First Molecular Identification of Calicophoron daubneyi (Dinnik, 1962) and Paramphistomum leydeni (Nasmark, 1937) in Wild Ruminants from Romania
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. PCR Assay
2.3. Sequencing
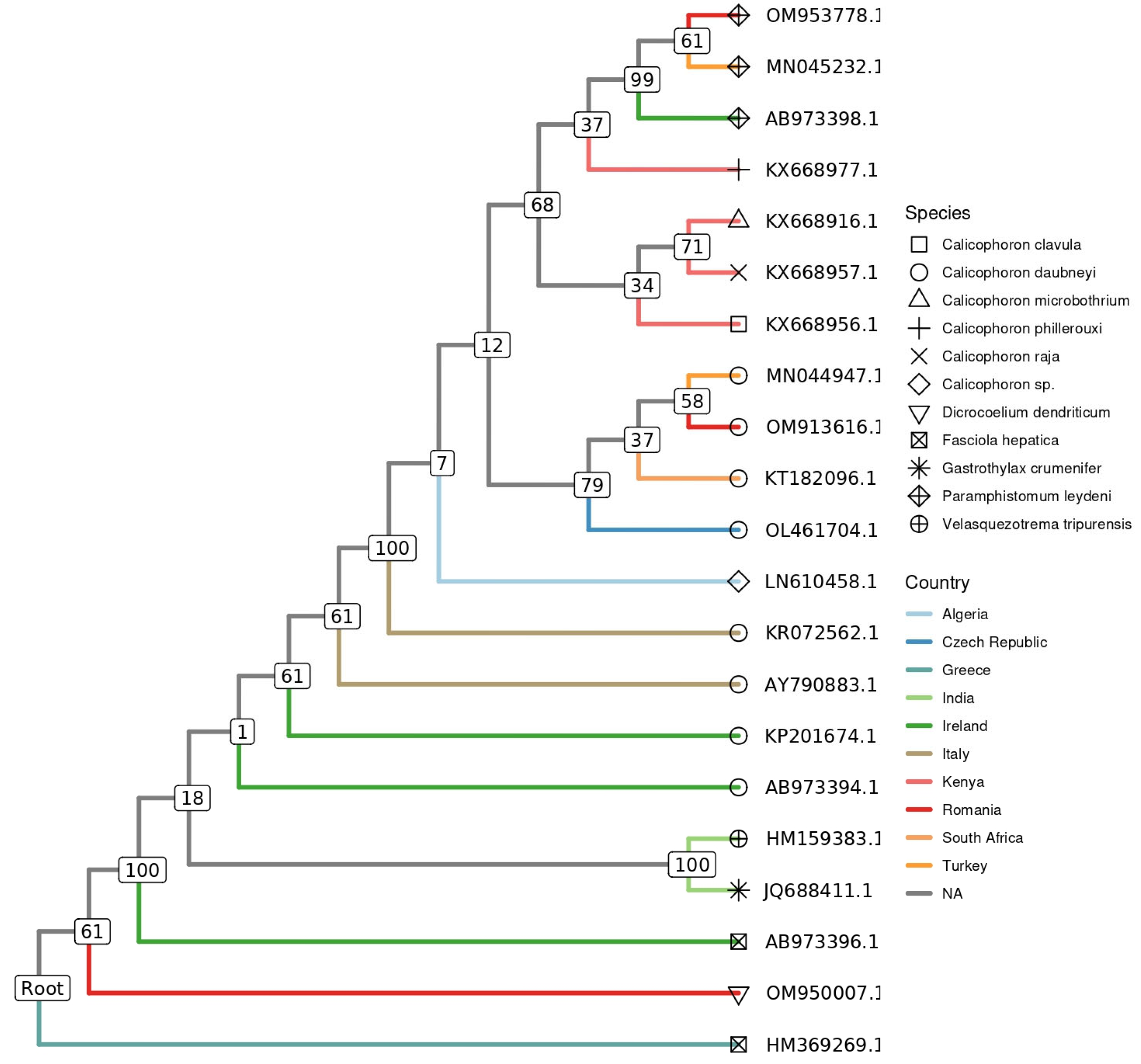
2.4. Phylogenetic Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kahl, A.; von Samson-Himmelstjerna, G.; Krucken, J.; Ganter, M. Chronic wasting due to liver and rumen flukes in sheep. Animals 2021, 11, 549. [Google Scholar] [CrossRef] [PubMed]
- Atcheson, E.; Skuce, P.J.; Oliver, N.A.M.; McNeilly, T.N.; Robinson, M.W. Calicophoron daubneyi—The path toward understanding its pathogenicity and host interactions. Front. Vet. Sci. 2020, 7, 606. [Google Scholar] [CrossRef] [PubMed]
- Deplazes, P.; Eckert, J.; Mathis, A.; Samson-Himmelstjerna, G.V.; Zahner, H. Parasitology in Veterinary Medicine, 1st ed.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2016. [Google Scholar]
- Dinnik, J. Paramphistomum daubneyi sp.nov. from cattle and its snail host in the Kenya Highlands. Parasitology 1962, 52, 143–151. [Google Scholar] [CrossRef]
- Bargues, M.D.; Vigo, M.; Horak, P.; Dvorak, J.; Patzner, R.A.; Pontier, J.P.; Jackiewicz, M.; Meier-Brook, C.; Mas-Coma, S. European Lymnaeidae (Mollusca: Gastropoda), intermediate hosts of trematodiases, based on nuclear ribosomal DNA ITS-2 sequences. Infect. Genet. Evol. 2001, 1, 85–107. [Google Scholar] [CrossRef]
- Rondelaud, D.; Vignoles, P.; Dreyfuss, G. Parasite development and visceral pathology in Galba truncatula co-infected with Fasciola hepatica and Paramphistomum Daubneyi. Parasitoloy 2007, 81, 317–322. [Google Scholar]
- Martinez-Ibeas, A.M.; Munita, M.P.; Lawlor, K.; Sekiya, M.; Mulcahy, G.; Sayers, R. Rumen fluke in Irish sheep: Prevalence, risk factors and molecular identification of two paramphistome species. BMC Vet. Res. 2016, 12, 143. [Google Scholar] [CrossRef]
- Iglesias-Piñeiro, J.; González-Warleta, M.; Castro-Hermida, J.A.; Córdoba, M.; González-lanza, C.; Manga-González, Y.; Mezo, M. Transmission of Calicophoron daubneyi and Fasciola hepatica in Galicia (Spain): Temporal follow-up in the intermediate and definitive hosts. Parasites Vectors 2016, 9, 610. [Google Scholar] [CrossRef]
- Gonzalez-Warleta, M.; Llladosa, S.; Castro-Hermida, J.A.; Martinez-Ibeas, A.M.; Conesa, D.; Munoz, F.; Lopez-quilez, A.; Manga-Gonzalez, Y.; Mezo, M. Bovine paramphistomosis in Galicia (Spain): Prevalence, intensity, aetiology and geospatial distribution of the infection. Vet. Parasitol. 2013, 191, 252–263. [Google Scholar] [CrossRef]
- Mage, C.; Bourgne, H.; Toullieu, J.-M.; Rondelaud, D.; Dreyfuss, G. Fasciola hepatica and Paramphistomum daubneyi: Changes in prevalences of natural infections in cattle and in Lymnaea truncatula from central France over the past 12 years. Vet. Res. 2002, 33, 439–447. [Google Scholar] [CrossRef]
- Malrait, K.; Verschave, S.; Skuce, P.; Van Loo, H.; Vercruysse, J.; Charlier, J. Novel insights into the pathogenic importance, diagnosis and treatment of the rumen fluke (Calicophoron daubneyi) in cattle. Vet. Parasitol. 2015, 207, 134–139. [Google Scholar] [CrossRef]
- Gordon, D.K.; Roberts, L.C.P.; Lean, N.; Zadoks, R.N.; Sargison, N.D.; Skuce, P.J. Identification of the rumen fluke, Calicophoron daubneyi, in GB livestock: Possible implications for liver fluke diagnosis. Vet. Parasitol. 2013, 195, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.A.; Brophy, P.M.; Mitchell, E.S.; Williams, H.W.Y.N. Rumen fluke (Calicophoron daubneyi) on Welsh farms: Prevalence, risk factors and observations on co-infection with Fasciola hepatica. Parasitology 2017, 144, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Ferreras, M.C.; Gonzalez-Lanza, C.; Perez, V.; Fuertes, M.; Benavides, J.; Mezo, M. Calicophoron daubneyi (Paramphistomidae) in slaughtered cattle in Castilla y Leon (Spain). Vet. Parasitol. 2014, 199, 268–271. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, L.; Perugini, A.G.; Capuano, F.; Fenizia, D.; Musella, V.; Veneziano, V.; Cringoli, G. Characterization of the second internal transcribed spacer of ribosomal DNA of Calicophoron daubneyi from various hosts and locations in southern Italy. Vet. Parasitol. 2005, 131, 247–253. [Google Scholar] [CrossRef]
- Toolan, D.P.; Mitchell, G.; Searle, K.; Sheehan, M.; Skuce, P.J.; Zadoks, R.N. Bovine and ovine rumen fluke in Ireland—Prevalence, risk factors and species identity based on passive veterinary surveillance and abattoir findings. Vet. Parasitol. 2015, 212, 168–174. [Google Scholar] [CrossRef]
- Rolfe, P.F.; Boray, J.C.; Nichols, P.; Collins, G.H. Epidemiology of paramphistomosis in cattle. Int. J. Parasitol. 1991, 21, 813–819. [Google Scholar] [CrossRef]
- Ma, J.; He, J.J.; Liu, G.H.; Zhou, D.H.; Liu, J.Z.; Liu, Y. Mitochondrial and nuclear ribosomal DNA dataset supports that Paramphistomum leydeni (Trematoda: Digenea) is a distinct rumen fluke species. Parasites Vectors 2015, 8, 201. [Google Scholar] [CrossRef]
- Sindičić, M.; Martinković, F.; Strišković, T.; Špehar, M.; Štimac, I.; Bujanić, M.; Konjević, D. Molecular identification of the rumen flukes Paramphistomum leydeni and Paramphistomum cervi in a concurrent infection of the red deer (Cervus elaphus). J. Helminthol. 2017, 91, 637–641. [Google Scholar] [CrossRef]
- Kotrlá, B.; Kotrlý, A. The incidence of flukes of the genus Paramphistomum in Czechoslovakia. Vet. Med. 1982, 27, 483–490. [Google Scholar]
- Ploeger, H.W.; Ankum, L.; Moll, L.; van Doorn, D.C.K.; Mitchell, G.; Skuce, P.J. Presence and species identity of rumen flukes in cattle and sheep in the Netherlands. Vet. Parasitol. 2017, 243, 42–46. [Google Scholar] [CrossRef]
- Nikander, S.; Saari, S. Notable seasonal variation observed in the morphology of the reindeer rumen fluke (Paramphistomum leydeni) in Finland. Rangifer 2007, 27, 47–58. [Google Scholar] [CrossRef]
- O’Shaughnessy, J.; Garcia-Campos, A.; McAloon, C.G.; Fagan, S.; de Waal, T.; McElroy, M.; Casey, M.; Good, B.; Mulcahy, G.; Fagan, J.; et al. Epidemiological investigation of a severe rumen fluke outbreak on an Irish dairy farm. Parasitology 2017, 145, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Sey, O. Examination of rumen flukes (Trematoda: Paramphistomata) of cattle in Rumania. Parasit. Hung. 1978, 11, 23–25. [Google Scholar]
- Tudor, G.; Anton, E. Research on paramphistomiasis in ruminants in the Tulcea area. Rev. Zooteh. Med. Vet. 1968, 18, 72–82. [Google Scholar]
- Wenzel, C.; Kuchler, A.; Strube, C.; Knubben-Schweizer, G. Paramphistomidosis—An overview on epidemiology and clinical signs. Tierarztl. Prax. Ausg. G Grosstiere/Nutztiere 2019, 47, 184–191. [Google Scholar]
- Jones, A. Techniques for hand-sectioning thick bodied Platyhelminths. Syst. Parasitol. 1990, 15, 211–218. [Google Scholar] [CrossRef]
- Tandon, V.; Roy, B. SEM Pictorial Guide to Trematodes of Live Stock in India; Regency Publication: New Delhi, India, 2002. [Google Scholar]
- Sharma, S.; Lyngdoh, D.; Roy, B. Differential diagnosis and molecular characterization of Hymenolepis nana and Hymenolepis diminuta (Cestoda: Cyclophyllidea: Hymenolepididae) based on nuclear rDNA ITS2 gene marker. Parasitol. Res. 2016, 5, 4293–4298. [Google Scholar] [CrossRef]
- Lotfy, W.M.; Brant, S.V.; Ashmawy, K.I. A molecular approach for identification of paramphistomes from Africa and Asia. Vet. Parasitol. 2010, 174, 234–240. [Google Scholar] [CrossRef]
- Itagaki, T.; Tsumagari, N.; Tsutsumi, K. Discrimination of three amphistome species by PCR-RFLP based on rDNA ITS2 markers. J. Vet. Med. Sci. 2003, 65, 931–933. [Google Scholar] [CrossRef]
- Arias, M.S.; Sanchis, J.; Franciso, I.; Francisco, R.; Pineiro, P.; Cazapal-Monteiro, C.; Cortinas, F.J.; Suarez, J.L.; Sanchez-Andrade, R.; Paz-Silva, A. The efficacy of four anthelmintics against Calicophoron daubneyi in naturally infected dairy cattle. Vet. Parasitol. 2013, 197, 126–129. [Google Scholar] [CrossRef]
- Imre, K.; Luca, C.; Costache, M.; Sala, C.; Morar, A.; Morariu, S.; Ilie, M.S.; Imre, M.; Dărăbuş, G. Zoonotic Cryptosporidium parvum in Romanian newborn lambs (Ovis aries). Vet. Parasitol. 2013, 191, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Imre, K.; Morariu, S.; Ilie, M.S.; Imre, M.; Ferrari, N.; Genchi, C.; Dărăbuş, G. Serological survey of Neospora caninum infection in cattle herds from western Romania. J. Parasitol. 2012, 98, 683–685. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dărăbuş, G.; Afrenie, M.; Hotea, I.; Imre, M.; Morariu, S. Endoparasites in mammals from seven zoological gardens in Romania. J. Zoo Wildl. Med. 2014, 45, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Imre, K.; Dărăbuș, G.; Tîrziu, E.; Morariu, S.; Imre, M.; Plutzer, J.; Boldea, M.V.; Morar, A. Sarcocystis spp. in Romanian slaughtered cattle: Molecular characterization and epidemiological significance of the findings. Biomed. Res. Int. 2019, 2019, 4123154. [Google Scholar] [CrossRef] [PubMed]
- Dărăbuș, G.; Cosoroabă, I.; Oprescu, I.; Morariu, S. Epidemiology of cryptosporidiosis in animals in the Western Romania. Rev. Med. Vet. 2001, 152, 399–404. [Google Scholar]
- Sîrbu, C.B.; Dărăbuș, G.; Oprescu, I.; Mederle, N.; Ilie, M.S.; Imre, M.; Suici, T.; Luca, I.; Morariu, S. Electron-microscopic investigations on the morphology of the recrudescent Paramphistomum (Calicophoron) daubneyi trematode. Rev. Rom. Med. Vet. 2020, 30, 47–52. [Google Scholar]
- Sîrbu, C.B.; Imre, K.; Dărăbuș, G.; Suici, T.; Mates, B.; Morariu, S. Prevalence of gastrointestinal parasitic infections in cattle and sheep in two regions of Romania. Turk. J. Vet. Anim. Sci. 2020, 44, 581–587. [Google Scholar] [CrossRef]
- Morariu, S.; Bulacu, A.; Morariu, F.; Hagatis, A.; Darabus, G.; Suici, T.; Sirbu, C.; Mederle, N.; Imre, M.; Ilie, M.S. Parasites—An important obstacle in acclimatization and conservation of the European bison (Bison bonasus bonasus) in Romanian Armenis-Plopu reserve. Rev. Rom. Med. Vet. 2021, 31, 54–58. [Google Scholar]
- Hora, F.S.; Genchi, C.; Ferrari, N.; Morariu, S.; Mederle, N.; Dărăbuș, G. Frequency of gastrointestinal and pulmonary helminth infections in wild deer from western Romania. Vet. Parasitol. Reg. Stud. 2018, 8, 75–77. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Schliep, K.P. Phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef]
- Yu, G. Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Hardstaff, J.L.; Marion, G.; Hutchings, M.R.; White, P.C.L. Evaluating the tuberculosis hazard posed to cattle from wildlife across Europe. Res. Vet. Sci. 2013, 97, S86–S93. [Google Scholar] [CrossRef]
- Ruiz-Fons, F.; Sánchez-Matamoros, A.; Gortázar, C.; Sánchez-Vizcaíno, J.M. The role of wildlife in bluetongue virus maintenance in Europe: Lessons learned after the natural infection in Spain. Virus. Res. 2014, S182, 50–58. [Google Scholar] [CrossRef]
- Zintl, A.; Garcia-Campos, A.; Trudgett, A.; Chryssafidis, A.L.; Talavera-Arce, S.; Fu, Y. Bovine paramphistomes in Ireland. Vet. Parasitol. 2014, 204, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Sanabria, R.; More, G.; Romero, J. Molecular characterization of the ITS-2 fragment of Paramphistomum leydeni (Trematoda: Paramphistomidae). Vet. Parasitol. 2011, 177, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Ates, C.; Umur, S. Paramphistome species in water buffaloes and intermediate hosts in the Kizilirmak Delta in Samsun province, Turkey. Acta Parasitol. 2021, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Bazsalovicsová, E.; Králová-Hromadová, I.; Špakulová, M.; Reblánová, M.; Oberhauserová, K. Determination of ribosomal internal transcribed spacer 2 (ITS2) interspecific markers in Fasciola hepatica, Fascioloides magna, Dicrocoelium dendriticum and Paramphistomum cervi (Trematoda), parasites of wild and domestic ruminants. Helminthologia 2010, 47, 76–82. [Google Scholar] [CrossRef]
- Pavlović, I.; Savić, B.; Ivanović, S.; Ćirović, D. First occurrence of Paramphistomum microbothrium (Fischoeder 1901) in roe deer (Capreolus capreolus) in Serbia. J. Wildl. Dis. 2012, 48, 520–522. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huson, K.M.; Oliver, N.A.M.; Robinson, M.W. Paramphistomosis of ruminants: An emerging parasitic disease in Europe. Trends Parasitol. 2017, 33, 836–844. [Google Scholar] [CrossRef]
- Millar, M.; Colloff, A.; Scholes, S. Disease associated with immature paramphistome infection. Vet. Rec. 2012, 171, 509–510. [Google Scholar] [PubMed]
- Brutovska, A.B.; Vogalova, P.; Rost, M.; Sak, B.; Kvac, M. Calicophoron daubneyi (Dinnik, 1962) (Digenea) in beef and dairy cattle in the Czech Republic: Prevalence and drug efficiency. Folia Parasitol. 2023, 70, 1–7. [Google Scholar]
- Forstmaier, T.; Knubben-Schweizer, G.; Strube, C.; Zablotski, Y.; Wenzel, C. Rumen (Calicophoron/Paramphistomum spp.) and liver flukes (Fasciola hepatica) in cattle—Prevalence, distribution, and impact of management factors in Germany. Animals 2021, 11, 2727. [Google Scholar] [CrossRef] [PubMed]
- Sey, O. Revision of the amphistomes of European ruminants. Parasit. Hung. 1980, 13, 13–25. [Google Scholar]
- Capuano, F.; Rinaldi, L.; Maurelli, M.P.; Perugini, A.G.; Musella, V.; Cringoli, G. A comparison of five methods for DNA isolation from liver and rumen flukes to perform ITS-2+ amplification. Parasitologia 2007, 49, 27–31. [Google Scholar]
- Mitchell, G.; Zadoks, R.N.; Skuce, P.J. A universal approach to molecular identification of rumen fluke species across hosts, continents and sample types. Front. Vet. Sci. 2021, 7, 605259. [Google Scholar] [CrossRef]
- Alstedt, U.; Voigt, K.; Jager, M.C.; Knubben-Schweizer, G.; Zablotski, Y.; Strube, C.; Wenzel, C. Rumen and liver fluke infections in sheep and goats in Northern and Southern Germany. Animals 2022, 12, 876. [Google Scholar] [CrossRef]
- O’Toole, A.; Browne, J.A.; Hogan, S.; Bassiere, T.; DeWaal, T.; Mulcahy, G.; Zintl, A. Identity of rumen fluke in deer. Parasitol. Res. 2014, 113, 4097–4103. [Google Scholar] [CrossRef]
- Cauquil, L.; Hue, T.; Hurlin, J.C.; Mitchell, G.; Searle, K.; Skuce, P.; Zadoks, R. Prevalence and sequence-based identity of rumen fluke in cattle and deer in New Caledonia. PLoS ONE 2016, 11, e0152603. [Google Scholar] [CrossRef]
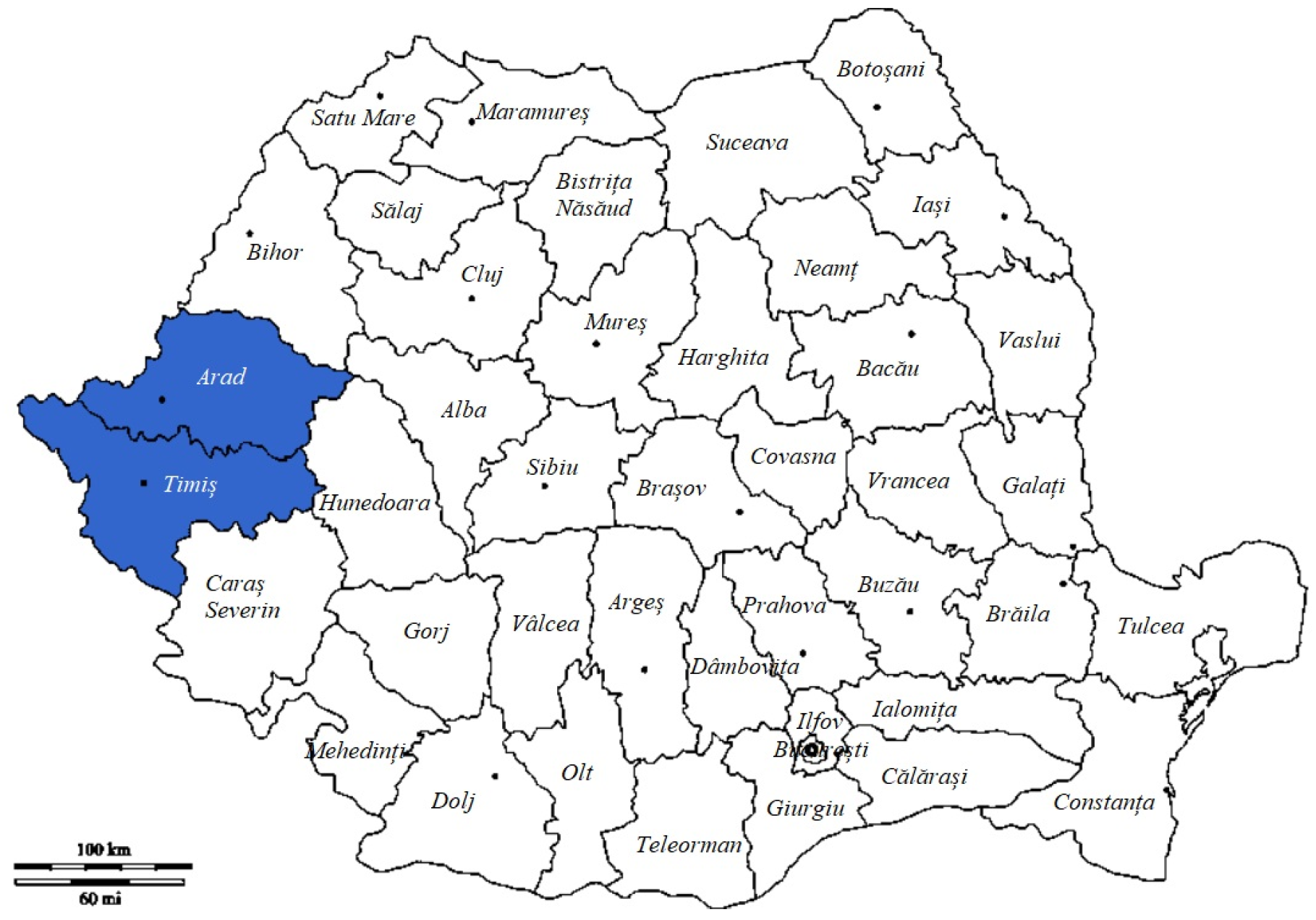


| Crt.No | County | Hunting Ground | Total Samples | Positive Samples (%) | Negative Samples (%) |
|---|---|---|---|---|---|
| 1. | Timiș | Bethausen | 9 | 2 (22.22%) | 7 (77.78%) |
| Valea Lungă | 4 | - | 4 | ||
| Făget | 1 | - | 1 | ||
| Ohaba | 7 | 1 (14.28%) | 6 (85.72%) | ||
| Surduc | 4 | - | 4 | ||
| Traian Vuia | 5 | - | 5 | ||
| Nevrincea | 3 | - | 3 | ||
| 2. | Arad | Săvârșin | 5 | 1 (20%) | 4 (80%) |
| Dumbrava | 3 | - | 3 | ||
| Moneasa | 1 | - | 1 | ||
| Lipova | 3 | - | 3 | ||
| Petriș | 2 | - | 2 | ||
| Șimand | 4 | - | 4 | ||
| Fiac | 1 | - | 1 | ||
| TOTAL | 52 | 4 | 48 | ||
| Sample No. | Sample Identification | Codon | Similar Sequence | Species | |
|---|---|---|---|---|---|
| Start | End | ||||
| 1. | 254 | 10 | 1310 | KJ995531.1 | Paramphistomum leydeni |
| KJ995529.1 | |||||
| AB973398.1 | |||||
| HM209064.1 | |||||
| 2. | 255 | 10 | 1040 | KP201674.1 | Calicophoron daubneyi |
| AY790883.1 | |||||
| 3. | 256 | 10 | 920 | AB973394.1 | Calicophoron daubneyi |
| 4. | 259 | 10 | 850 | KT182100.1 | Calicophoron daubneyi |
| KT182096.1 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morariu, S.; Sîrbu, C.B.; Tóth, A.G.; Dărăbuș, G.; Oprescu, I.; Mederle, N.; Ilie, M.S.; Imre, M.; Sîrbu, B.A.-M.; Solymosi, N.; et al. First Molecular Identification of Calicophoron daubneyi (Dinnik, 1962) and Paramphistomum leydeni (Nasmark, 1937) in Wild Ruminants from Romania. Vet. Sci. 2023, 10, 603. https://doi.org/10.3390/vetsci10100603
Morariu S, Sîrbu CB, Tóth AG, Dărăbuș G, Oprescu I, Mederle N, Ilie MS, Imre M, Sîrbu BA-M, Solymosi N, et al. First Molecular Identification of Calicophoron daubneyi (Dinnik, 1962) and Paramphistomum leydeni (Nasmark, 1937) in Wild Ruminants from Romania. Veterinary Sciences. 2023; 10(10):603. https://doi.org/10.3390/vetsci10100603
Chicago/Turabian StyleMorariu, Sorin, Cătălin Bogdan Sîrbu, Adrienn Gréta Tóth, Gheorghe Dărăbuș, Ion Oprescu, Narcisa Mederle, Marius Stelian Ilie, Mirela Imre, Beatrice Ana-Maria Sîrbu, Norbert Solymosi, and et al. 2023. "First Molecular Identification of Calicophoron daubneyi (Dinnik, 1962) and Paramphistomum leydeni (Nasmark, 1937) in Wild Ruminants from Romania" Veterinary Sciences 10, no. 10: 603. https://doi.org/10.3390/vetsci10100603
APA StyleMorariu, S., Sîrbu, C. B., Tóth, A. G., Dărăbuș, G., Oprescu, I., Mederle, N., Ilie, M. S., Imre, M., Sîrbu, B. A.-M., Solymosi, N., Florea, T., & Imre, K. (2023). First Molecular Identification of Calicophoron daubneyi (Dinnik, 1962) and Paramphistomum leydeni (Nasmark, 1937) in Wild Ruminants from Romania. Veterinary Sciences, 10(10), 603. https://doi.org/10.3390/vetsci10100603











