Abstract
The genome of Bacillus thuringiensis strain INTA 103-23 was sequenced, revealing a high-quality draft assembly comprising 243 contigs with a total size of 6.30 Mb and a completeness of 99%. Phylogenetic analysis classified INTA 103-23 within the Bacillus cereus sensu stricto cluster. Genome annotation identified 6993 genes, including 2476 hypothetical proteins. Screening for pesticidal proteins unveiled 10 coding sequences with significant similarity to known pesticidal proteins, showcasing a potential efficacy against various insect orders. AntiSMASH analysis predicted 13 biosynthetic gene clusters (BGCs), including clusters with 100% similarity to petrobactin and anabaenopeptin NZ857/nostamide A. Notably, fengycin exhibited a 40% similarity within the identified clusters. Further exploration involved a comparative genomic analysis with ten phylogenetically closest genomes. The ANI values, calculated using fastANI, confirmed the closest relationships with strains classified under Bacillus cereus sensu stricto. This comprehensive genomic analysis of B. thuringiensis INTA 103-23 provides valuable insights into its genetic makeup, potential pesticidal activity, and biosynthetic capabilities. The identified BGCs and pesticidal proteins contribute to our understanding of the strain’s biocontrol potential against diverse agricultural pests.
Dataset: The raw genome sequencing data were submitted to the NCBI with BioSample number SAMN39459532, under BioProject PRJNA1065439. The assembled genome is available in the NCBI WGS project, under accession number JAYWUX000000000.
Dataset License: CC-BY license
1. Summary
Bacillus thuringiensis, an adept Gram-positive bacterium with spore-forming capabilities, has established itself as a predominant resident in diverse ecosystems, particularly thriving in soil environments [1]. Renowned for its remarkable protein synthesis machinery, B. thuringiensis synthesizes a diverse array of insecticidal proteins, including Cry, Cyt, Vip, Vpa/Vpb, and Sip, each exhibiting a high specificity against various insect orders [2]. The integration of genes encoding these insecticidal proteins into crops, known as Bt crops, imparts targeted resistance against insect pests [3].
Bacillus thuringiensis proteins exhibit toxicity upon ingestion, primarily targeting the host’s midgut epithelium [4]. In the form of crystalline inclusions, such as the bacterial parasporal crystal containing Cry and Cyt proteins, they are solubilized to release protoxins. These protoxins are activated by midgut proteases and then cross the peritrophic matrix and bind to specific receptors on the midgut epithelial membrane. The oligomerization of B. thuringiensis proteins forms pores, each likely consisting of four toxin monomers assembled into a pore-forming oligomer, observed in both Cry and Vip3 proteins [5,6]. Upon insertion into the epithelial membrane, these pores disrupt it, allowing gut bacteria invasion into the hemolymph, ultimately resulting in septicemia and insect mortality [7].
In addition to its well-established insecticidal prowess, select strains of B. thuringiensis showcase additional advantageous activities, manifesting nematicidal, bactericidal, fungicidal, and plant growth-promoting activities in specific contexts [8,9]. Notably, the environmentally friendly characteristics of B. thuringiensis have fueled their widespread adoption as integral components in global crop pest control initiatives [10].
The utility of these proteins has catalyzed ongoing efforts to explore novel B. thuringiensis isolates, with the goal of identifying and characterizing insecticidal proteins possessing unique specificities. Addressing the challenge of insect genetic resistance, particularly in transgenic crops expressing B. thuringiensis genes, necessitates the exploration of alternative strategies. The advent of revolutionary next-generation sequencing technology has facilitated the sequencing of numerous genomes from B. thuringiensis, aiming to unveil new invertebrate toxic proteins applicable to the biological control of agricultural pests and disease vectors (mosquitoes) [11,12,13].
This paper aims to bridge this knowledge gap by presenting the first sequenced and analyzed genome of the Argentine B. thuringiensis strain INTA 103-23, shedding light on its genetic makeup and potential applications in biocontrol strategies. Through elucidating the genomic underpinnings of this strain, we aim to contribute to the advancement of sustainable pest management practices and the development of environmentally friendly solutions for agricultural and public health challenges.
2. Data Description
2.1. Isolation, Morphological Characterization, and Insecticidal Activity of Bacillus thuringiensis INTA 103-23
Bacillus thuringiensis INTA 103-23, isolated in 2005 from a waterlogged soil sample in Rafaela, Santa Fe, Argentina, exhibits distinctive characteristics when cultivated on a sporulation BM agar medium [14]. After 72 h at 29 °C, the strain forms round, elevated, opaque colonies with a matte texture, displaying a creamy white color (Figure 1A). The vegetative cells are rod-shaped and can form subterminal spores. Upon completion of the sporulation phase, ovoid parasporal crystals are released from the sporangia (Figure 1B).
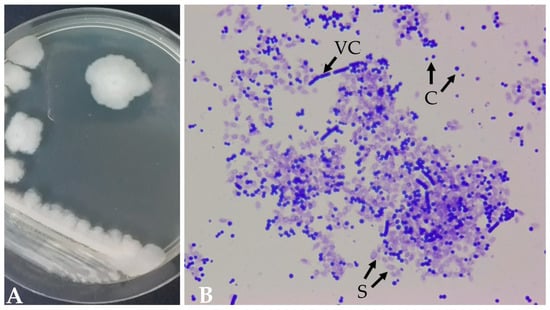
Figure 1.
Morphology of B. thuringiensis INTA 103-23 colonies (A) after 72 h of cultivation on an BM agar plate at 29 °C [14] and (B) a sporulating culture stained with Coomassie brilliant blue (VC, vegetative cells; S, spores; C, crystals) (100× objective).
INTA 103-23 was preliminary classified as B. thuringiensis based on its ability to produce parasporal crystals with insecticidal activity against second instar ladybird beetle (Epilachna paenulata, Coleoptera: Coccinellidae) [15] and neonate cotton boll weevil (Anthonomus grandis, Coleoptera: Curculionidae) larvae [14]. Furthermore, INTA 103-23 is characterized by the absence of thuringiensin (β-exotoxin) production [14].
2.2. Genome Assembly and Annotation
The genome of the INTA 103-23 strain was sequenced using the Illumina HiSeq X platform, generating high-quality paired-end reads (150 bp). Quality control with the FastQC v0.12.1 [16] program confirmed this, showing uniform quality scores, GC content, and acceptable N content across all bases for both raw and adapter-trimmed DNA libraries. SPAdes v3.15.4 [17] assembled the genome into 243 contigs with a total size of 6.30 Mb and an N50 of 0.92 Mb. Notably, it achieved 99% completeness and only 1% contamination, as assessed by CheckM v1.0.18 [18]. Additional details about the assembly are provided in Table 1.

Table 1.
Key characteristics of the draft genome assembly of INTA 103-23 obtained using QUAST v4.4 [19] and CheckM v1.0.18 [18].
When employing the BUSCO v5.5.0 program [20], our analysis revealed that the quantity of fully assembled single-copy orthologs reached a minimum of 99.8%, compared to the Bacillales_odb10 dataset (Table 2). These findings unequivocally attest to the high quality and comprehensiveness of the assembled genome.

Table 2.
Assessment of BUSCO v5.5.0 marker [20] presence in protein-coding genes within the assembly. The table displays the count of orthologs identified in the assembly alongside their corresponding percentages.
A phylogenetic investigation performed with the Type Strain Genome Server (TYGS) [21] revealed that INTA 103-23 formed a distinct cluster alongside strain types B. thuringiensis ATCC 10792 and B. cereus ATCC 14579 (Figure 2). In accordance with the recently suggested classification, this cluster, referred to as Bacillus cereus sensu stricto, includes the majority of biovar Thuringiensis strains [22].
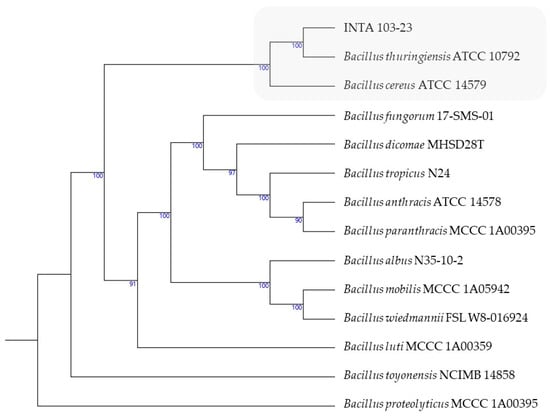
Figure 2.
GBDP phylogeny tree based on whole-genome data using the TYGS (average branch support of 98.0%). The utilization of the gray color serves to emphasize the clustering of INTA 103-23 with the type strains.
Subsequently, we selected ten bacterial genome assemblies from the Bacillaceae family available in the NCBI RefSeq database [23], focusing on those with the closest phylogenetic relationships. This selection process relied on the calculation of average nucleotide identity (ANI) values using the fastANI v0.1.3 tool [24], as detailed in Table 3. The strains displaying the highest degree of similarity in nucleotide sequences among all the genomes were identified as members of Bacillus cereus sensu stricto, thus affirming the accurate taxonomic classification of the INTA 103-23 strain (Table 3).

Table 3.
Phylogenetically closest assemblies to the genome of INTA 103-23: ANI values calculated with the FastANI v0.1.3 software [24] and taxonomic assignment through the TYGS [21].
The annotation using RASTtk—v1.073 revealed that the genome of INTA 103-23 contains 6993 genes, with 6736 being coding sequences and 2476 of them marked as hypothetical proteins (Supplementary Data S1). The draft genome sequence of INTA 103-23 harbors potential pesticidal proteins and virulence factors associated with insect pathogenesis. Eleven coding sequences (CDS) showed significant BlastX [25] similarity to known pesticidal proteins such as Mpp46, Spp1, Xpp22, Cry73, Cry54, Cry4, Tpp49, and Tpp36, which have demonstrated efficacy against a broad spectrum of insects, spanning five different orders (Table 4). Additionally, INTA 103-23 contains other coding sequences encoding ten putative chitinases and six chitin-binding proteins. As expected, genes associated with thuringiensin synthesis, such as thuE, were absent [26]. The demonstrated insecticidal efficacy of INTA 103-23 against E. paenulata [15] and A. grandis [16] may be attributed to coleoptericidal proteins encoded by genes identified within its genome (Table 4). Furthermore, the presence of genes encoding pesticidal toxins known to target mosquito and lepidopteran larvae in the genome of INTA 103-23 suggests a potential, broader insecticidal activity (Table 4). However, further validation through bioassays is warranted to confirm and characterize this association.

Table 4.
Pesticidal proteins repertoire identified in the INTA103-23 genome. Experimentally derived data from the Bacterial Pesticidal Protein Resource Center (BPPRC) specificity database for the target species [27].
The application of the antiSMASH v6.1.1 tool [28] identified a total of 13 biosynthetic gene clusters (BGCs), as outlined in Table 5. Notably, among the identified BGCs, the ones bearing the highest similarity were associated with well-known entities such as petrobactin and anabaenopeptin NZ857/nostamide A, exhibiting 100% similarity. Additionally, it is noteworthy that fengycin showed a 40% similarity within the identified clusters.

Table 5.
Predicted biosynthetic gene clusters in the INTA 103-23 genome with the antiSMASH v6.1.1 [28] program.
The present study unveils the genetic makeup and potential applications of the Argentine B. thuringiensis INTA 103-23 strain. By leveraging next-generation sequencing technology, we sequenced and analyzed the genome of INTA 103-23, shedding light on its genomic characteristics and insecticidal potential. Our findings contribute to the broader understanding of B. thuringiensis and its role in pest management strategies.
In conclusion, the sequencing and analysis of the INTA 103-23 genome offers valuable insights into its genetic composition and potential applications. The presence of genes encoding pesticidal proteins and other virulence factors underscores its potential as a biocontrol agent against agricultural pests and disease vectors. Moreover, the absence of genes associated with thuringiensin synthesis suggests alternative mechanisms for insecticidal activity. The elucidation of the genomic underpinnings of INTA 103-23 paves the way for the further exploration and utilization of this strain in sustainable pest management practices. This research contributes to the advancement of knowledge regarding B. thuringiensis and its applications in agriculture and public health.
3. Methods
3.1. DNA Extraction, Library Construction, and Massive Genome Sequencing
DNA, comprising both chromosome and plasmids, was extracted utilizing the Wizard genomic DNA purification kit (Promega, Madison, WI, USA). Subsequently, the extracted DNA underwent electrophoresis, was stained with SYBR Safe, and quantified through a Multiskan SkyHigh µDrop Plate spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA). A comprehensive Illumina library was then constructed using the purified DNA and subjected to high-throughput Illumina sequencing at the Unidad Operativa Centro Nacional de Genómica y Bioinformática (ANLIS Malbrán, Santiago del Estero, Argentina) in the paired-end mode, producing reads with a length of 2 × 150 bp. Finally, the short nucleotide reads underwent quality control using FastQC v0.12.1 [16].
3.2. Genome Assembly, Sequence Analysis, and Annotation
Genome assembly was performed de novo using the SPAdes v3.15.4 genome assembler [13], and the resulting assembly underwent a rigorous quality control assessment with CheckM v1.0.18 [18] and QUAST v4.4 [19]. To evaluate the taxonomy-wise completeness, the percentage of one-copy orthologs from the “Bacillales_odb10” database was determined using BUSCO v5.5.0 [20]. Species delimitation was performed using the TYGS [21].
In the subsequent phase, we employed fastANI v1.33 [24] to identify the ten phylogenetically closest genomes downloaded from the NCBI RefSeq database [23], selecting those with the highest ANI values when compared to our assembly. Initial genome annotation was carried out using the NCBI Prokaryotic Genome Annotation Pipeline (2023 release) and RASTtk—v1.073 [29].
Following annotation, a targeted BLAST analysis was conducted to identify genes encoding pesticidal proteins, utilizing a customized non-redundant insecticidal protein database. The putative susceptible insect species affected by the identified virulence factors were obtained from the Bacterial Pesticidal Protein Resource Center (BPPRC) specificity database [27]. The prediction and characterization of biosynthetic gene clusters were accomplished using antiSMASH v6.1.1 [29].
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/data9030040/s1: Supplementary Data S1: The annotation results in the GBK format made using RASTtk—v1.073.
Author Contributions
Conceptualization, L.P. and D.S.; software, L.P. and D.S.; methodology, L.P., L.O., J.N. and D.S.; validation, L.P., M.B. and D.S.; formal analysis, L.P. and D.S.; investigation, L.P., M.B. and D.S.; resources, D.S.; data curation, L.P. and D.S.; writing—original draft preparation, D.S.; writing—review and editing, L.P., L.O., J.N., M.B. and D.S.; visualization, D.S.; supervision, L.P. and D.S.; project administration, D.S.; funding acquisition, D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by INTA, with grants number 2023-PD-L01-I087 and 2023-PE-L04-I073.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw genome sequencing data were submitted to the NCBI with BioSample number SAMN39459532, under BioProject PRJNA1065439. The assembled genome is available in the NCBI WGS project, under JAYWUX000000000.
Acknowledgments
Leopoldo Palma would like to express gratitude to the Spanish Government-Universities Ministry and the NextGenerationEU and Recovery, Transformation, and Resilience plans for funding his awarded María Zambrano contract.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Martin, P.A.; Travers, R.S. Worldwide Abundance and Distribution of Bacillus thuringiensis Isolates. Appl. Environ. Microbiol. 1989, 55, 2437–2442. [Google Scholar] [CrossRef] [PubMed]
- Crickmore, N.; Berry, C.; Panneerselvam, S.; Mishra, R.; Connor, T.R.; Bonning, B.C. A structure-based nomenclature for Bacillus thuringiensis and other bacteria-derived pesticidal proteins. J. Invertebr. Pathol. 2021, 186, 107438. [Google Scholar] [CrossRef] [PubMed]
- Arthur, B.P.; Suh, C.P.; McKnight, B.M.; Parajulee, M.N.; Yang, F.; Kerns, D.L. Field Evaluation of Cotton Expressing Mpp51Aa2 as a Management Tool for Cotton Fleahoppers, Pseudatomoscelis seriatus (Reuter). Toxins 2023, 15, 644. [Google Scholar] [CrossRef] [PubMed]
- Jurat-Fuentes, J.L.; Crickmore, N. Specificity determinants for Cry insecticidal proteins: Insights from their mode of action. J. Invertebr. Pathol. 2017, 142, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Gómez, I.; Sánchez, J.; Miranda, R.; Bravo, A.; Soberón, M. Cadherin-like receptor binding facilitates proteolytic cleavage of helix α-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett. 2002, 513, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Núñez-Ramírez, R.; Huesa, J.; Bel, Y.; Ferré, J.; Casino, P.; Arias-Palomo, E. Molecular architecture and activation of the insecticidal protein Vip3Aa from Bacillus thuringiensis. Nat. Commun. 2020, 11, 3974. [Google Scholar] [CrossRef] [PubMed]
- Caccia, S.; Di Lelio, I.; La Storia, A.; Marinelli, A.; Varricchio, P.; Franzetti, E.; Banyuls, N.; Tettamanti, G.; Casartelli, M.; Giordana, B.; et al. Midgut microbiota and host immunocompetence underlie Bacillus thuringiensis killing mechanism. Proc. Natl. Acad. Sci. USA 2016, 113, 9486–9491. [Google Scholar] [CrossRef] [PubMed]
- Unzue, A.; Caballero, C.J.; Villanueva, M.; Fernández, A.B.; Caballero, P. Multifunctional Properties of a Bacillus thuringiensis Strain (BST-122): Beyond the Parasporal Crystal. Toxins 2022, 14, 768. [Google Scholar] [CrossRef]
- Sauka, D.H.; Piccinetti, C.F.; Vallejo, D.A.; Onco, M.I.; Pérez, M.P.; Benintende, G.B. New entomopathogenic strain of Bacillus thuringiensis is able to solubilize different sources of inorganic phosphates. Appl. Soil Ecol. 2021, 160, 103839. [Google Scholar] [CrossRef]
- Lacey, L.A.; Grzywacz, D.; Shapiro-Ilan, D.I.; Frutos, R.; Brownbridge, M.; Goettel, M.S. Insect Pathogens as Biological Control Agents: Back to the Future. J. Invertebr. Pathol. 2015, 132, 1–41. [Google Scholar] [CrossRef]
- He, J.; Wang, J.; Yin, W.; Shao, X.; Zheng, H.; Li, M.; Zhao, Y.; Sun, M.; Wang, S.; Yu, Z. Complete genome sequence of Bacillus thuringiensis subsp. chinensis strain CT-43. J. Bacteriol. 2011, 193, 3407–3408. [Google Scholar] [PubMed]
- Liu, J.; Li, L.; Peters, B.M.; Li, B.; Chen, D.; Xu, Z.; Shirtliff, M.E. Complete genome sequence and bioinformatics analyses of Bacillus thuringiensis strain BM-BT15426. Microb. Pathog. 2017, 108, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Fayad, N.; Barssoum, R.; Marsaud, N.; Nasseredine, R.; Abdelmalek, N.; Rouis, S.; Teste, M.A.; Pailler, V.; Gautier, V.; Belmonte, E.; et al. Complete genome sequences of two Bacillus thuringiensis serovar kurstaki strains isolated from Lebanon and Tunisia, highly toxic against lepidopteran larvae. Microbiol. Resour. Announc. 2023, 12, e0006023. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.P.; Sauka, D.H.; Onco, M.I.; Berretta, M.F.; Benintende, G.B. Selection of Bacillus thuringiensis strains toxic to cotton boll weevil (Anthonomus grandis, Coleoptera: Curculionidae) larvae. Rev. Argent. Microbiol. 2017, 49, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Sauka, D.H.; Monella, R.H.; Benintende, G.B. Induced-feeding bioassays for detection of Bacillus thuringiensis insecticidal activity against Epilachna paenulata (Coleoptera). Pesqui. Agropecuária Bras. 2010, 45, 430–432. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 January 2024).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinfomatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Carroll, L.M.; Wiedmann, M.; Kovac, J. Proposal of a Taxonomic Nomenclature for the Bacillus cereus Group Which Reconciles Genomic Definitions of Bacterial Species with Clinical and Industrial Phenotypes. mBio 2020, 11, 1–15. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ruan, L.; Peng, D.; Li, L.; Sun, M.; Yu, Z. Thuringiensin: A thermostable secondary metabolite from Bacillus thuringiensis with insecticidal activity against a wide range of insects. Toxins 2014, 6, 2229–2238. [Google Scholar] [CrossRef] [PubMed]
- Crickmore, N.; Berry, C.; Panneerselvam, S.; Mishra, R.; Connor, T.R.; Bonning, B.C. Bacterial Pesticidal Protein Resource Center. Data. Available online: https://www.bpprc.org (accessed on 10 January 2024).
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).