Abstract
Here we provide all datasets and details applied in the construction of a composite protein database required for the proteogenomic analyses of the article “Putative Antimicrobial Peptides of the Posterior Salivary Glands from the Cephalopod Octopus vulgaris Revealed by Exploring a Composite Protein Database”. All data, subdivided into six datasets, are deposited at the Mendeley Data repository as follows. Dataset_1 provides our composite database “All_Databases_5950827_sequences.fasta” derived from six smaller databases composed of (i) protein sequences retrieved from public databases related to cephalopods’ salivary glands, (ii) proteins identified with Proteome Discoverer software using our original data obtained by shotgun proteomic analyses of posterior salivary glands (PSGs) from three Octopus vulgaris specimens (provided as Dataset_2) and (iii) a non-redundant antimicrobial peptide (AMP) database. Dataset_3 includes the transcripts obtained by de novo assembly of 16 transcriptomes from cephalopods’ PSGs using CLC Genomics Workbench. Dataset_4 provides the proteins predicted by the TransDecoder tool from the de novo assembly of 16 transcriptomes of cephalopods’ PSGs. Further details about database construction, as well as the scripts and command lines used to construct them, are deposited within Dataset_5 and Dataset_6. The data provided in this article will assist in unravelling the role of cephalopods’ PSGs in feeding strategies, toxins and AMP production.
Dataset (DOI): 10.17632/df8w8dct3b.1 (Dataset_1); 10.17632/hrydnjz937.1 (Dataset_2); 10.17632/fjnnjv6nnn.1 (Dataset_3); 10.17632/h94v3bk4j6.1 (Dataset_4); 10.17632/p6vnj6ssrf.1 (Dataset_5); 10.17632/x73ff3n744.1 (Dataset_6).
Dataset License: CC BY 4.0.
1. Summary
In this work, we provide all datasets and detailed description about the construction of a composite protein database used for proteogenomic analyses in the article “Putative Antimicrobial Peptides of the Posterior Salivary Glands from the Cephalopod Octopus vulgaris Revealed by Exploring a Composite Protein Database”. This protein database compiles information of cephalopods’ salivary apparatus and provides a composite protein database in a unique FASTA file, which can be used by researchers as reference to discover new proteins in the salivary apparatus of cephalopods or for comparison purposes. The composite database, All_Databases_5950827_sequences.fasta, made available in this work was elaborated to provide researchers with an extended database for the identification of proteins from cephalopods (e.g., cephalopod posterior salivary glands (PSGs) proteome), detailing at the same time the entire methodological approach employed for the creation of a composite database that can be useful for several research purposes. This knowledge may help researchers to analyse their proteomic data obtained from cephalopod PSGs. This database helps to explore with confidence a wide range of compounds produced in the PSGs, giving new insights on the presence of toxins and antimicrobial peptides (AMPs), which still remain underexplored. Indeed, we identified a total of 10,075 proteins clustered in 1868 proteinGroups with this composite database [1], with a false discovery rate (FDR) of 1.5%, whereas the proportion values of false positives for individual databases at the set FDR 1% were as follows: (Dataset_1—Databases A, B, C, D, E and F) 0.33% for Database A; NA (not applicable—no false positive found) for Database B; 0.87% for Database C; 0.32% for Database D; 4.71% for Database E; and 1.44% for Database F. Therefore, it represents an interesting resource to recover some information that is usually discarded in the proteogenomic analyses of the PSGs from cephalopods.
The composite protein database gathered public information and proteins identified through our own original data obtained by shotgun proteomic analyses of PSGs from three Octopus vulgaris specimens. All data, subdivided into six datasets (deposited at the Mendeley Data repository), comprise protein sequences coded from 16 transcriptomes of cephalopods’ PSGs, a published proteome from O. vulgaris, a non-redundant antimicrobial protein database, as well as the proteins identified with Proteome Discoverer software v2.2.0.388, using our 12 original raw files obtained through the shotgun proteomics analyses of the PSGs from three specimens of O. vulgaris. The database built constitutes a valuable resource that could facilitate and improve the protein identification process of samples derived from cephalopods’ salivary glands. Moreover, these data contain relevant information for researchers interested in the study of cephalopods’ salivary apparatus, cephalopods’ ecology, feeding strategies, toxins and AMPs production.
2. Data Description
2.1. Dataset Description
Herein, we provide all the datasets and scripts contemplated in the construction of the composite protein database used for the proteogenomic analyses performed in the article referenced as [1]. This includes proteins from public databases, combined with proteins identified by shotgun proteomics from original data.
The composite database, named “All_Databases_5950827_sequences.fasta”, is provided in Dataset_1. This database contains protein sequences retrieved from public databases related to cephalopods’ salivary glands and proteins identified from our original data. The composite database comprises a total of 5,950,827 protein sequences and, in turn, it is composed of six smaller databases, named with capital letters from A to F (Dataset_1—Databases A, B, C, D, E and F). Each one of these databases, within Dataset_1, contains data from several sources, i.e., Database A—protein database from proteogenomic analyses of the O. vulgaris salivary apparatus, built by Fingerhut et al. (2018) [2]; Database B—antimicrobial peptides from a non-redundant database [3]; Database C—proteins identified with Proteome Discoverer using our 12 raw files against the UniProt database for the Metazoan taxonomic selection (2018_07 release); Database D—proteins identified from de novo transcriptome assemblies of 16 cephalopods’ PSGs by TransDecoder; Database E—proteins identified from de novo transcriptome assemblies of 16 cephalopods’ PSGs using a six-frame translation tool, which are not included in Database D; Database F—proteins obtained using a six-frame translation tool using the transcripts profiled in the transcriptome of O. vulgaris [2], but not included by the authors in Database A.
Of the six smaller databases (Databases A, B, C, D, E and F) that make up our composite database, three of them (i.e., Databases A, B and C) present in their constitution sequences from the same source—the UniProt public database. Therefore, considering the inclusion of sequences from a common source in these three smaller databases, some sequence overlap could be present in our composite database (i.e., with the same assession number). In order to assess the percentage of possible redundancy present in our composite database, the above-mentioned three databases were compared to each other (Figure 1).
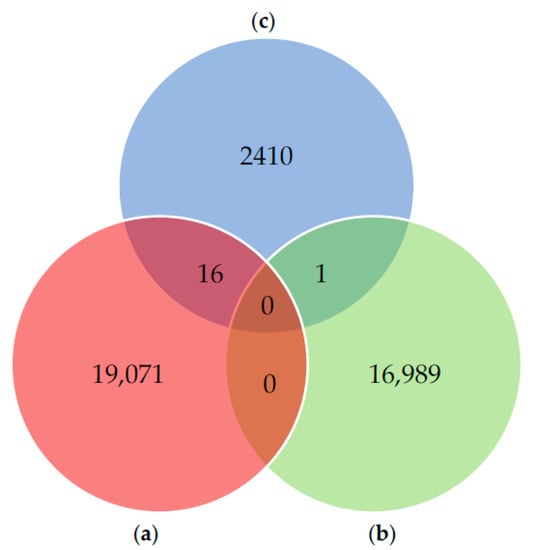
Figure 1.
Pairwise sequence comparison between the three smaller databases containing protein sequences from the UniProt database: (a) Database A (in red); (b) Database B (in green); (c) Database C (in blue). Identical overlap—number of identical sequences shared between databases: Database A vs. Database B (0 sequences); Database A vs. Database C (16 sequences: tr|Q9TWW9, tr|U5XKL5, tr|Q9NL91, tr|Q9NL93, tr|Q9BLF6, tr|Q60HA9, tr|K7NCQ5, tr|Q2V0V0, tr|Q3L636, tr|H2B4T3, tr|A6YM28, tr|C5MRD9, tr|A0A1S7J1Y5, tr|A0A1S7J204, sp|P81431, A0A1B4X9A8); Database B vs. Database C (1 sequence: BPT1_BOVIN); Database A vs. Database B vs. Database C (0 sequences).
The previous analysis revealed that, in total, there are just 17 overlapping or redundant sequences among these smaller databases, thus corresponding to only ~0.00029% of redundancy within the composite database. Considering the low redundancy level and in order to preserve the original number of sequences present in each of these smaller databases—some of which are already published (Databases A and B)—as well as the output from the computational strategy used in this work to build Database C, the 17 mentioned sequences were kept, for the sake of the analyses presented in the research article [1].
Additionally, we made available in the following datasets (Dataset_2, Dataset_3, Dataset_4, Dataset_5 and Dataset_6) the files, scripts and command lines used to construct some of the smaller databases that integrate our composite database “All_Databases_5950827_sequences.fasta”.
Specifically, in Dataset_2 are provided the output files from the Proteome Discoverer v2.2.0.388 analysis of our original data obtained by shotgun proteomic analyses of PSGs from three O. vulgaris specimens. In Dataset_3 are provided the de novo assemblies of 16 transcriptomes from cephalopods’ PSGs using CLC Genomics Workbench v11.0.1, as well as all the assemblies integrated into one unique FASTA file that were used to generate both Databases D and E, as well as the list of adapters and possible contaminants previously used for trimming the raw data before assembly. In Dataset_4 are provided the proteins predicted by the TransDecoder v5.5.0 tool from the de novo assembly of 16 cephalopods’ PSG transcriptomes, used to construct Database D. Finally, in Dataset_5 and Dataset_6 are provided the scripts and command lines used to construct Databases E and F, respectively.
2.2. Tables
Detailed information about the above-mentioned datasets, deposited at the Mendeley Data repository (dataset name, file composition, type of files and DOI) are summarized in Table 1.

Table 1.
Datasets provided in this article and deposited at the Mendeley Data repository.
All statistics for the 16 transcriptomes from cephalopods’ PSGs assembled with the CLC Genomics Workbench v11.0.1 are provided in Table 2.

Table 2.
Summary statistics for de novo assembly and further TransDecoder v5.5.0 and six-frame translation analyses of 16 posterior salivary glands transcriptomes of cephalopods.
3. Materials and Methods
3.1. Database Construction for Proteogenomic Analyses
For proteogenomic analyses, a composite database named “All_Databases_5950827_sequences.fasta” provided in Dataset_1 was built, composed of a total of 5,950,827 protein sequences, which includes the sequences from six smaller databases named with capital letters from A to F (Databases A, B, C, D, E and F) in order to simplify the exposition of the source of each protein sequence and the methodology used to construct the composite database. More details about the composition of each database and software versions can be found below.
3.1.1. Database A: Protein Sequences from Fingerhut et al. (2018)
Database A (Database_A_19087_sequences.fasta) included in Dataset_1 is the same comprehensive database deposited by Fingerhut et al. (2018) under the identifier PXD010298 at ProteomeXchange via PRIDE (OVulgarisMQ_20172206.fasta) [2]. Briefly, this database was provided as a FASTA file (5.9 MB) accounting for a total of 19,087 protein sequences. This database included 18,536 protein sequences (called “known” by the authors) predicted by TransDecoder, embedded in Trinity v3.0.1, and 354 protein sequences (called “novel” by the authors) obtained with the six-frame translation tool and validated at the proteomic level (using the script “sixframe.rb” available as part of the Protk toolkit [4]). Both, “known” and “novel” proteins were obtained from the transcriptome of the PSGs of O. vulgaris [2]. Moreover, the database included 197 protein sequences of cephalopods retrieved UniProt, inferred from the O. vulgaris saliva proteome. The detailed description of how this database was constructed can be found in Fingerhut et al. (2018) [2].
3.1.2. Database B: Antimicrobial Peptides (AMPs)
Database B (Database_B_16990_sequences.fasta) included in Dataset_1 contains one of the most comprehensive collections of non-redundant AMPs, which comprises 16,990 AMPs carefully gathered by Aguilera-Mendoza et al. (2015) from 25 AMPs databases [3]. Some of these proteins came from the UniProt database. Details related to this AMPs database construction can be found in the original article [3]. In order to perform MaxQuant analyses, the names of these sequences were edited through the removal of all the characters located after the first space detected.
3.1.3. Database C: Proteins Identified with Proteome Discoverer
Database C (Database_C_2427_sequences.fasta), provided in Dataset_1, contains original PSGs proteomes from three O. vulgaris specimens comprising 2427 protein sequences identified with Proteome Discoverer software v2.2.0.388 (Thermo Scientific, Waltham, MA, USA) against the Orbitrap raw data deposited at the Mendeley Data repository (http://dx.doi.org/10.17632/csc8shzkwc.1; http://dx.doi.org/10.17632/787g95ppwv.1; http://dx.doi.org/10.17632/8fhx775zdf.1; http://dx.doi.org/10.17632/d6wxyt22kx.1; http://dx.doi.org/10.17632/zbtkf2nsvh.1 and http://dx.doi.org/10.17632/df4cbg73tx.1) [1]. All these protein sequences have a name composed of the UniProt accession followed by “_PD”, indicating that those sequences came from the Proteome Discoverer analysis (e.g., sp|P18499_PD). Details about sample preparation for LC–MS/MS analysis and further protein identification using Proteome Discoverer for the construction of this database can be found in the two steps described below (STEP 1 and STEP 2).
- STEP 1: Sample preparation and LC–MS/MS analysis
Briefly, protein samples from PSGs comprising three biological replicates of O. vulgaris caught in the eastern Atlantic (Portuguese waters) were processed in duplicate following two distinct protocols (i.e., total of six protein samples for each protocol) as follows: filter-aided sample preparation (FASP) [5] and in-solution digestion (ISD) using RapiGest SF Surfactant according to the manufacturer’s specifications (Waters Corporation, Milford, MA, USA). Protein samples prepared according to FASP and ISD protocols were processed using a nano LC–MS/MS, composed of an Ultimate 3000 liquid chromatography system coupled to a Q-Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany).
- STEP 2: Protein identification using Proteome Discoverer
The raw data from LC–MS/MS analysis corresponding to 12 Orbitrap files of PSGs from O. vulgaris were processed using Proteome Discoverer software v2.2.0.388 (Thermo-Fisher, Waltham, MA, USA) and searched against the UniProt database for the Metazoan taxonomic selection (https://www.uniprot.org/taxonomy/33208; 2018_07 release) [6]. The Sequest HT search engine was used for protein identification. The ion mass tolerance was 10 ppm for precursor ions and 0.02 Da for fragment ions. The maximum number of missing cleavage sites allowed was set to 2. Cysteine carbamidomethylation was defined as a constant modification. Methionine oxidation and protein N-terminus acetylation were defined as variable modifications. Peptide confidence was set to high. The processing node Percolator was enabled with the following settings: maximum delta Cn 0.05, decoy database search target FDR 1%, validation was based on q-value. The output files from this analysis were provided in Dataset_2.
3.1.4. Databases D and E: Proteins Identified from the de novo Transcriptome Assemblies of Cephalopods’ PSGs
Briefly, the workflow to construct Databases D (Database_D_84778_sequences.fasta) and E (Database_E_5106635_sequences.fasta), both included in Dataset_1, consisted of the following steps: (i) a first analysis of all the resulting contigs (272,704 contigs) from the de novo assembly of the cephalopods’ PSGs transcriptomes with the TransDecoder v5.5.0 tool [7], predicting a total of 84,778 protein sequences grouped in Database D; and (ii) an analysis with the six-frame translation tool (“sixframe.rb” script [4]) of those contigs that did not produce protein-coding sequences when analysed by the TransDecoder v5.5.0 tool (i.e., 187,926 contigs discarded by TransDecoder when considering default parameters and not included in Database D), whose results compose Database E (5,106,635 protein sequences). Details about the construction of these databases can be found in Table 2 and in the three steps described below (STEP 1 to STEP 3).
- STEP 1: Search and de novo assembly of cephalopods’ PSGs transcriptomes
In order to obtain the available data from transcriptomes of the cephalopods’ PSGs, a search at the Sequence Read Archive (SRA) of National Center for Biotechnology Information (NCBI) was first performed. This search was based on the following criteria: (i) the term “Cephalopoda” was considered [8], all returned results were selected, sent to “Run selector” tool and posteriorly downloaded in a Tab delimited format (SraRunTable.txt); (ii) the SraRunTable file was then inspected to select all the SRA run accessions (SRR#) associated with any of these terms, namely “poison gland”, “posterior venom gland” and “posterior salivary gland/glands”, recovering reads from seven transcriptomes (SRR6349992, SRR2047107, SRR3105321, SRR3105558, SRR7130741, SRR5204441 and SRR5204442). Then, to get additional information, a search with the same term “Cephalopoda” was performed at the Sequence Set Browser from NCBI [9], which after being filtered by “posterior venom gland” retrieved 10 results (i.e., BioProject accessions: PRJNA188569, PRJNA188570, PRJNA188571, PRJNA188572, PRJNA188573, PRJNA188575, PRJNA188576, PRJNA188577, PRJNA188658 and PRJNA188659). These BioProject accessions were searched at the SRA of NCBI in order to obtain their corresponding SRR#, which were compiled together with the previous ones, resulting in a total of 17 transcriptomes from cephalopods’ PSGs. From these 17 transcriptomes, SRR7130741 was discarded in order to avoid redundancy in further MaxQuant analyses since it corresponded to the O. vulgaris PSGs transcripts [2] already included within Database A. Then, the FASTQ files of each one of the remaining 16 transcriptomes (SRR6349992, SRR2047107, SRR3105321, SRR3105558, SRR5204441, SRR5204442, SRR680047, SRR725938, SRR725597, SRR725937, SRR725936, SRR684223, SRR725779, SRR684167, SRR725935 and SRR725780) [10] were downloaded from the European Nucleotide Archive [11] for further assembly. The FASTQ files of each transcriptome were imported to CLC Genomics Workbench v11.0.1 [12] for adapter trimming and further de novo assembly. The reads present within the FASTQ files were first trimmed considering a list of adapters and possible contaminants, provided within Dataset_3 (Table_S1.xlsx), in case of being paired reads, or by removing terminal nucleotides of each read (i.e., 10 nucleotides from the 5′ and 3′ ends), in case of being single reads, and finally assembled using default parameters (Table 2). All the resulting assemblies were provided as Dataset_3.
- STEP 2: Database D—proteins identified by TransDecoder
All the contigs resulting from the CLC de novo assemblies (total of 272,704 contigs) provided in Dataset_3 (272704_contigs_from_16_cephalopods_PSGs_transcriptome_assemblies.fasta) were then conducted to the TransDecoder v5.5.0 tool [7], using default parameters, in order to predict protein-coding regions in transcripts. The TransDecoder v5.5.0 tool extracted open reading frames (ORFs) that were at least 100 amino acids (AA) long regardless of coding potential and then predicted which of them were likely to be coding (Table 2). The resulting proteins predicted by TransDecoder from the de novo assembly of 16 transcriptomes of PSGs from cephalopods were provided within Dataset_4. All the files provided in Dataset_4 were compiled into a single FASTA file (total of 84,778 protein sequences) using Geneious v11.1.2 [13], and the sequence names were edited with the following format: “SRR#_c#_g#”, where “#” is a number, “c” means contig and “g” means gene, thus creating Database D provided in Dataset_1 (Database_D_84778_sequences.fasta).
- STEP 3: Database E—proteins identified by the six-frame translation tool
In order to prevent the loss of relevant peptides (such as antimicrobial peptides) shorter than the TransDecoder minimum protein length threshold of 100 AA (i.e., default parameters), the contigs not included in Database D (i.e., 187,926 contigs) were analysed by the six-frame translation tool (“sixframe.rb” script [4]). The details of this approach can be found below and in Table 2.
All the files mentioned in the paragraph below are available in Dataset_5.
First, two tables in a csv (comma-separated values) format were prepared. One of the tables was entitled as “cases.csv”, consisting of one column with the header “Name” listing all the SRR# corresponding to the sequences present within Database D (i.e., 84,778 contigs in the format, e.g., SRR684167_c1). The other table was named as “transcripts.csv” and contains two columns, “Name” and “Sequence”, with all the de novo assembled transcript names (SRR#_c#) and corresponding nucleotide sequences deposited in Dataset_3 (i.e., 272,704 contigs). Then, a database (DB.db) was created using the DB Browser for SQLite v3.11.2 [14] and the two tables in csv format were imported. After executing the SQL command (SQL_command.txt), the resulting contigs not included in Database D were exported as a csv table (187926_contigs_not_included_in_Database_D.csv) and posteriorly converted to a FASTA file (187926_contigs_not_included_in_Database_D.fasta) using Geneious v11.1.2 [13]. Furthermore, the “187926_contigs_not_included_in_Database_D.fasta” file was analysed by the six-frame translation tool (“sixframe.rb” script [4]), using default parameters with the exception of the “--min-len” parameter that was set to 10. In this way, 5,106,635 protein sequences with sequence length greater than or equal to 10 AA (six-frame_translation_of_187926_contigs_not_included_in_Database_D.fasta) were obtained. Finally, the names of the sequences within the “six-frame_translation_of_187926_contigs_not_included_in_Database_D.fasta” file were edited by removing everything after the space in order to obtain the final database for MaxQuant analysis, and it was deposited in Dataset_1 (Database_E_5106635_sequences.fasta).
3.1.5. Database F: O. vulgaris Proteins Identified by the Six-Frame Translation Tool
Database F contains 720,910 protein sequences provided in Dataset_1 (Database_F_720910_sequences.fasta) corresponding to the ORFs from O. vulgaris PSGs transcriptomes not included in Database A. Details for the construction of this database can be found below.
First, the files “Database_A_19087_sequences.fasta” (provided in Dataset_1) and “GGNR01.1.fsa_nt.gz” (46,490 contigs deposited under accession PRJNA464423) were opened with Geneious v11.1.2 [13], and the sequence names within both files were edited with the following format: “TRINITY_DN0_c0_g1_i1”. Then, two csv files, each one from the previously edited files, were saved and made available in Dataset_6.
Hereafter, all the mentioned files are available in Dataset_6. Specifically, for the preparation of these csv files, we followed two steps: (i) from the “Database_A_19087_sequences.fasta” file, all the names with the “TRINITY” term included were selected and saved as a list, making a total of 18,890 sequence names (cases.csv: consisting of one column with the header “Name”); and (ii) from the “GGNR01.1.fsa_nt.gz” file, a list with all the nucleotide sequences was saved (transcripts.csv: consisting of two columns with the headers “Name” and “Sequence”). Thus, there was a match between the header “Name” of these two saved files. Then, a database (DB1.db) was created using the DB Browser for SQLite v3.11.2 [14] and the two lists in csv format were imported. After executing the SQL command (SQL_command1.txt), the resulting contigs not included in Database A were exported as a csv table (31661_contigs_not_included_in_Database_A.csv) and posteriorly converted to a FASTA file (31661_contigs_not_included_in_Database_A.fasta) using Geneious v11.1.2. The “31661_contigs_not_included_in_Database_A.fasta” file was analysed by the six-frame translation tool (sixframe.rb [4]), using default parameters with the exception of the “--min-len” parameter that was set to 10. In this way, 720,910 protein sequences with sequence length greater than or equal to 10 AA (six-frame_translation_of_31661_contigs_not_included_in_Database_A.fasta) were obtained. Finally, the names of the sequences within the “six-frame_translation_of_31661_contigs_not_included_in_Database_A.fasta” file were edited by removing everything after the space in order to obtain the final database for MaxQuant analysis, and it was deposited in Dataset_1 (Database_F_720910_sequences.fasta).
Author Contributions
Conceptualization, D.A., D.D.-P., G.A.-C. and A.A.; methodology, D.A. and D.D.-P.; software, D.A., D.D.-P., Y.C. and A.A.; validation, A.M., V.V., A.C., G.A.-C. and A.A.; formal analysis, D.A., D.D.-P. and Y.C.; resources, V.V., A.C. and A.A.; data curation, D.A., D.D.-P. and A.M.; writing—original draft preparation, D.A., D.D.-P. and A.M.; writing—review and editing, D.A., D.D.-P., A.M., G.A.-C. and A.A.; funding acquisition, V.V., A.C. and A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Strategic Funding UIDB/04423/2020 and UIDP/04423/2020 through national funds provided by the Foundation for Science and Technology (FCT) and the European Regional Development Fund (ERDF) in the framework of the program PT2020, by the European Structural and Investment Funds (ESIF) through the Competitiveness and Internationalization Operational Program—COMPETE 2020 and by National Funds through the FCT under the project PTDC/CTA-AMB/31774/2017 (POCI-01-0145-FEDER/031774/2017).
Acknowledgments
We are grateful to Robert Carcasses (Full Stack developer at Kenkou GmbH, Germany) for language programming advice and assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Almeida, D.; Domínguez-Pérez, D.; Matos, A.; Agüero-Chapin, G.; Osório, H.; Vasconcelos, V.; Campos, A.; Antunes, A. Putative antimicrobial peptides of the posterior salivary glands from the cephalopod Octopus vulgaris revealed by exploring a composite protein database. Antibiotics 2020, 9, 757. [Google Scholar] [CrossRef] [PubMed]
- Fingerhut, L.C.H.W.; Strugnell, J.M.; Faou, P.; Labiaga, Á.R.; Zhang, J.; Cooke, I.R. Shotgun Proteomics Analysis of Saliva and Salivary Gland Tissue from the Common Octopus Octopus vulgaris. J. Proteome Res. 2018, 17, 3866–3876. [Google Scholar] [CrossRef] [PubMed]
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; Tellez-Ibarra, R.; Llorente-Quesada, M.T.; Salgado, J.; Barigye, S.J.; Liu, J. Overlap and diversity in antimicrobial peptide databases: Compiling a non-redundant set of sequences. Bioinformatics 2015, 31, 2553–2559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Proteomics Toolkit (Protk). Available online: https://github.com/iracooke/protk (accessed on 14 April 2019).
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Sequence Read Archive of National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/sra/?term=Cephalopoda (accessed on 26 October 2018).
- Sequence Set Browser from National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/Traces/wgs/?page=1&view=TSA&search=Cephalopoda (accessed on 26 October 2018).
- Ruder, T.; Sunagar, K.; Undheim, E.A.B.; Ali, S.A.; Wai, T.-C.; Low, D.H.W.; Jackson, T.N.W.; King, G.F.; Antunes, A.; Fry, B.G. Molecular Phylogeny and Evolution of the Proteins Encoded by Coleoid (Cuttlefish, Octopus, and Squid) Posterior Venom Glands. J. Mol. Evol. 2013, 76, 192–204. [Google Scholar] [CrossRef] [PubMed]
- European Nucleotide Archive. Available online: https://www.ebi.ac.uk/ena (accessed on 16 November 2018).
- CLC Genomics Workbench 11.0.1. Available online: https://www.qiagenbioinformatics.com/ (accessed on 16 November 2018).
- Geneious. Available online: https://www.geneious.com (accessed on 16 November 2018).
- DB Browser for SQLite. Available online: https://sqlitebrowser.org/ (accessed on 16 November 2018).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).