor analysis. Psychometrika 1965, 30, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Garnier, S.; Ross, N.; Rudis, B.; Filipovic-Pierucci, A.; Galili, T.; Greenwell, B.; Sievert, C.; Harris, D.J.; Chen, J.J. Sjmgarnier/Viridis: Viridis 0.6.0 (Pre-CRAN Release). 2021. Available online: https://zenodo.org/record/4679424#.ZAqmOh9Bx7M (accessed on 11 November 2022). [CrossRef]
- Galyean, M.L.; Perino, L.J.; Duff, G.C. Interaction of cattle health/immunity and nutrition. J. Anim. Sci. 1999, 77, 1120–1134. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.W.; Meek, A.H.; Davis, D.G.; Johnson, J.A.; Curtis, R.A. Factors associated with mortality and treatment costs in feedlot calves: The Bruce County Beef Project, years 1978, 1979, 1980. Can. J. Comp. Med. 1982, 46, 341–349. [Google Scholar]
- Scott, M.A.; Woolums, A.R.; Swiderski, C.E.; Perkins, A.D.; Nanduri, B.; Smith, D.R.; Karisch, B.B.; Epperson, W.B.; Blanton, J.R., Jr. Whole blood transcriptomic analysis of beef cattle at arrival identifies potential predictive molecules and mechanisms that indicate animals that naturally resist bovine respiratory disease. PLoS ONE 2020, 15, e0227507. [Google Scholar] [CrossRef]
- Scott, M.A.; Woolums, A.R.; Swiderski, C.E.; Perkins, A.D.; Nanduri, B.; Smith, D.R.; Karisch, B.B.; Epperson, W.B.; Blanton, J.R. Comprehensive at-arrival transcriptomic analysis of post-weaned beef cattle uncovers type I interferon and antiviral mechanisms associated with bovine respiratory disease mortality. PLoS ONE 2021, 16, e0250758. [Google Scholar] [CrossRef]
- Scott, M.A.; Woolums, A.R.; Swiderski, C.E.; Finley, A.; Perkins, A.D.; Nanduri, B.; Karisch, B.B. Hematological and gene co-expression network analyses of high-risk beef cattle defines immunological mechanisms and biological complexes involved in bovine respiratory disease and weight gain. PLoS ONE 2022, 17, e0277033. [Google Scholar] [CrossRef]
- Scott, M.A.; Woolums, A.R.; Swiderski, C.E.; Perkins, A.D.; Nanduri, B.; Smith, D.R.; Karisch, B.B.; Epperson, W.B.; Blanton, J.R. Multipopulational transcriptome analysis of post-weaned beef cattle at arrival further validates candidate biomarkers for predicting clinical bovine respiratory disease. Sci. Rep. 2021, 11, 23877. [Google Scholar] [CrossRef]
- Sun, H.-Z.; Srithayakumar, V.; Jiminez, J.; Jin, W.; Hosseini, A.; Raszek, M.; Orsel, K.; Guan, L.L.; Plastow, G. Longitudinal blood transcriptomic analysis to identify molecular regulatory patterns of bovine respiratory disease in beef cattle. Genomics 2020, 112, 3968–3977. [Google Scholar] [CrossRef]
- Jiminez, J.; Timsit, E.; Orsel, K.; van der Meer, F.; Guan, L.L.; Plastow, G. Whole-Blood Transcriptome Analysis of Feedlot Cattle With and Without Bovine Respiratory Disease. Front. Genet. 2021, 12, 627623. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Mukiibi, R.; Jiminez, J.; Wang, Z.; Akanno, E.C.; Timsit, E.; Plastow, G.S. Applying multi-omics data to study the genetic background of bovine respiratory disease infection in feedlot crossbred cattle. Front. Genet. 2022, 13, 1046192. [Google Scholar] [CrossRef] [PubMed]
- Hasankhani, A.; Bahrami, A.; Sheybani, N.; Fatehi, F.; Abadeh, R.; Farahani, H.G.M.; Behzadi, M.R.B.; Javanmard, G.; Isapour, S.; Khadem, H.; et al. Integrated Network Analysis to Identify Key Modules and Potential Hub Genes Involved in Bovine Respiratory Disease: A Systems Biology Approach. Front. Genet. 2021, 12, 753839. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Furuta, H.; Kondo, Y.; Mukoyama, H. Association of BoLA-DRB3 alleles with mastitis resistance and susceptibility in Japanese Holstein cows: BoLA-DRB3 alleles and cow mastitis. Anim. Sci. J. 2011, 83, 359–366. [Google Scholar] [CrossRef]
- Hayashi, T.; Mekata, H.; Sekiguchi, S.; Kirino, Y.; Mitoma, S.; Honkawa, K.; Horii, Y.; Norimine, J. Cattle with the BoLA class II DRB3*0902 allele have significantly lower bovine leukemia proviral loads. J. Vet. Med. Sci. 2017, 79, 1552–1555. [Google Scholar] [CrossRef]
- Gowane, G.; Sharma, A.; Sankar, M.; Narayanan, K.; Das, B.; Subramaniam, S.; Pattnaik, B. Association of BoLA DRB3 alleles with variability in immune response among the crossbred cattle vaccinated for foot-and-mouth disease (FMD). Res. Veter.-Sci. 2013, 95, 156–163. [Google Scholar] [CrossRef]
- Fukunaga, K.; Yamashita, Y.; Yagisawa, T. Copy number variations in BOLA-DQA2, BOLA-DQB, and BOLA-DQA5 show the genomic architecture and haplotype frequency of major histocompatibility complex class II genes in Holstein cows. Hla 2020, 96, 601–609. [Google Scholar] [CrossRef]
- Gelhaus, A.; Förster, B.; Wippern, C.; Horstmann, R.D. Evidence for an additional cattle DQA locus, BoLA-DQA5. Immunogenetics 1999, 49, 321–327. [Google Scholar] [CrossRef]
- Ren, K.; Lv, Y.; Zhuo, Y.; Chen, C.; Shi, H.; Guo, L.; Yang, G.; Hou, Y.; Tan, R.X.; Li, E. Suppression of IRG-1 Reduces Inflammatory Cell Infiltration and Lung Injury in Respiratory Syncytial Virus Infection by Reducing Production of Reactive Oxygen Species. J. Virol. 2016, 90, 7313–7322. [Google Scholar] [CrossRef]
- Tallam, A.; Perumal, T.M.; Antony, P.M.; Jäger, C.; Fritz, J.V.; Vallar, L.; Balling, R.; del Sol, A.; Michelucci, A. Gene Regulatory Network Inference of Immunoresponsive Gene 1 (IRG1) Identifies Interferon Regulatory Factor 1 (IRF1) as Its Transcriptional Regulator in Mammalian Macrophages. PLoS ONE 2016, 11, e0149050. [Google Scholar] [CrossRef]
- Chen, F.; Elgaher, W.A.M.; Winterhoff, M.; Büssow, K.; Waqas, F.H.; Graner, E.; Pires-Afonso, Y.; Perez, L.C.; de la Vega, L.; Sahini, N.; et al. Citraconate inhibits ACOD1 (IRG1) catalysis, reduces interferon responses and oxidative stress, and modulates inflammation and cell metabolism. Nat. Metab. 2022, 4, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Yang, H.; Lin, Y.; Zhang, Y.; Pan, C.; Feng, P.; Yu, Y.; Chen, X. LncRNA and mRNA profiling during activation of tilapia macrophages by HSP70 and Streptococcus agalactiae antigen. Oncotarget 2017, 8, 98455–98470. [Google Scholar] [CrossRef] [PubMed]
- Yang-Chun, F.; Sen-Yu, W.; Yuan, Z.; Yan-Chun, H. Genome-Wide Profiling of Human Papillomavirus DNA Integration into Human Genome and Its Influence on PD-L1 Expression in Chinese Uygur Cervical Cancer Women. J. Immunol. Res. 2020, 2020, 6284960. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Xu, M.; Li, J.; Liu, L.; Xu, M.; Zhou, C.; Shen, L.; Tang, Z.; Wan, F. Mas-related G protein-coupled receptor D participates in inflammatory pain by promoting NF-κB activation through interaction with TAK1 and IKK complex. Cell. Signal. 2020, 76, 109813. [Google Scholar] [CrossRef]
- He, X.; Ashbrook, A.W.; Du, Y.; Wu, J.; Hoffmann, H.-H.; Zhang, C.; Xia, L.; Peng, Y.-C.; Tumas, K.C.; Singh, B.K.; et al. RTP4 inhibits IFN-I response and enhances experimental cerebral malaria and neuropathology. Proc. Natl. Acad. Sci. USA 2020, 117, 19465–19474. [Google Scholar] [CrossRef]
- Malone, D.; Lardelli, R.M.; Li, M.; David, M. Dephosphorylation activates the interferon-stimulated Schlafen family member 11 in the DNA damage response. J. Biol. Chem. 2019, 294, 14674–14685. [Google Scholar] [CrossRef]
- Radjabova, V.; Mastroeni, P.; Skjødt, K.; Zaccone, P.; de Bono, B.; Goodall, J.C.; Chilvers, E.R.; Juss, J.K.; Jones, D.C.; Trowsdale, J.; et al. TARM1 Is a Novel Leukocyte Receptor Complex–Encoded ITAM Receptor That Costimulates Proinflammatory Cytokine Secretion by Macrophages and Neutrophils. J. Immunol. 2015, 195, 3149–3159. [Google Scholar] [CrossRef]
- Scott, M.A.; Woolums, A.R.; Swiderski, C.E.; Perkins, A.D.; Nanduri, B. Genes and regulatory mechanisms associated with experimentally-induced bovine respiratory disease identified using supervised machine learning methodology. Sci. Rep. 2021, 11, 22916. [Google Scholar] [CrossRef]
- Tizioto, P.C.; Kim, J.; Seabury, C.M.; Schnabel, R.D.; Gershwin, L.J.; Van Eenennaam, A.; Toaff-Rosenstein, R.; Neibergs, H.L.; Taylor, J.F. Bovine Respiratory Disease Complex Coordinated Agricultural Project Research Team Immunological Response to Single Pathogen Challenge with Agents of the Bovine Respiratory Disease Complex: An RNA-Sequence Analysis of the Bronchial Lymph Node Transcriptome. PLoS ONE 2015, 10, e0131459. [Google Scholar] [CrossRef]
- Behura, S.K.; Tizioto, P.C.; Kim, J.; Grupioni, N.V.; Seabury, C.M.; Schnabel, R.D.; Gershwin, L.J.; Van Eenennaam, A.L.; Toaff-Rosenstein, R.; Neibergs, H.L.; et al. Tissue Tropism in Host Transcriptional Response to Members of the Bovine Respiratory Disease Complex. Sci. Rep. 2017, 7, 17938. [Google Scholar] [CrossRef]
- Johnston, D.; Earley, B.; McCabe, M.S.; Kim, J.; Taylor, J.F.; Lemon, K.; Duffy, C.; McMenamy, M.; Cosby, S.L.; Waters, S.M. Messenger RNA biomarkers of Bovine Respiratory Syncytial Virus infection in the whole blood of dairy calves. Sci. Rep. 2021, 11, 9392. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.A.; Woolums, A.R.; Swiderski, C.E.; Thompson, A.C.; Perkins, A.D.; Nanduri, B.; Karisch, B.B.; Goehl, D.R. Use of nCounter mRNA profiling to identify at-arrival gene expression patterns for predicting bovine respiratory disease in beef cattle. BMC Veter-Res. 2022, 18, 77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gong, Y.; Xiao, J.; Chai, Y.; Lei, J.; Huang, H.; Xiang, T.; Shen, W. A COL1A1 Promoter-Controlled Expression of TGF-β Soluble Receptor Inhibits Hepatic Fibrosis Without Triggering Autoimmune Responses. Dig. Dis. Sci. 2018, 63, 2662–2672. [Google Scholar] [CrossRef] [PubMed]
- Tsitoura, E.; Trachalaki, A.; Vasarmidi, E.; Mastrodemou, S.; Margaritopoulos, G.A.; Kokosi, M.; Fanidis, D.; Galaris, A.; Aidinis, V.; Renzoni, E.; et al. Collagen 1a1 Expression by Airway Macrophages Increases In Fibrotic ILDs and Is Associated With FVC Decline and Increased Mortality. Front. Immunol. 2021, 12, 645548. [Google Scholar] [CrossRef]
- Li, W.; Duan, X.; Zhu, C.; Liu, X.; Jeyarajan, A.J.; Xu, M.; Tu, Z.; Sheng, Q.; Chen, D.; Zhu, C.; et al. Hepatitis B and Hepatitis C Virus Infection Promote Liver Fibrogenesis through a TGF-β1–Induced OCT4/Nanog Pathway. J. Immunol. 2022, 208, 672–684. [Google Scholar] [CrossRef]
- Nuyttens, B.P.; Thijs, T.; Deckmyn, H.; Broos, K. Platelet adhesion to collagen. Thromb. Res. 2011, 127 (Suppl. S2), S26–S29. [Google Scholar] [CrossRef]
- Hu, H.; Zhu, L.; Huang, Z.; Ji, Q.; Chatterjee, M.; Zhang, W.; Li, N. Platelets enhance lymphocyte adhesion and infiltration into arterial thrombus. Thromb. Haemost. 2010, 104, 1184–1192. [Google Scholar] [CrossRef]
- Chebbo, M.; Duez, C.; Alessi, M.C.; Chanez, P.; Gras, D. Platelets: A potential role in chronic respiratory diseases? Eur. Respir. Rev. 2021, 30, 210062. [Google Scholar] [CrossRef]
- Avra, T.D.; Abell, K.M.; Shane, D.D.; Theurer, M.E.; Larson, R.L.; White, B.J. A retrospective analysis of risk factors associated with bovine respiratory disease treatment failure in feedlot cattle1. J. Anim. Sci. 2017, 95, 1521–1527. [Google Scholar] [CrossRef]
- White, B.J.; Amrine, D.E.; Goehl, D.R. Determination of value of bovine respiratory disease control using a remote early disease identification system compared with conventional methods of metaphylaxis and visual observations1. J. Anim. Sci. 2015, 93, 4115–4122. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Green, M.M.; Woolums, A.R.; Karisch, B.B.; Harvey, K.M.; Capik, S.F.; Scott, M.A. Influence of the At-Arrival Host Transcriptome on Bovine Respiratory Disease Incidence during Backgrounding. Vet. Sci. 2023, 10, 211. https://doi.org/10.3390/vetsci10030211
Green MM, Woolums AR, Karisch BB, Harvey KM, Capik SF, Scott MA. Influence of the At-Arrival Host Transcriptome on Bovine Respiratory Disease Incidence during Backgrounding. Veterinary Sciences. 2023; 10(3):211. https://doi.org/10.3390/vetsci10030211
Chicago/Turabian StyleGreen, Mollie M., Amelia R. Woolums, Brandi B. Karisch, Kelsey M. Harvey, Sarah F. Capik, and Matthew A. Scott. 2023. "Influence of the At-Arrival Host Transcriptome on Bovine Respiratory Disease Incidence during Backgrounding" Veterinary Sciences 10, no. 3: 211. https://doi.org/10.3390/vetsci10030211
APA StyleGreen, M. M., Woolums, A. R., Karisch, B. B., Harvey, K. M., Capik, S. F., & Scott, M. A. (2023). Influence of the At-Arrival Host Transcriptome on Bovine Respiratory Disease Incidence during Backgrounding. Veterinary Sciences, 10(3), 211. https://doi.org/10.3390/vetsci10030211





