Data Employed in the Construction of a Composite Protein Database for Proteogenomic Analyses of Cephalopods Salivary Apparatus
Abstract
1. Summary
2. Data Description
2.1. Dataset Description
2.2. Tables
3. Materials and Methods
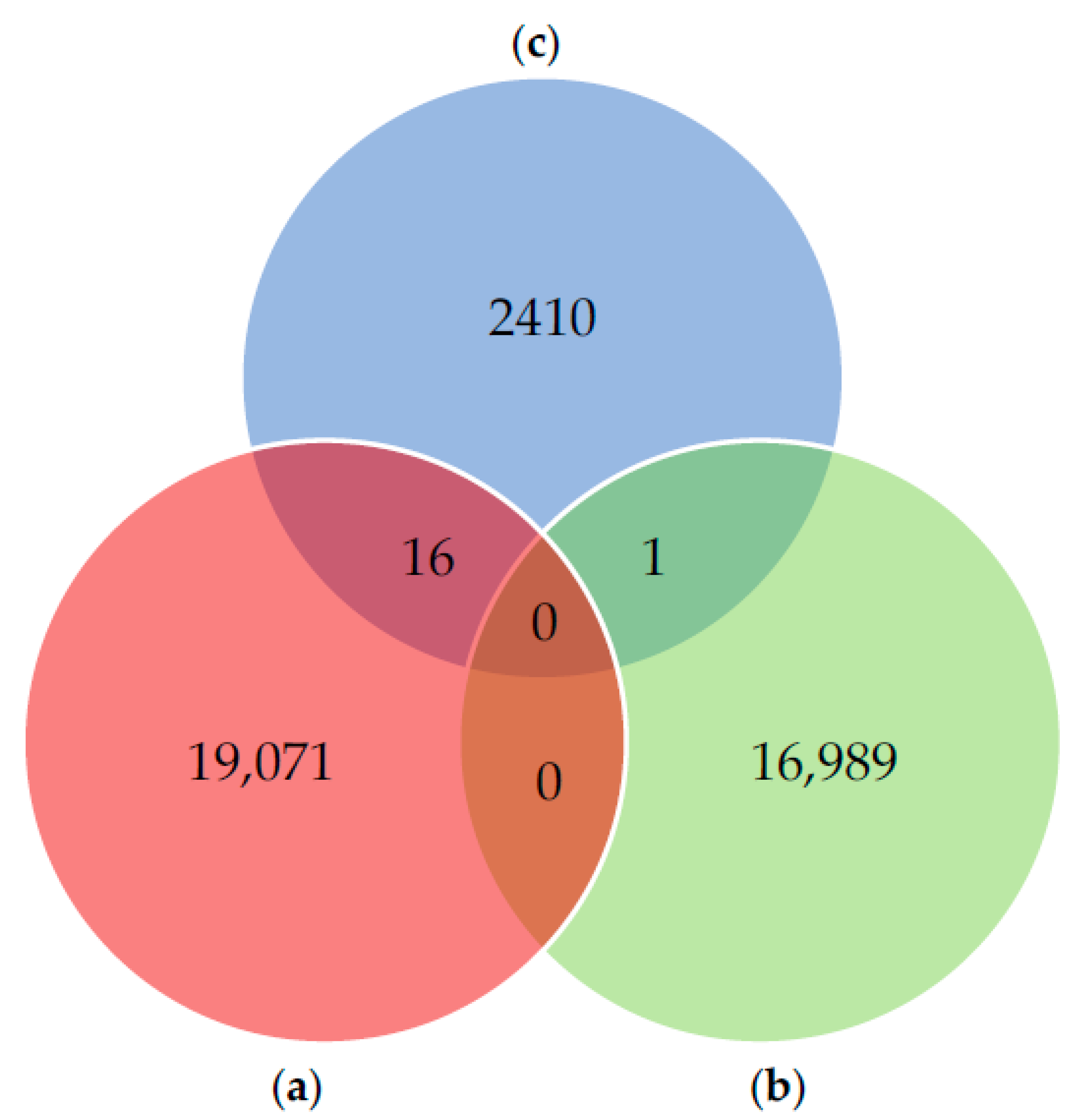
3.1. Database Construction for Proteogenomic Analyses
3.1.1. Database A: Protein Sequences from Fingerhut et al. (2018)
3.1.2. Database B: Antimicrobial Peptides (AMPs)
3.1.3. Database C: Proteins Identified with Proteome Discoverer
- STEP 1: Sample preparation and LC–MS/MS analysis
- STEP 2: Protein identification using Proteome Discoverer
3.1.4. Databases D and E: Proteins Identified from the de novo Transcriptome Assemblies of Cephalopods’ PSGs
- STEP 1: Search and de novo assembly of cephalopods’ PSGs transcriptomes
- STEP 2: Database D—proteins identified by TransDecoder
- STEP 3: Database E—proteins identified by the six-frame translation tool
3.1.5. Database F: O. vulgaris Proteins Identified by the Six-Frame Translation Tool
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Almeida, D.; Domínguez-Pérez, D.; Matos, A.; Agüero-Chapin, G.; Osório, H.; Vasconcelos, V.; Campos, A.; Antunes, A. Putative antimicrobial peptides of the posterior salivary glands from the cephalopod Octopus vulgaris revealed by exploring a composite protein database. Antibiotics 2020, 9, 757. [Google Scholar] [CrossRef] [PubMed]
- Fingerhut, L.C.H.W.; Strugnell, J.M.; Faou, P.; Labiaga, Á.R.; Zhang, J.; Cooke, I.R. Shotgun Proteomics Analysis of Saliva and Salivary Gland Tissue from the Common Octopus Octopus vulgaris. J. Proteome Res. 2018, 17, 3866–3876. [Google Scholar] [CrossRef] [PubMed]
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; Tellez-Ibarra, R.; Llorente-Quesada, M.T.; Salgado, J.; Barigye, S.J.; Liu, J. Overlap and diversity in antimicrobial peptide databases: Compiling a non-redundant set of sequences. Bioinformatics 2015, 31, 2553–2559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Proteomics Toolkit (Protk). Available online: https://github.com/iracooke/protk (accessed on 14 April 2019).
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Sequence Read Archive of National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/sra/?term=Cephalopoda (accessed on 26 October 2018).
- Sequence Set Browser from National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/Traces/wgs/?page=1&view=TSA&search=Cephalopoda (accessed on 26 October 2018).
- Ruder, T.; Sunagar, K.; Undheim, E.A.B.; Ali, S.A.; Wai, T.-C.; Low, D.H.W.; Jackson, T.N.W.; King, G.F.; Antunes, A.; Fry, B.G. Molecular Phylogeny and Evolution of the Proteins Encoded by Coleoid (Cuttlefish, Octopus, and Squid) Posterior Venom Glands. J. Mol. Evol. 2013, 76, 192–204. [Google Scholar] [CrossRef] [PubMed]
- European Nucleotide Archive. Available online: https://www.ebi.ac.uk/ena (accessed on 16 November 2018).
- CLC Genomics Workbench 11.0.1. Available online: https://www.qiagenbioinformatics.com/ (accessed on 16 November 2018).
- Geneious. Available online: https://www.geneious.com (accessed on 16 November 2018).
- DB Browser for SQLite. Available online: https://sqlitebrowser.org/ (accessed on 16 November 2018).
| Dataset Name | File Name | File Type | DOI |
|---|---|---|---|
| Dataset_1 | All_Databases_5950827_sequences | FASTA | 10.17632/df8w8dct3b.1 |
| Database_A_19087_sequences | FASTA | ||
| Database_B_16990_sequences | FASTA | ||
| Database_C_2427_sequences | FASTA | ||
| Database_D_84778_sequences | FASTA | ||
| Database_E_5106635_sequences | FASTA | ||
| Database_F_720910_sequences | FASTA | ||
| Dataset_2 | DA_summary_Proteome_Discoverer_ISD | XLSX | 10.17632/hrydnjz937.1 |
| DA_summary_Proteome_Discoverer_FASP | XLSX | ||
| Dataset_3 | 272704_contigs_from_16_cephalopods_PSGs_transcriptome_assemblies | FASTA | 10.17632/fjnnjv6nnn.1 |
| SRR680047_assembly | FASTA | ||
| SRR684167_assembly | FASTA | ||
| SRR684223_assembly | FASTA | ||
| SRR725597_assembly | FASTA | ||
| SRR725779_assembly | FASTA | ||
| SRR725780_assembly | FASTA | ||
| SRR725935_assembly | FASTA | ||
| SRR725936_assembly | FASTA | ||
| SRR725937_assembly | FASTA | ||
| SRR725938_assembly | FASTA | ||
| SRR2047107_assembly | FASTA | ||
| SRR3105321_assembly | FASTA | ||
| SRR3105558_assembly | FASTA | ||
| SRR5204441_assembly | FASTA | ||
| SRR5204442_assembly | FASTA | ||
| SRR6349992_assembly | FASTA | ||
| Table_S1 | XLSX | ||
| Dataset_4 | SRR680047_assembly.fasta.transdecoder.pep | FASTA | 10.17632/h94v3bk4j6.1 |
| SRR684167_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR684223_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR725597_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR725779_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR725780_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR725935_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR725936_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR725937_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR725938_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR2047107_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR3105321_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR3105558_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR5204441_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR5204442_assembly.fasta.transdecoder.pep | FASTA | ||
| SRR6349992_assembly.fasta.transdecoder.pep | FASTA | ||
| Dataset_5 | cases | CSV | 10.17632/p6vnj6ssrf.1 |
| transcripts | CSV | ||
| DB | DB | ||
| SQL_command | TXT | ||
| 187926_contigs_not_included_in_Database_D | CSV | ||
| 187926_contigs_not_included_in_Database_D | FASTA | ||
| a sixframe.rb | RB | ||
| six-frame_translation_of_187926_contigs_not_included_in_Database_D | FASTA | ||
| Dataset_6 | cases | CSV | 10.17632/x73ff3n744.1 |
| transcripts | CSV | ||
| DB1 | DB | ||
| SQL_command1 | TXT | ||
| 31661_contigs_not_included_in_Database_A | CSV | ||
| 31661_contigs_not_included_in_Database_A | FASTA | ||
| a sixframe.rb | RB | ||
| six-frame_translation_of_31661_contigs_not_included_in_Database_A | FASTA |
| Instrument Platform (Library Layout) | Species | CLC Genomics Workbench de novo Assembly a | TransDecoder Analysis a,b | Six-Frame Translation Tool Analysis a,c | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SRA Run Accession d | Number of Reads | Matched e | Contig Count | Contig Average Length | Reads Mapped in Pairs f | Reads Mapped in Broken Pairs g | N50 h | N75 i | # of Contigs Analyzed j | # of Proteins Identified k | # of Contigs Analyzed l | # of ORFs Identified m | ||
| Illumina (paired) | Sepia officinalis (female) | SRR5204441 | 34,623,104 | 31,510,916 | 47,489 | 686 | 23,187,508 | 8,323,408 | 1005 | 425 | 47,489 | 14,583 | 32,906 | 870,077 |
| Sepia officinalis (male) | SRR5204442 | 21,428,980 | 18,038,146 | 40,778 | 675 | 14,141,858 | 3,896,288 | 929 | 426 | 40,778 | 14,056 | 26,722 | 691,205 | |
| Callistoctopus minor | SRR6349992 | 69,681,384 | 52,377,156 | 58,327 | 703 | 39,695,532 | 12,681,624 | 1072 | 440 | 58,327 | 15,365 | 42,962 | 1,164,790 | |
| Hapalochlaena maculosa | SRR3105558 | 16,128,360 | 13,948,566 | 36,755 | 636 | 12,399,458 | 1,549,108 | 832 | 410 | 36,755 | 13,695 | 23,060 | 580,147 | |
| Octopus kaurna | SRR3105321 | 46,268,294 | 40,764,402 | 33,936 | 584 | 37,224,454 | 3,539,948 | 718 | 379 | 33,936 | 10,965 | 22,971 | 572,048 | |
| Octopus bimaculoides | SRR2047107 | 71,186,024 | 65,629,243 | 50,286 | 875 | 58,627,142 | 7,002,101 | 1606 | 582 | 50,286 | 14,267 | 36,019 | 1,145,961 | |
| LS454 (single) | Abdopus aculeatus | SRR680047 | 33,464 | 21,627 | 774 | 526 | N.A. | N.A. | 529 | 411 | 774 | 331 | 443 | 11,133 |
| Hapalochlaena maculosa | SRR725938 | 55,955 | 49,003 | 528 | 475 | N.A. | N.A. | 494 | 378 | 528 | 154 | 374 | 9310 | |
| Loliolus noctiluca | SRR725597 | 72,031 | 67,299 | 200 | 552 | N.A. | N.A. | 545 | 436 | 200 | 93 | 107 | 2724 | |
| Octopus cyanea | SRR725937 | 55,039 | 40,899 | 964 | 503 | N.A. | N.A. | 521 | 396 | 964 | 352 | 612 | 15,328 | |
| Pareledone turqueti | SRR725936 | 64,419 | 60,295 | 231 | 500 | N.A. | N.A. | 522 | 404 | 231 | 101 | 130 | 3024 | |
| Octopus kaurna | SRR684223 | 61,953 | 55,831 | 491 | 497 | N.A. | N.A. | 497 | 394 | 491 | 164 | 327 | 7985 | |
| Sepia latimanus | SRR725779 | 49,960 | 42,657 | 434 | 461 | N.A. | N.A. | 459 | 361 | 434 | 83 | 351 | 8693 | |
| Adelieledone polymorpha | SRR684167 | 71,506 | 69,025 | 116 | 528 | N.A. | N.A. | 474 | 397 | 116 | 37 | 79 | 1847 | |
| Sepia pharaonis | SRR725935 | 45,677 | 36,088 | 492 | 489 | N.A. | N.A. | 480 | 395 | 492 | 166 | 326 | 7756 | |
| Sepioteuthis australis | SRR725780 | 68,851 | 60,037 | 903 | 562 | N.A. | N.A. | 563 | 448 | 903 | 366 | 537 | 14,607 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, D.; Domínguez-Pérez, D.; Matos, A.; Agüero-Chapin, G.; Castaño, Y.; Vasconcelos, V.; Campos, A.; Antunes, A. Data Employed in the Construction of a Composite Protein Database for Proteogenomic Analyses of Cephalopods Salivary Apparatus. Data 2020, 5, 110. https://doi.org/10.3390/data5040110
Almeida D, Domínguez-Pérez D, Matos A, Agüero-Chapin G, Castaño Y, Vasconcelos V, Campos A, Antunes A. Data Employed in the Construction of a Composite Protein Database for Proteogenomic Analyses of Cephalopods Salivary Apparatus. Data. 2020; 5(4):110. https://doi.org/10.3390/data5040110
Chicago/Turabian StyleAlmeida, Daniela, Dany Domínguez-Pérez, Ana Matos, Guillermin Agüero-Chapin, Yuselis Castaño, Vitor Vasconcelos, Alexandre Campos, and Agostinho Antunes. 2020. "Data Employed in the Construction of a Composite Protein Database for Proteogenomic Analyses of Cephalopods Salivary Apparatus" Data 5, no. 4: 110. https://doi.org/10.3390/data5040110
APA StyleAlmeida, D., Domínguez-Pérez, D., Matos, A., Agüero-Chapin, G., Castaño, Y., Vasconcelos, V., Campos, A., & Antunes, A. (2020). Data Employed in the Construction of a Composite Protein Database for Proteogenomic Analyses of Cephalopods Salivary Apparatus. Data, 5(4), 110. https://doi.org/10.3390/data5040110











