Generation of Transgenic Sperm Expressing GFP by Lentivirus Transduction of Spermatogonial Stem Cells In Vivo in Cynomolgus Monkeys
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Ethics
2.2. EGFP Lentivirus
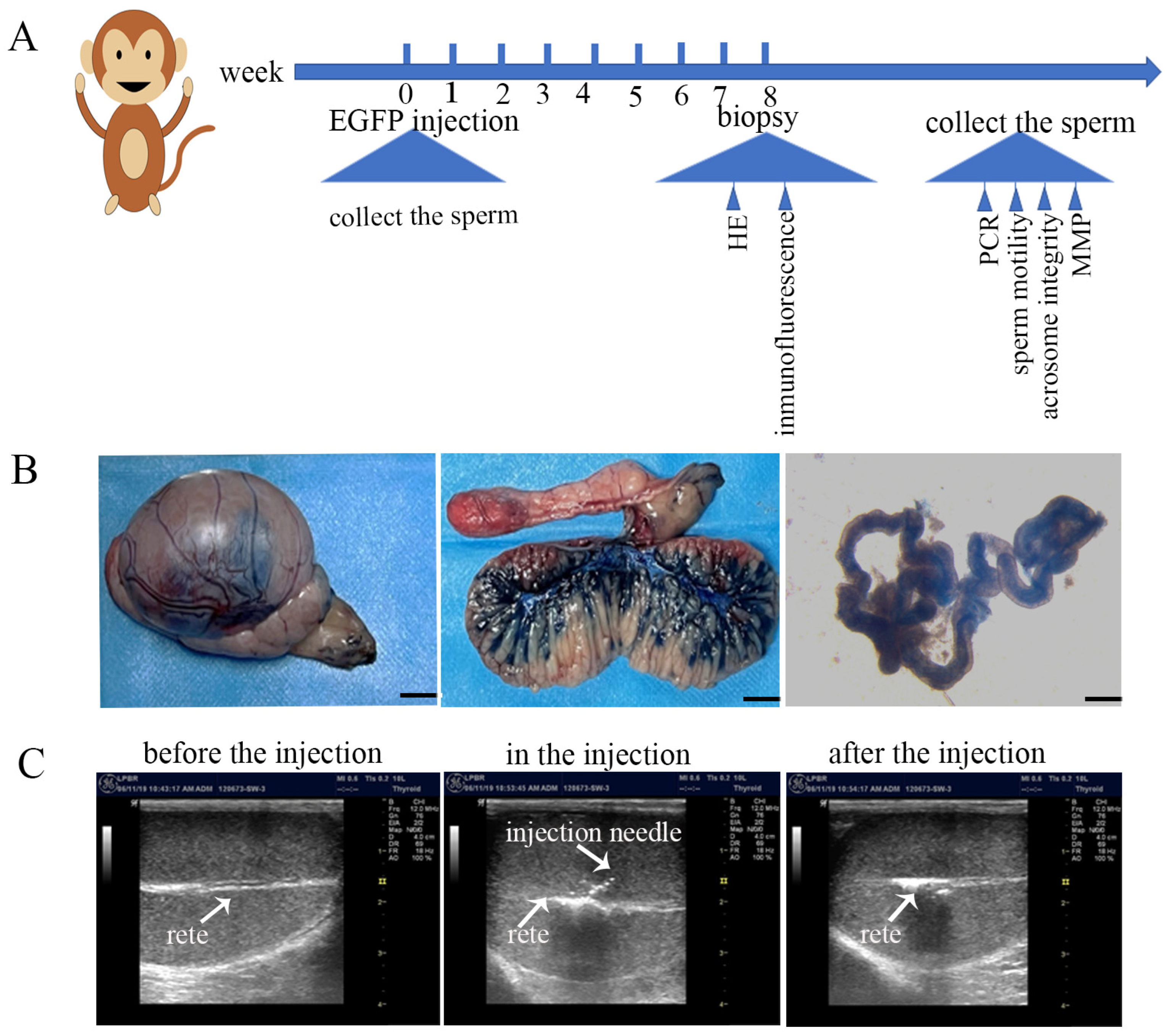
2.3. Injection of EGFP Lentivirus into the Rete of Cynomolgus Macaque Testicles
2.4. Pathology of Testicular Tissue
2.5. Immunofluorescence Examination
2.6. Semen Collection, Evaluation and Cryopreservation
2.7. Sperm DNA Isolation, q-PCR and Sequencing
2.8. Statistical Analysis
2.9. Data Availability
3. Results
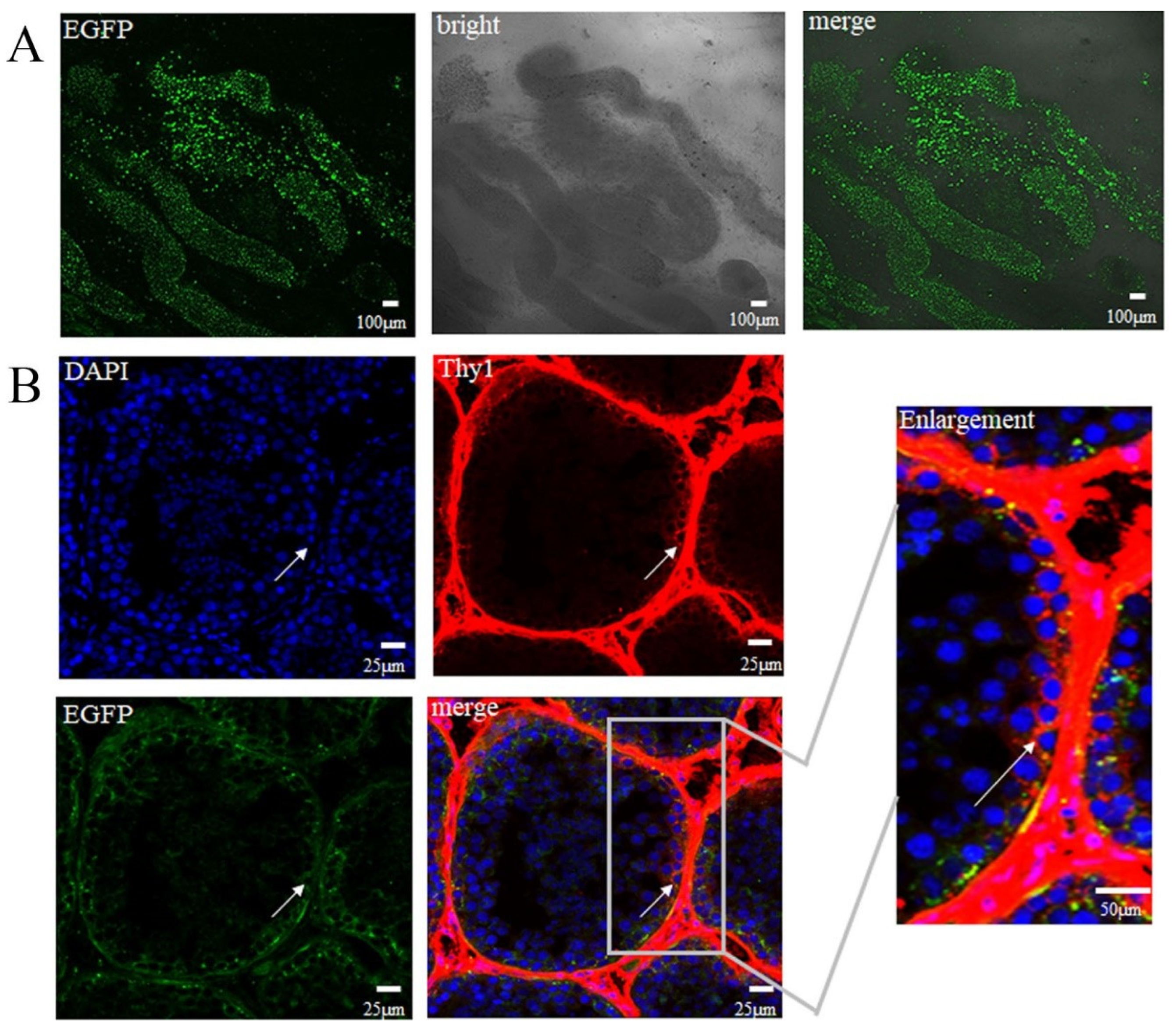
3.1. Validation of Rete Testis Injection Method and Introduction of EGFP Lentiviruses
3.2. The Injection of EGFP Lentivirus Did Not Affect Spermatogenesis
3.3. EGFP Expressed in SSCs
3.4. Confirmation of the Generation of Transgenic Sperm and Sperm Cryopreservation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zheng, Y.; Liu, X.; Le, W.; Xie, L.; Li, H.; Wen, W.; Wang, S.; Ma, S.; Huang, Z.; Ye, J.; et al. A human circulating immune cell landscape in aging and COVID-19. Protein Cell 2020, 11, 740–770. [Google Scholar] [CrossRef]
- Palliyaguru, D.L.; Shiroma, E.J.; Nam, J.K.; Duregon, E.; Vieira Ligo Teixeira, C.; Price, N.L.; Bernier, M.; Camandola, S.; Vaughan, K.L.; Colman, R.J.; et al. Fasting blood glucose as a predictor of mortality: Lost in translation. Cell Metab. 2021, 33, 2189–2200.e3. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Shen, B.; Cui, Y.; Chen, Y.; Wang, J.; Wang, L.; Kang, Y.; Zhao, X.; Si, W.; Li, W.; et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, J.; Qiu, S.; Duan, Y.; Si, W. The Lumenal Microbiota Varies Biogeographically in the Gastrointestinal Tract of Rhesus Macaques. Microbiol. Spectr. 2022, 10, e0034322. [Google Scholar] [CrossRef] [PubMed]
- Lois, C.; Hong, E.J.; Pease, S.; Brown, E.J.; Baltimore, D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 2002, 295, 868–872. [Google Scholar] [CrossRef] [PubMed]
- Michalkiewicz, M.; Michalkiewicz, T.; Geurts, A.M.; Roman, R.J.; Slocum, G.R.; Singer, O.; Weihrauch, D.; Greene, A.S.; Kaldunski, M.; Verma, I.M.; et al. Efficient transgenic rat production by a lentiviral vector. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, H881–H894. [Google Scholar] [CrossRef]
- Hiripi, L.; Negre, D.; Cosset, F.L.; Kvell, K.; Czömpöly, T.; Baranyi, M.; Gócza, E.; Hoffmann, O.; Bender, B.; Bosze, Z. Transgenic rabbit production with simian immunodeficiency virus-derived lentiviral vector. Transgenic Res. 2010, 19, 799–808. [Google Scholar] [CrossRef]
- Sasaki, E.; Suemizu, H.; Shimada, A.; Hanazawa, K.; Oiwa, R.; Kamioka, M.; Tomioka, I.; Sotomaru, Y.; Hirakawa, R.; Eto, T.; et al. Generation of transgenic non-human primates with germline transmission. Nature 2009, 459, 523–527. [Google Scholar] [CrossRef]
- Liu, Z.; Li, X.; Zhang, J.T.; Cai, Y.J.; Cheng, T.L.; Cheng, C.; Wang, Y.; Zhang, C.C.; Nie, Y.H.; Chen, Z.F.; et al. Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature 2016, 530, 98–102. [Google Scholar] [CrossRef]
- Yang, S.H.; Cheng, P.H.; Banta, H.; Piotrowska-Nitsche, K.; Yang, J.J.; Cheng, E.C.; Snyder, B.; Larkin, K.; Liu, J.; Orkin, J.; et al. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature 2008, 453, 921–924. [Google Scholar] [CrossRef]
- Gama Sosa, M.A.; De Gasperi, R.; Elder, G.A. Animal transgenesis: An overview. Brain Struct. Funct. 2010, 214, 91–109. [Google Scholar] [CrossRef]
- Zhou, Y.; Sharma, J.; Ke, Q.; Landman, R.; Yuan, J.; Chen, H.; Hayden, D.S.; Fisher, J.W., 3rd; Jiang, M.; Menegas, W.; et al. Atypical behaviour and connectivity in SHANK3-mutant macaques. Nature 2019, 570, 326–331. [Google Scholar] [CrossRef]
- Martin, N.P.; Myers, P.; Goulding, E.; Chen, S.H.; Walker, M.; Porter, T.M.; Van Gorder, L.; Mathew, A.; Gruzdev, A.; Romeo, C. En masse lentiviral gene delivery to mouse fertilized eggs via laser perforation of zona pellucida. Transgenic Res. 2018, 27, 39–49. [Google Scholar] [CrossRef]
- Byrne, J.A.; Pedersen, D.A.; Clepper, L.L.; Nelson, M.; Sanger, W.G.; Gokhale, S.; Wolf, D.P.; Mitalipov, S.M. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature 2007, 450, 497–502. [Google Scholar] [CrossRef]
- Tachibana, M.; Amato, P.; Sparman, M.; Gutierrez, N.M.; Tippner-Hedges, R.; Ma, H.; Kang, E.; Fulati, A.; Lee, H.S.; Sritanaudomchai, H.; et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 2013, 153, 1228–1238. [Google Scholar] [CrossRef]
- Meng, L.; Ely, J.J.; Stouffer, R.L.; Wolf, D.P. Rhesus monkeys produced by nuclear transfer. Biol. Reprod. 1997, 57, 454–459. [Google Scholar] [CrossRef]
- Chapman, K.M.; Medrano, G.A.; Jaichander, P.; Chaudhary, J.; Waits, A.E.; Nobrega, M.A.; Hotaling, J.M.; Ober, C.; Hamra, F.K. Targeted Germline Modifications in Rats Using CRISPR/Cas9 and Spermatogonial Stem Cells. Cell Rep. 2015, 10, 1828–1835. [Google Scholar] [CrossRef]
- Brinster, R.L. Male germline stem cells: From mice to men. Science 2007, 316, 404–405. [Google Scholar] [CrossRef]
- Li, C.H.; Yan, L.Z.; Ban, W.Z.; Tu, Q.; Wu, Y.; Wang, L.; Bi, R.; Ji, S.; Ma, Y.H.; Nie, W.H.; et al. Long-term propagation of tree shrew spermatogonial stem cells in culture and successful generation of transgenic offspring. Cell Res. 2017, 27, 241–252. [Google Scholar] [CrossRef]
- Wang, S.; Duan, Y.; Yan, Y.; Adar, C.; Braslavsky, I.; Chen, B.; Huang, T.; Qiu, S.; Li, X.; Inglis, B.M.; et al. Improvement of sperm cryo-survival of cynomolgus macaque (Macaca fascicularis) by commercial egg-yolk-free freezing medium with type III antifreeze protein. Anim. Reprod. Sci. 2019, 210, 106177. [Google Scholar] [CrossRef]
- Takehashi, M.; Kanatsu-Shinohara, M.; Inoue, K.; Ogonuki, N.; Miki, H.; Toyokuni, S.; Ogura, A.; Shinohara, T. Adenovirus-mediated gene delivery into mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 2596–2601. [Google Scholar] [CrossRef]
- Guo, Y.; Hai, Y.; Gong, Y.; Li, Z.; He, Z. Characterization, isolation, and culture of mouse and human spermatogonial stem cells. J. Cell. Physiol. 2014, 229, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Kanatsu-Shinohara, M.; Ogonuki, N.; Iwano, T.; Lee, J.; Kazuki, Y.; Inoue, K.; Miki, H.; Takehashi, M.; Toyokuni, S.; Shinkai, Y.; et al. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development 2005, 132, 4155–4163. [Google Scholar] [CrossRef] [PubMed]
- Hamra, F.K.; Chapman, K.M.; Nguyen, D.M.; Williams-Stephens, A.A.; Hammer, R.E.; Garbers, D.L. Self renewal, expansion, and transfection of rat spermatogonial stem cells in culture. Proc. Natl. Acad. Sci. USA 2005, 102, 17430–17435. [Google Scholar] [CrossRef] [PubMed]
- Goodyear, S.; Brinster, R. Culture and Expansion of Primary Undifferentiated Spermatogonial Stem Cells. Cold Spring Harb. Protoc. 2017. [Google Scholar] [CrossRef]
- Sinha, N.; Pilder, S.; Vijayaraghavan, S. Significant expression levels of transgenic PPP1CC2 in testis and sperm are required to overcome the male infertility phenotype of Ppp1cc null mice. PLoS ONE 2012, 7, e47623. [Google Scholar] [CrossRef]
- Antonangeli, F.; Petrungaro, S.; Coluccia, P.; Filippini, A.; Ziparo, E.; Giampietri, C. Testis atrophy and reduced sperm motility in transgenic mice overexpressing c-FLIP(L). Fertil. Steril. 2010, 93, 1407–1414. [Google Scholar] [CrossRef]
- Lipták, N.; Bősze, Z.; Hiripi, L. GFP transgenic animals in biomedical research: A review of potential disadvantages. Physiol. Res. 2019, 68, 525–530. [Google Scholar] [CrossRef]
- Hermann, B.P.; Sukhwani, M.; Winkler, F.; Pascarella, J.N.; Peters, K.A.; Sheng, Y.; Valli, H.; Rodriguez, M.; Ezzelarab, M.; Dargo, G.; et al. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell 2012, 11, 715–726. [Google Scholar] [CrossRef]
- Amann, R.P.; Johnson, L.; Thompson, D.L., Jr.; Pickett, B.W. Daily spermatozoal production, epididymal spermatozoal reserves and transit time of spermatozoa through the epididymis of the rhesus monkey. Biol. Reprod. 1976, 15, 586–592. [Google Scholar] [CrossRef]
- Hermann, B.P.; Sukhwani, M.; Hansel, M.C.; Orwig, K.E. Spermatogonial stem cells in higher primates: Are there differences from those in rodents? Reproduction 2010, 139, 479–493. [Google Scholar] [CrossRef]
- Sankai, T.; Terao, K.; Yanagimachi, R.; Cho, F.; Yoshikawa, Y. Cryopreservation of spermatozoa from cynomolgus monkeys (Macaca fascicularis). J. Reprod. Fertil. 1994, 101, 273–278. [Google Scholar] [CrossRef]
- Chen, B.; Li, S.; Yan, Y.; Duan, Y.; Chang, S.; Wang, H.; Ji, W.; Wu, X.; Si, W. Cryopreservation of cynomolgus macaque (Macaca fascicularis) sperm with glycerol and ethylene glycol, and its effect on sperm-specific ion channels—CatSper and Hv1. Theriogenology 2017, 104, 37–42. [Google Scholar] [CrossRef]
- Gould, K.G.; Mann, D.R. Comparison of electrostimulation methods for semen recovery in the rhesus monkey (Macaca mulatto). J. Med. Primatol. 1988, 17, 95–103. [Google Scholar] [CrossRef]
- Kanatsu-Shinohara, M.; Toyokuni, S.; Shinohara, T. Genetic selection of mouse male germline stem cells in vitro: Offspring from single stem cells. Biol. Reprod. 2005, 72, 236–240. [Google Scholar] [CrossRef]
- Kim, B.J.; Kim, Y.H.; Oh, M.G.; Kim, K.J.; Jung, S.E.; Jin, J.H.; Kim, S.U.; Min, K.S.; Ryu, B.Y. Direct modification of spermatogonial stem cells using lentivirus vectors in vivo leads to efficient generation of transgenic rats. Asian J. Androl. 2019, 21, 190–195. [Google Scholar] [CrossRef]
- Moran, S.P.; Chi, T.; Prucha, M.S.; Agca, Y.; Chan, A.W. Cryotolerance of Sperm from Transgenic Rhesus Macaques (Macaca mulatta). J. Am. Assoc. Lab. Anim. Sci. JAALAS 2016, 55, 520–524. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Duan, Y.; Chen, B.; Qiu, S.; Huang, T.; Si, W. Generation of Transgenic Sperm Expressing GFP by Lentivirus Transduction of Spermatogonial Stem Cells In Vivo in Cynomolgus Monkeys. Vet. Sci. 2023, 10, 104. https://doi.org/10.3390/vetsci10020104
Wang S, Duan Y, Chen B, Qiu S, Huang T, Si W. Generation of Transgenic Sperm Expressing GFP by Lentivirus Transduction of Spermatogonial Stem Cells In Vivo in Cynomolgus Monkeys. Veterinary Sciences. 2023; 10(2):104. https://doi.org/10.3390/vetsci10020104
Chicago/Turabian StyleWang, Shengnan, Yanchao Duan, Bingbing Chen, Shuai Qiu, Tianzhuang Huang, and Wei Si. 2023. "Generation of Transgenic Sperm Expressing GFP by Lentivirus Transduction of Spermatogonial Stem Cells In Vivo in Cynomolgus Monkeys" Veterinary Sciences 10, no. 2: 104. https://doi.org/10.3390/vetsci10020104
APA StyleWang, S., Duan, Y., Chen, B., Qiu, S., Huang, T., & Si, W. (2023). Generation of Transgenic Sperm Expressing GFP by Lentivirus Transduction of Spermatogonial Stem Cells In Vivo in Cynomolgus Monkeys. Veterinary Sciences, 10(2), 104. https://doi.org/10.3390/vetsci10020104





