Molecular Methods for Detecting Microorganisms in Beverages
Abstract
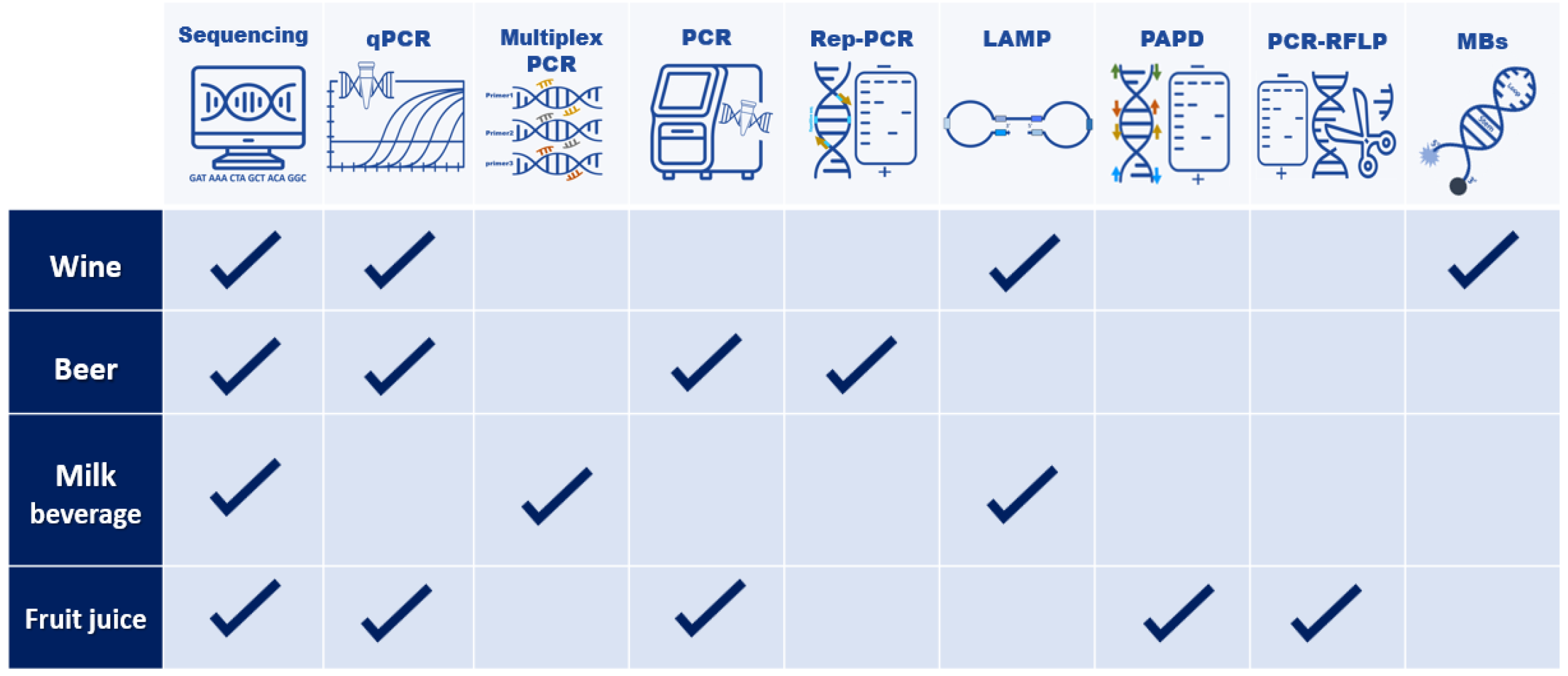
1. Introduction
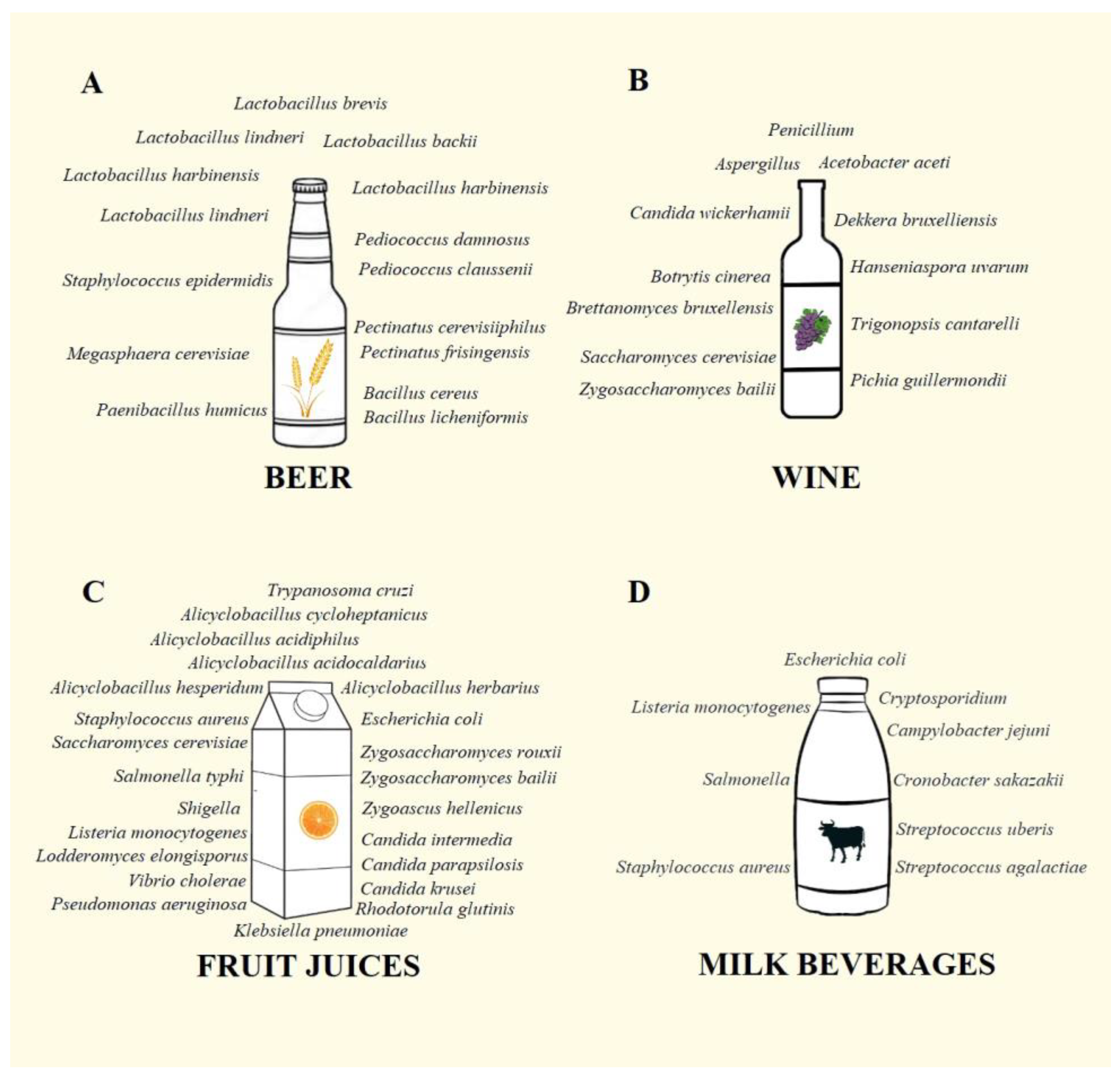
2. Microorganisms in Beer
3. Microorganisms in Wine
4. Microorganisms in Fruit Juices
5. Microorganisms in Dairy Beverages
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, M.; Wu, J.; Shi, Z.; Cao, A.; Fang, W.; Yan, D.; Wang, Q.; Li, Y. Molecular methods for identification and quantification of foodborne pathogens. Molecules 2022, 27, 8262. [Google Scholar] [CrossRef] [PubMed]
- Skowron, K.; Budzyńska, A.; Grudlewska-Buda, K.; Wiktorczyk-Kapischke, N.; Andrzejewska, M.; Wałecka-Zacharska, E.; Gospodarek-Komkowska, E. Two faces of fermented foods—The benefits and threats of its consumption. Front. Microbiol. 2022, 13, 845166. [Google Scholar] [CrossRef] [PubMed]
- Avîrvarei, A.C.; Salanță, L.C.; Pop, C.R.; Mudura, E.; Pasqualone, A.; Anjos, O.; Barboza, N.; Usaga, J.; Dărab, C.P.; Burja-Udrea, C.; et al. Fruit-based fermented beverages: Contamination sources and emerging technologies applied to assure their safety. Foods 2023, 12, 838. [Google Scholar] [CrossRef] [PubMed]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef] [PubMed]
- Franz, C.M.A.P.; den Besten, H.M.W.; Böhnlein, C.; Gareis, M.; Zwietering, M.H.; Fusco, V. Reprint of: Microbial food safety in the 21st century: Emerging challenges and foodborne pathogenic bacteria. Trends Food Sci. Technol. 2019, 84, 34–37. [Google Scholar] [CrossRef]
- Juvonen, R.; Virkajärvi, V.; Priha, O.; Laitila, A. Microbiological spoilage and safety risks in non-beer beverages. VTT Tied. Res. Notes 2011, 2599, 107. [Google Scholar] [CrossRef]
- Azeredo, D.R.; Alvarenga, V.; Sant’Ana, A.S.; Srur, A.U.S. An over- view of microorganisms and factors contributing for the micro-bial stability of carbonated soft drinks. Food Res. Int. 2016, 82, 136–144. [Google Scholar] [CrossRef]
- Zhao, X.; Lin, C.W.; Wang, J.; Oh, D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Li, W.L.; Wu, A.; Li, Z.C.; Zhang, G.; Yu, W.Y. A new calibration method between an optical sensor and a rotating platform in turbine blade inspection. Meas. Sci. Technol. 2017, 28, 035009. [Google Scholar] [CrossRef]
- Zhong, J.; Zhao, X. Detection of viable but non-culturable Escherichia coli O157:H7 by PCR in combination with propidium monoazide. 3 Biotechnology 2018, 8, 28. [Google Scholar] [CrossRef]
- Fusco, V.; Quero, G.M. Culture-dependent and culture-independent nucleic-acid-based methods used in the microbial safety assessment of milk and dairy products. Compr. Rev. Food Sci. Food Saf. 2014, 13, 493–537. [Google Scholar] [CrossRef] [PubMed]
- Zeng, P.; Guan, Q.; Zhang, Q. SERS detection of foodborne pathogens in beverage with Au nanostars. Microchim. Acta 2024, 191, 28. [Google Scholar] [CrossRef] [PubMed]
- Länge, K. Bulk and surface acoustic wave biosensors for milk analysis. Biosensors 2022, 12, 602. [Google Scholar] [CrossRef] [PubMed]
- da Silva Campos, J.; Júnior, A.C.V.; Boniek, D.; da Silva, D.L.; de Paula Lana, U.G.; Fernandes, J.V.; de Resende Stoianoff, M.A.; Andrade, V.S. Identification and evaluation of thermotolerance of yeasts from milk in natura exposed to high temperature and slow and fast pasteurization. Braz. J. Microbiol. 2023, 54, 1075–1082. [Google Scholar] [CrossRef]
- Grützke, J.; Gwida, M.; Deneke, C.; Brendebach, H.; Projahn, M.; Schattschneider, A.; Hofreuter, D.; El-Ashker, M.; Malorny, B.; Al Dahouk, S. Direct identification and molecular characterization of zoonotic hazards in raw milk by metagenomics using Brucella as a model pathogen. Microb. Genet. 2021, 7, 000552. [Google Scholar] [CrossRef]
- Jaudou, S.; Deneke, C.; Tran, M.L.; Schuh, E.; Goehler, A.; Vorimore, F.; Malorny, B.; Fach, P.; Grützke, J.; Delannoy, S. A step forward for Shiga toxin-producing Escherichia coli identification and characterization in raw milk using long-read metagenomics. Microb. Genet. 2022, 8, mgen000911. [Google Scholar] [CrossRef] [PubMed]
- Palomino-Camargo, C.; Gonzalez-Munoz, Y. Molecular techniques for detection and identification of pathogens in food: Advantages and limitations. Rev. Peru. Med. Exp. Y Salud Publica 2014, 31, 535–546. [Google Scholar]
- Mangal, M.; Bansal, S.; Sharma, S.K.; Gupta, R.K. Molecular detection of foodborne pathogens: A rapid and accurate answer to food safety. Crit. Rev. Food Sci. Nutr. 2016, 56, 1568–1584. [Google Scholar] [CrossRef]
- Jiyeon, H.; Ingyun, H.; Hyosun, K.; Chankyu, P.; Insoo, C.; Kunho, S. Evaluation of PCR inhibitory effect of enrichment broths and comparison of DNA extraction methods for detection of Salmonella enteritidis using real-time PCR assay. J. Vet. Sci. 2010, 11, 143–149. [Google Scholar] [CrossRef][Green Version]
- Löfström, C.; Hansen, F.; Hoorfar, J. Validation of a 20-h real-time PCR method for screening of Salmonella in poultry faecal samples. Vet. Microbiol. 2010, 144, 511–514. [Google Scholar] [CrossRef]
- Cantekin, C.; Ergun, Y.; Solmaz, H.; Özmen, G.Ö.; Demir, M.; Saidi, R. PCR assay with host specific internal control for Staphylococcus aureus from bovine milk samples. Maced. Vet. Rev. 2015, 38, 97–100. [Google Scholar] [CrossRef][Green Version]
- Forghani, F.; Wei, S.; Oh, D.H. A rapid multiplex real-time PCR high-resolution melt curve assay for the simultaneous detection of Bacillus cereus, Listeria monocytogenes, and Staphylococcus aureus in Food. J. Food Prot. 2016, 79, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Elizaquível, P.; Aznar, R. A multiplex RTi-PCR reaction for simultaneous detection of Escherichia coli O157:H7, Salmonella spp. and Staphylococcus aureus on fresh, minimally processed vegetables. Food Microbiol. 2008, 25, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, S.; Fratamico, P.M.; Horikoshi, N.; Okada, Y.; Takeshita, K.; Sameshima, T.; Kawamoto, S. Multiplex real-time polymerase chain reaction assay for simultaneous detection and quantification of Salmonella species, Listeria monocytogenes, and Escherichia coli O157:H7 in ground pork samples. Foodborne Pathog. Dis. 2010, 7, 549–554. [Google Scholar] [CrossRef]
- Singh, J.; Batish, V.K.; Grover, S. Simultaneous detection of Listeria monocytogenes and Salmonella spp. in dairy products using real time PCR-melt curve analysis. J. Food Sci. Technol. 2012, 49, 234–239. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bastam, M.M.; Jalili, M.; Pakzad, I.; Maleki, A.; Ghafourian, S. Pathogenic bacteria in cheese, raw and pasteurised milk. J. Vet. Med. Sci. 2021, 7, 2445–2449. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.J.; Farber, J.N.; Beuchat, L.R.; Parish, M.E.; Suslow, T.V.; Garrett, E.H.; Busta, F.F. Outbreaks associated with fresh produce: Incidence, growth, and survival of pathogens in fresh and fresh-cut produce. Compr. Rev. Food Sci. Food Saf. 2003, 2, 78–141. [Google Scholar] [CrossRef]
- Grumezescu, A.; Holban, A.M. Preservatives and Preservation Approaches in Beverages; Academic Press: Cambridge, MA, USA, 2019; Volume 15, pp. 540–562. [Google Scholar]
- Aneja, K.R.; Dhiman, R.; Aggarwal, N.K.; Kumar, V.; Kaur, M. Microbes associated with freshly prepared juices of citrus and carrots. Int. J. Food Sci. 2014, 2014, 408085. [Google Scholar] [CrossRef] [PubMed]
- Kregiel, D.; James, S.A.; Rygala, A.; Berlowska, J.; Antolak, H.; Pawlikowska, E. Consortia formed by yeasts and acetic acid bacteria Asaia spp. in soft drinks. Antonie Leeuwenhoek 2018, 111, 373–383. [Google Scholar] [CrossRef]
- Kregiel, D. Health safety of soft drinks: Contents, containers, and microorganisms. BioMed Res. Int. 2015, 2015, 128697. [Google Scholar] [CrossRef]
- Bintsis, T. Lactic acid bacteria: Their applications in foods. J. Bacteriol. Mycol. 2018, 6, 89–94. [Google Scholar] [CrossRef]
- Hernández, A.; Pérez-Nevado, F.; Ruiz-Moyano, S.; Serradilla, M.J.; Villalobos, M.C.; Martín, A.; Córdoba, M.G. Spoilage yeasts: What are the sources of contamination of foods and beverages? Int. J. Food Microbiol. 2018, 286, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Coskun, F.A. Traditional turkish fermented non-alcoholic grape-based beverage, “Hardaliye”. Beverages 2017, 3, 2. [Google Scholar] [CrossRef]
- Romero-Luna, H.E.; Peredo-Lovillo, A.; Davila-Ortiz, G. Tepache: A pre-hispanic fermented beverage as a potential source of probiotic yeasts. In Chemistry of Fermented Foods; American Chemical Society: Washington, DC, USA, 2022; pp. 135–147. [Google Scholar] [CrossRef]
- Motlhanka, K.; Lebani, K.; Boekhout, T.; Zhou, N. Fermentative microbes of khadi, a traditional alcoholic beverage of Botswana. Fermentation 2020, 6, 51. [Google Scholar] [CrossRef]
- Azam, M.S.; Ahmed, S.; Islam, M.N.; Maitra, P.; Islam, M.M.; Yu, D. Critical assessment of mycotoxins in beverages and their control measures. Toxins 2021, 13, 323. [Google Scholar] [CrossRef]
- Larcher, R.; Nicolini, G. Survey of ochratoxin A in musts, concentrated musts and wines produced or marketed in Trentino (Italy). J. Commod. Sci. 2001, 40, 69–78. [Google Scholar]
- Yazdanpanah, H. Mycotoxin contamination of foodstuffs and feedstuffs in Iran. Iran. J. Pharm. Res. 2006, 5, 9–16. [Google Scholar] [CrossRef]
- Mule, G.; Susca, A.; Logrieco, A.; Stea, G.; Visconti, A. Development of a quantitative real-time PCR assay for the detection of Aspergillus carbonarius in grapes. Int. J. Food Microbiol. 2006, 111, 28–34. [Google Scholar] [CrossRef]
- Rani, H.; Bhardwaj, R.D. Quality attributes for barley malt: “The backbone of beer”. J. Food Sci. 2021, 86, 3322–3340. [Google Scholar] [CrossRef]
- Zugravu, C.A.; Medar, C.; Manolescu, L.S.C.; Constantin, C. Beer and Microbiota: Pathways for a Positive and Healthy Interaction. Nutrients 2023, 15, 844. [Google Scholar] [CrossRef]
- Kordialik-Bogacka, E. Biopreservation of beer: Potential and constraints. Biotechnol. Adv. 2022, 58, 107910. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Konings, W.N. Beer spoilage bacteria and hop resistance. Int. J. Food Microbiol. 2003, 89, 105–124. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Luo, Y.; Mao, Y.; Peng, R.; Chen, J.; Soteyome, T.; Bai, C.; Chen, L.; Liang, Y.; Su, J.; et al. Spoilage lactic acid bacteria in the brewing industry. World J. Microbiol. Biotechnol. 2020, 30, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Tsekouras, G.; Tryfinopoulou, P.; Panagou, E. Detection and identification of lactic acid bacteria in semi-finished beer products using molecular techniques. In Proceedings of the 2nd International Electronic Conference on Foods—“Future Foods and Food Technologies for a Sustainable World”, Virtual, 15–30 October 2021. [Google Scholar] [CrossRef]
- Satokari, R.; Juvonen, R.; Wright, A.; Haikara, A. Detection of Pectinatus beer spoilage bacteria by using the polymerase chain reaction. J. Food Prot. 1997, 60, 1571–1573. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Hill, A.E. 7—Gram-positive spoilage bacteria in brewing. In Technology and Nutrition; Woodhead Publishing: Sawston, UK, 2015; pp. 141–173. [Google Scholar] [CrossRef]
- Garofalo, C.; Osimani, A.; Milanović, V.; Taccari, M.; Aquilanti, L.; Clementi, F. The occurrence of beer spoilage lactic acid bacteria in craft beer production. J. Food Sci. 2015, 80, M2845-52. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Huys, G.; Swings, J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 2001, 205, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Deng, Y.; Xu, Z.; Liu, J.; Dong, J.; Yin, H.; Yu, J.; Chang, Z.; Wang, D. Development of a propidium monoazide-polymerase chain reaction assay for detection of viable Lactobacillus brevis in beer. Braz. J. Microbiol. 2017, 48, 740–746. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Latorre, M.; Bruzone, M.C.; de Garcia, V.; Libkind, D. Microbial contaminants in bottled craft beer of Andean Patagonia, Argentina. Rev. Argent. Microbiol. 2023, 55, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Geissler, A.J.; Behr, J.; Vogel, R.F. Multiple genome sequences of important beer-spoiling lactic acid bacteria. Genome Announc. 2016, 4, e01077-16. [Google Scholar] [CrossRef]
- Kiousi, D.E.; Bucka-Kolendo, J.; Wojtczak, A.; Sokołowska, B.; Doulgeraki, A.I.; Galanis, A. Genomic analysis and in vitro investigation of the hop resistance phenotype of two novel Loigolactobacillus backii strains, isolated from spoiled beer. Microorganisms 2023, 11, 280. [Google Scholar] [CrossRef]
- Asano, S.; Shimokawa, M.; Suzuki, K. PCR analysis methods for detection and identification of beer-spoilage lactic acid bacteria. Methods Mol. Biol. 2019, 1887, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Haakensen, M.; Ziola, B. Identification of novel horA-harbouring bacteria capable of spoiling beer. Can. J. Microbiol. 2008, 54, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Satokari, R.; Juvonen, R.; Mallison, K.; von Wright, A.; Haikara, A. Detection of beer spoilage bacteria Megasphaera and Pectinatus by polymerase chain reaction and colorimetric microplate hybridization. Int. J. Food Microbiol. 1998, 45, 119–127. [Google Scholar] [PubMed]
- Juvonen, R.; Koivula, T.; Haikara, A. Group-specific PCR-RFLP and real-time PCR methods for detection and tentative discrimination of strictly anaerobic beer-spoilage bacteria of the class Clostridia. Int. J. Food Microbiol. 2008, 125, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Sanzani, S.M.; Miazzi, M.M.; di Rienzo, V.; Fanelli, V.; Gambacorta, G.; Taurino, M.R.; Montemurro, C. A rapid assay to detect toxigenic Penicillium spp. contamination in wine and musts. Toxins 2016, 8, 235. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, K. Occurrence of ochratoxin A in commodities and processed food—A review of EU occurrence data. Food Addit. Contam. 2005, 1, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Majerus, P.; Hain, J.; Kölb, C. Patulin in grape must and new, still fermenting wine (Federweißer). Mycotoxin Res. 2008, 24, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Sanzani, S.M.; Reverberi, M.; Fanelli, C.; Ippolito, A. Detection of ochratoxin A using molecular beacons and real-time PCR thermal cycler. Toxins 2015, 7, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Sanzani, S.M.; Montemurro, C.; Di Rienzo, V.; Solfrizzo, M.; Ippolito, A. Genetic structure and natural variation associated with host of origin in Penicillium expansum strains causing blue mould. Int. J. Food Microbiol. 2013, 165, 111–120. [Google Scholar] [CrossRef]
- Silva, P. First science & wine world congress. J. Agric. Food Chem. 2020, 68, 3299–3301. [Google Scholar] [CrossRef]
- López-Barajas, M.; López-Tamames, E.; Buxaderas, S.; Tomás, X.; de La Torre, M.C. Prediction of wine foaming. J. Agric. Food Chem. 1999, 47, 3743–3748. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.H.; Vrigneau, C.; Salmon, T.; Hoang, D.A.; Boulet, J.C.; Jégou, S.; Marchal, R. Influence of grape berry maturity on juice and base wine composition and foaming properties of sparkling wines from the champagne region. Molecules 2018, 23, 1372. [Google Scholar] [CrossRef]
- Amaro, F.; Almeida, J.; Oliveira, A.S.; Furtado, I.; Bastos, M.L.; Guedes de Pinho, P.; Pinto, J. Impact of cork closures on the volatile profile of sparkling wines during bottle aging. Foods 2022, 11, 293. [Google Scholar] [CrossRef]
- Marchal, R.; Warchol, M.; Cilindre, C.; Jeandet, P. Evidence for protein degradation by Botrytis cinerea and relationships with alteration of synthetic wine foaming properties. J. Agric. Food Chem. 2006, 54, 5157–5165. [Google Scholar] [CrossRef]
- Marchal, R.; Salmon, T.; Gonzalez, R.; Kemp, B.; Vrigneau, C.; Williams, P.; Doco, T. Impact of Botrytis cinerea contamination on the characteristics and foamability of yeast macromolecules released during the alcoholic fermentation of a model grape juice. Molecules 2020, 25, 472. [Google Scholar] [CrossRef] [PubMed]
- Echeverrigaray, S.; Randon, M.; da Silva, K.; Zacaria, J.; Longaray, A.P. Delamare Identification and characterization of non-saccharomyces spoilage yeasts isolated from Brazilian wines. World J. Microbiol. Biotechnol. 2013, 29, 1019–1027. [Google Scholar] [CrossRef]
- Millet, V.; Lonvaud-Funel, A. The viable but non-culturable state of wine micro-organisms during storage. Lett. Appl. Microbiol. 2000, 30, 136–141. [Google Scholar] [CrossRef]
- Caruso, M.; Fiore, C.; Contursi, M.; Salzano, G.; Paparella, A.; Romano, P. Formation of biogenic amines as criteria for the selection of wine yeasts. World J. Microbiol. Biotechnol. 2002, 18, 159–163. [Google Scholar] [CrossRef]
- Hierro, N.; Esteve-Zarzoso, B.; González, A.; Mas, A.; Guillamón, J.M. Real-time quantitative PCR (QPCR) and reverse transcription-QPCR for detection and enumeration of total yeasts in wine. Appl. Environ. Microbiol. 2006, 72, 7148–7155. [Google Scholar] [CrossRef]
- Willenburg, E.; Divol, B. Quantitative PCR: An appropriate tool to detect viable but not culturable Brettanomyces bruxellensis in wine. Int. J. Food Microbiol. 2012, 160, 131–136. [Google Scholar] [CrossRef]
- Phister, T.G.; Mills, D.A. Real-time PCR assay for detection and enumeration of Dekkera bruxellensis in wine. Appl. Environ. Microbiol. 2003, 69, 7430–7434. [Google Scholar] [CrossRef] [PubMed]
- Delaherche, A.; Claisse, O.; Lonvaud-Funel, A. Detection and quantification of Brettanomyces bruxellensis and ‘ropy’ Pediococcus damnosus strains in wine by real-time polymerase chain reaction. J. Appl. Microbiol. 2004, 97, 910–915. [Google Scholar] [CrossRef] [PubMed]
- Tessonniere, H.; Vidal, S.; Barnavon, L.; Alexandre, H.; Remize, F. Design and performance testing of a real-time PCR assay for sensitive and reliable direct quantification of Brettanomyces in wine. Int. J. Food Microbiol. 2009, 129, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Tofalo, R.; Schirone, M.; Corsetti, A.; Suzzi, G. Detection of Brettanomyces spp. in red wines using real-time PCR. J. Food Sci. 2012, 77, M545-9. [Google Scholar] [CrossRef] [PubMed]
- Palma, M.; Sa-Correia, I. Physiological genomics of the highly weak-acid-tolerant food spoilage yeasts of Zygosaccharomyces bailii sensu lato. Prog. Mol. Subcell. Biol. 2019, 58, 85–109. [Google Scholar] [CrossRef]
- Rawsthorne, H.; Phister, T.G. A real-time PCR assay for the enumeration and detection of Zygosaccharomyces bailii from wine and fruit juices. Int. J. Food Microbiol. 2006, 112, 1–7. [Google Scholar] [CrossRef] [PubMed]
- González-Muñoz, B.; Garrido-Vargas, F.; Pavez, C.; Osorio, F.; Chen, J.; Bordeu, E.; O’Brien, J.A.; Brossard, N. Wine astringency: More than just tannin-protein interactions. J. Sci. Food Agric. 2022, 102, 1771–1781. [Google Scholar] [CrossRef]
- Bartowsky, E.J.; Henschke, P.A. Acetic acid bacteria spoilage of bottled red wine—A review. Int. J. Food Microbiol. 2008, 125, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, L.; Shi, L.; Chen, X.; Liang, M.; Zhao, L. Development and application of a real-time loop-mediated isothermal amplification method for quantification of Acetobacter aceti in red wine. FEMS Microbiol. Lett. 2020, 367, fnaa152. [Google Scholar] [CrossRef]
- Valera, M.J.; Torija, M.J.; Mas, A.; Mateo, E. Acetic acid bacteria from biofilm of strawberry vinegar visualized by microscopy and detected by complementing culture-dependent and culture-independent techniques. Food Microbiol. 2015, 46, 452–462. [Google Scholar] [CrossRef]
- Longin, C.; Guilloux-Benatier, M.; Alexandre, H. Design and performance testing of a DNA extraction assay for sensitive and reliable quantification of acetic acid bacteria directly in red wine using real time PCR. Front. Microbiol. 2016, 7, 831. [Google Scholar] [CrossRef] [PubMed]
- Mandappa, I.M.; Basavaraj, K.; Manonmani, H.K. Analysis of Mycotoxins in Fruit Juices; Academic Press: Cambridge, MA, USA, 2018; pp. 763–777. [Google Scholar] [CrossRef]
- Karasawa, M.M.G.; Mohan, C. Fruits as prospective reserves of bioactive compounds: A review. Nat. Prod. Bioprospect. 2018, 8, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Dhalaria, R.; Verma, R.; Kumar, D.; Puri, S.; Tapwal, A.; Kumar, V. Bioactive compounds of edible fruits with their anti-aging properties: A comprehensive review to prolong human life. Antioxidants 2020, 9, 1123. [Google Scholar] [CrossRef]
- Raybaudi-Massilia, R.M.; Mosqueda-Melgar, J.; Soliva-Fortuny, R.; Martin-Belloso, O. Control of pathogenic and spoilage microorganisms in fresh-cut fruits and fruit juices by traditional and alternative natural antimicrobials. Compr. Rev. Food Sci. Food Saf. 2009, 8, 157–180. [Google Scholar] [CrossRef] [PubMed]
- Jimma, F.I.; Mohammed, A.; Adzaworlu, E.G.; Nzeh, J.; Quansah, L.; Dufailu, O.A. Microbial quality and antimicrobial residue of local and industrial processed fruit juice sold in Tamale, Ghana. Food Technol. 2022, 2, 26. [Google Scholar] [CrossRef]
- Salomao, B.D.C.M. Pathogens and Spoilage Microorganisms in Fruit Juice: An Overview; Academic Press: Cambridge, MA, USA, 2018; pp. 291–308. [Google Scholar] [CrossRef]
- Sharma, N.; Singh, K.; Toor, D.; Pai, S.S.; Chakraborty, R.; Khan, K.M. Antibiotic resistance in microbes from street fruit drinks and hygiene behavior of the vendors in Delhi, India. Int. J. Environ. Res. Public Health 2020, 17, 4829. [Google Scholar] [CrossRef] [PubMed]
- Tasnim, F., Jr.; Anwar Hossain, M.; Kamal Hossain, M.; Lopa, D.; Formuzul Haque, K.M. Quality assessment of industrially processed fruit juices available in Dhaka city, Bangladesh. Malays. J. Nutr. 2010, 16, 431–438. [Google Scholar] [PubMed]
- Keller, S.E.; Miller, A.J. Microbiological safety of fresh citrus and apple juices. In Microbiology of Fruits and Vegetables; CRC Press: Boca Raton, FL, USA, 2005; pp. 227–246. [Google Scholar]
- Jensen, N. Alicyclobacillus: A new challenge for the food industry. Food Aust. 1999, 51, 33–36. [Google Scholar]
- Cacho, P.; Danyluk, M.; Rouseff, R. GC–MS quantification and sensory thresholds of guaiacol in orange juice and its correlation with Alicyclobacillus spp. Food Chem. 2011, 129, 45–50. [Google Scholar] [CrossRef]
- Chang, S.S.; Kang, D.H. Alicyclobacillus spp. in the fruit juice industry: History, characteristics, and current isolation/detection procedures. Crit. Rev. Microbiol. 2004, 30, 55–74. [Google Scholar] [CrossRef]
- Molva, C.; Baysal, A.H. Evaluation of bioactivity of pomegranate fruit extract against Alicyclobacillus acidoterrestris DSM 3922 vegetative cells and spores in apple juice. LWT-Food Sci. Technol. 2015, 62, 989–995. [Google Scholar] [CrossRef]
- Hui, L.; Hong, C.; Bin, L.; Rui, C.; Nan, J.; Tianli, Y.; Zhouli, W. Establishment of quantitative PCR assays for the rapid detection of Alicyclobacillus spp. that can produce guaiacol in apple juice. Int. J. Food Microbiol. 2021, 360, 109329. [Google Scholar] [CrossRef]
- Osopale, B.A.; Adewumi, G.A.; Witthuhn, R.C.; Kuloyo, O.O.; Oguntoyinbo, F.A. A review of innovative techniques for rapid detection and enrichment of Alicyclobacillus during industrial processing of fruit juices and concentrates. Food Control 2019, 99, 146–157. [Google Scholar] [CrossRef]
- Sourri, P.; Tassou, C.C.; Nychas, G.E.; Panagou, E.Z. Fruit juice spoilage by Alicyclobacillus: Detection and control methods-a comprehensive review. Foods 2022, 11, 747. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, K.; Okubo, T.; Inoue, N.; Shinano, H. Randomly amplified polymorphic DNA (RAPD) for rapid identification of the spoilage bacterium Alicyclobacillus acidoterrestris. Biosci. Biotechnol. Biochem. 1997, 61, 1016–1018. [Google Scholar] [CrossRef]
- Sourri, P.; Doulgeraki, A.I.; Tassou, C.C.; Nychas, G.E. A single enzyme PCR-RFLP assay targeting V1-V3 region of 16S rRNA gene for direct identification of Alicyclobacillus acidoterrestris from other Alicyclobacillus species. J. Appl. Genet. 2019, 60, 225–229. [Google Scholar] [CrossRef]
- Connor, C.J.; Luo, H.; Gardener, B.B.; Wang, H.H. Development of a real-time PCR-based system targeting the 16S rRNA gene sequence for rapid detection of Alicyclobacillus spp. in juice products. Int. J. Food Microbiol. 2005, 99, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Tang, Q.; Zhang, X.; Zhao, G.; Hu, X.; Liao, X.; Xiang, H. Isolation and characterization of thermo-acidophilic endospore-forming bacteria from the concentrated apple juice-processing environment. Food Microbiol. 2006, 23, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Dekowska, A.; Niezgoda, J.; Sokołowska, B. Genetic heterogeneity of Alicyclobacillus strains revealed by RFLP analysis of vdc region and rpoB gene. BioMed Res. Int. 2018, 2018, 9608756. [Google Scholar] [CrossRef]
- Wang, Z.; Yue, T.; Yuan, Y.; Zhang, Y.; Gao, Z.; Cai, R. Targeting the vanillic acid decarboxylase gene for Alicyclobacillus acidoterrestris quantification and guaiacol assessment in apple juices using real time PCR. Int. J. Food Microbiol. 2021, 338, 109006. [Google Scholar] [CrossRef]
- Cai, R.; Wang, Z.; Yuan, Y.; Liu, B.; Wang, L.; Yue, T. Detection of Alicyclobacillus spp. in fruit juice by combination of immunomagnetic separation and a SYBR Green I Real-Time PCR assay. PLoS ONE 2015, 10, e0141049. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Shi, Y.; Zhang, J.; Han, Q.; Xia, X.S.; Zhang, A.M.; Song, Y. Rapid and simultaneous detection of five, viable, foodborne pathogenic bacteria by photoinduced PMAxx-coupled multiplex PCR in fresh juice. Foodborne Pathog. Dis. 2021, 18, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Corbett, K.M.; de Smidt, O. Culture-dependent diversity profiling of spoilage yeasts species by PCR-RFLP comparative analysis. Food Sci. Technol. Int. 2019, 25, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Casey, G.D.; Dobson, A.D.W. Potential of using real-time PCR-based detection of spoilage yeast in fruit juice—A preliminary study. Int. J. Food Microbiol. 2004, 91, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.C.; Seixas, A.S.; Sousa, C.P.; Souza, C.W. Microbiological evaluation of sugarcane juice sold at street stands and juice handling conditions in Sao Carlos, Sao Paulo, Brazil. Cad. Saude Publica 2006, 22, 1111–1114. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Dawar, S.; Tariq, M. Mycoflora associated with sugar cane juice in Karachi city. Pak. J. Bot. 2010, 42, 2955–2962. [Google Scholar]
- Abbas, S.R.; Sabir, S.M.; Ahmad, S.D.; Boligon, A.A.; Athayde, M.L. Phenolic profile, antioxidant potential and DNA damage protecting activity of sugarcane (Saccharum officinarum). Food Chem. 2014, 147, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Mattos, E.C.; Meira-Strejevitch, C.D.S.; Marciano, M.A.M.; Faccini, C.C.; Lourenço, A.M.; Pereira-Chioccola, V.L. Molecular detection of Trypanosoma cruzi in acai pulp and sugarcane juice. Acta Trop. 2017, 176, 311–315. [Google Scholar] [CrossRef]
- Rettedal, E.A.; Altermann, E.; Roy, N.C.; Dalziel, J.E. The Effects of unfermented and fermented cow and sheep milk on the gut microbiota. Front. Microbiol. 2019, 10, 458. [Google Scholar] [CrossRef]
- Paszczyk, B.; Czarnowska-Kujawska, M.; Klepacka, J.; Tońska, E. Health-promoting ingredients in goat’s milk and fermented goat’s milk drinks. Animals 2023, 13, 907. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. 2013, 37, 664–698. [Google Scholar] [CrossRef]
- Aliyo, A.; Teklemariam, Z. Assessment of Milk Contamination, Associated Risk Factors, and Drug Sensitivity Patterns among Isolated Bacteria from Raw Milk of Borena Zone, Ethiopia. J. Trop. Med. 2022, 2022, 3577715. [Google Scholar] [CrossRef]
- Koskinen, M.T.; Holopainen, J.; Pyorala, S.; Bredbacka, P.; Pitkala, A.; Barkema, H.W.; Bexiga, R.; Roberson, J.; Solverod, L.; Piccinini, R.; et al. Analytical specificity and sensitivity of a real-time polymerase chain reaction assay for identification of bovine mastitis pathogens. J. Dairy Sci. 2009, 92, 952–959. [Google Scholar] [CrossRef]
- Pang, L.; Pi, X.; Yang, X.; Song, D.; Qin, X.; Wang, L.; Man, C.; Zhang, Y.; Jiang, Y. Nucleic acid amplification-based strategy to detect foodborne pathogens in milk: A review. Crit. Rev. Food Sci. Nutr. 2022, 8, 1–16. [Google Scholar] [CrossRef]
- Cornelissen, J.B.W.J.; De Greeff, A.; Heuvelink, A.E.; Swarts, M.; Smith, H.E.; Van der Wal, F.J. Rapid detection of Streptococcus uberis in raw milk by loop-mediated isothermal amplification. J. Dairy Sci. 2016, 99, 4270–4281. [Google Scholar] [CrossRef]
- Alber, J.; El-Sayed, A.; Lammler, C.; Hassan, A.A.; Zschock, M. Polymerase chain reaction mediated identification of Streptococcus uberis and Streptococcus parauberis using species-specific sequences of the genes encoding superoxide dismutase A and chaperonin 60. J. Vet. Med. B Infect. Dis. Vet. Public Health 2004, 51, 180–184. [Google Scholar] [CrossRef]
- Gillespie, B.E.; Oliver, S.P. Simultaneous detection of mastitis pathogens, Staphylococcus aureus, Streptococcus uberis, and Streptococcus agalactiae by multiplex real-time polymerase chain reaction. J. Dairy Sci. 2005, 88, 3510–3518. [Google Scholar] [CrossRef]
- Bruno, S.J.; Phiri, B.M.; Hang’ombe, M.E.; Mubanga, M.; Maurischat, S.; Wichmann-Schauer, H.; Schaarschmidt, S.; Fetsch, A. Prevalence and diversity of Staphylococcus aureus in the Zambian dairy value chain: A public health concern. Int. J. Food Microbiol. 2022, 375, 109737. [Google Scholar] [CrossRef]
- Machado, G.P.; Silva, R.C.; Guimarães, F.F.; Salina, A.; Langoni, H. Detection of Staphylococcus aureus, Streptococcus agalactiae and Escherichia coli in Brazilian mastitic milk goats by multiplex-PCR. Pesqui. Vet. Bras. 2018, 38, 1358–1364. [Google Scholar] [CrossRef]
- Straub, J.A.; Hertel, C.; Hammes, W.P. A 23S rDNA-targeted polymerase chain reaction-based system for detection of Staphylococcus aureus in meat starter cultures and dairy products. J. Prot. 1999, 62, 1150–1156. [Google Scholar] [CrossRef]
- Riffon, R.; Sayasith, K.; Khalil, H.; Dubreuil, P.; Drolet, M.; Lagacé, J. Development of a rapid and sensitive test for identification of major pathogens in bovine mastitis by PCR. J. Clin. Microbiol. 2001, 39, 2584–2589. [Google Scholar] [CrossRef]
- Chotár, M.; Vidova, B.; Godany, A. Development of specific and rapid detection of bacterial pathogens in dairy products by PCR. Folia Microbiol. 2006, 51, 639–646. [Google Scholar] [CrossRef]
- Luciani, M.; di Febo, T.; Zilli, K.; di Giannatale, E.; Armillotta, G.; Manna, L. Rapid detection and isolation of Escherichia coli O104:H4 from milk using monoclonal antibody-coated magnetic beads. Front. Microbiol. 2016, 7, 942. [Google Scholar] [CrossRef]
- Bai, Y.; Cui, Y.; Suo, Y.; Shi, C.; Wang, D.; Shi, X. A Rapid method for detection of Salmonella in milk based on extraction of mRNA using magnetic capture probes and RT-qPCR. Front. Microbiol. 2019, 10, 770. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, S.; Xue, Y.; Zhang, Y.; Zhang, W.; Wang, S. Quantitative detection of viable but nonculturable Cronobacter sakazakii using photosensitive nucleic acid dye PMA combined with isothermal amplification LAMP in raw milk. Foods 2022, 11, 2653. [Google Scholar] [CrossRef]
- Dauga, C. Evolution of the gyrB gene and the molecular phylogeny of Enterobacteriaceae: A model molecule for molecular systematic studies. Int. J. Syst. Evol. Microbiol. 2002, 52, 531–547. [Google Scholar] [CrossRef]
- Urso, R.; Rantsiou, K.; Dolci, P.; Rolle, L.; Comi, G.; Cocolin, L. Yeast biodiversity and dynamics during sweet wine production as determined by molecular methods. FEMS Yeast Res. 2008, 8, 1053–1062. [Google Scholar] [CrossRef]
- Zendeboodi, F.; Jannat, B.; Sohrabvandi, S.; Khanniri, E.; Mortazavian, A.M.; Khosravi, K.; Gholian, M.M.; Sarmadi, B.; Javadi, N.H.S. Detection of non-alcoholic beer spoilage microorganisms at critical points of production by polymerase chain reaction. Biointerface Res. Appl. Chem. 2021, 11, 9658–9966. [Google Scholar] [CrossRef]
- Phattaraporn, S.; Rachnarin, N.; Issara, P.; Siwarutt, B. Identification of bacteria and yeast communities in a Thai sugary kefir by polymerase chain reaction-denaturing gradient gel electrophoresis (pcr-dgge) analyses. J. Ind. Technol. 2015, 11, 25–39. [Google Scholar]
| № | Object | Target | Method | Primer | Subsequence (5′-3′) | Literature |
|---|---|---|---|---|---|---|
| 1 | Beer | Lactobacillus | Rep-PCR | REP1R-I | IIIICGICGICATCIGGC | [46,50] |
| REP2-I | IIICGNCGNCATCNGGC | |||||
| BOXA1R | CTACGGCAAGGCGACGCTGACG | |||||
| PRIMER | (GTG)5 (5GTGGTGGTGGTGGTG3) | |||||
| 2 | Pectinatus | qPCR | F | GCTTTTAGCTGTCGCTTGGA | [47] | |
| R | TGCATCTCTGCATACGTCAA | |||||
| 3 | Bacteria | qPCR | 27F | GAGAGTTTGATCCTGGCTCAG | [53] | |
| 1495r | CTACGGCTACCTTGTTACGA | |||||
| 4 | Yeasts | qPCR | ITS1 | TCCGTAGGTGAACCTGCGG | [52,53] | |
| ITS4 | TCCTCCGCTTATTGATATGC | |||||
| 5 | Resistance gene HorA/HorC | qPCR | LbHC-1 | ATCCGGCGGTGGCAAATCA | [56] | |
| LbHC-2 | AATCGCCAATCGTTGGCG | |||||
| Lactobacillus brevis | qPCR | LBP2 | CTGATTTCAACAATGAAGC | |||
| UNP1 | CCGTCAATTCCTTTGAGTTT | |||||
| Pectinatus cerevisiiphilus | qPCR | 16C-F | CGTATGCAGAGATGCATATT | |||
| IC-R | CACTCTTACAAGTATCTAC | |||||
| Pediococcus damnosus/Pediococcus inopinatus | qPCR | PIDF1 | TGTGAGAGTAACTGCTCATG | |||
| PIDR8 | ACGCCTAATCTCTTTGGTTA | |||||
| Megasphaera cerevisiae | qPCR | Mc-f4 | ACCGAATACGATCTAAAG | |||
| Mc-r4 | TTAAGACCGACTTACCGA | |||||
| 6 | Clostridia | End-point PCR | An-0279f | ACGATCAGTAGCCGGT | [59] | |
| An-0603r | AGCCCCGCACTTTTAAG | |||||
| 7 | Megasphaera cerevisiae | qPCR | F | CACTGAATAGTCTATCGC | [58] | |
| R | AAGACCGACTTACCGAAC | |||||
| 1 | Wine | Penicillium expansum | qPCR | PE F | ATCGGCTGCGGATTGAAAG | [64] |
| PE R | AGTCACGGGTTTGGAGGGA | |||||
| 2 | Acetobacter aceti | qLAMP | F3 | AGGTGGGGATGACGTCAAG | [84] | |
| B3 | CGGGAACGTATTCACCGC | |||||
| FIP | CTAGCTTCCCACTGTCACCG TCCTCATGGCCCTTATGTC | |||||
| BIP | AACCGTCTCAGTTCGGATTGCATCCGCGATTACTAGCGATTC | |||||
| LF | AGCACGTGTGTAGCCCA | |||||
| LB | CTCTGCAACTCGAGTGCATG | |||||
| F | CGGAATGACTGGGCGTAAG | |||||
| R | CAGTAATGAGCCAGGTTGCC | |||||
| PROBE | 6FAMCGGGCTTAACCTGGGAGCTGCATTBHQ1 | |||||
| 3 | Acetic acid bacteria | qPCR | AAB F | TGAGAGGATGATCAGCCACACT | [85] | |
| AAB R | TCACACACGCGGCATTG | |||||
| 4 | Yeasts | qPCR | YEAST F | GAGTCGAGTTGTTTGGGAATGC | [74] | |
| YEAST R | TCTCTTTCCAAAGTTCTTTTCATCTTT | |||||
| 5 | Brettanomyces/Dekkera spp. | qPCR | DBRUX F | GGATGGGTGCACCTGGTTTACAC | [76,78,79] | |
| DBRUXR | GAAGGGCCACATTCACGAACCCCG | |||||
| 6 | Brettanomyces bruxellensis | qPCR | BRETT 1 | CGAAGAAGTTGAACGGCCGCATTTG | [77] | |
| BRETT 2 | TCTTCGATATGCCGTCCAAAAGCTC | |||||
| RAD 1 | GTTCACACAATCCCCTCGATCAAC | [78] | ||||
| RAD 2 | TGCCAACTGCCGAATGTTCTC | |||||
| ACT 1 | TGTCAGAGACATCAAGGAGAAGCT | [75] | ||||
| ACT 2 | CGTCTGCATTTCCTGGTCAA | |||||
| 7 | Zygosaccharomyces bailii | qPCR | ZB F1 | CATGGTGTTTTGCGCC | [81] | |
| ZB R1 | CGTCCGCCACGAAGTGGTAG A | |||||
| 8 | Ochratoxin A | Molecular Beacon Method | APTABEACON | 6FAMCGCGCTGGATCGGGTGTGGGTGGCGTAAAGGGAGCATCGGACACAGCGCGBHQ1 | [63] | |
| 1 | Fruit juices | Alicyclobacillus acidoterrestris | PCR | A1-92-3 F | TCGCAACCTGCTTCTCCA | [100] |
| A1-92-3 R | TGGTGGACGGGATTGTTT | |||||
| Alicyclobacillus acidiphilus | PCR | A2-16S-1 F | ATGCAAGTCGAGCGAAC | |||
| A2-16S-1 R | GCAACTTTCCTCAACGG | |||||
| Alicyclobacillus cycloheptanicus | PCR | A3-16S-3 F | TGCAAATGCACCGCAGAT | |||
| A3-16S-3 R | GGCTTTCCACTCCCCTTG | |||||
| Alicyclobacillus herbarius | PCR | A4-5472 F | TGAGTCGCTTCTTCGTTCTT | |||
| A4-5472 R | CTACGGGATGACGGAAGC | |||||
| 2 | Alicyclobacillus acidoterrestris | RAPD | Ba-lO | AACGCGCAAC | [103] | |
| F -61 | CCTGTGATGGGC | |||||
| F-64 | GCCGCGCCAGTA | |||||
| 3 | Alicyclobacillus acidoterrestris | PCR-RFLP Restriction endonuclease: HhaI | P1 | GCGGCGTGCCTAATACATGC | [104] | |
| P4 | ATCTACGCATTTCACCGCTAC | |||||
| 4 | Alicyclobacillus | qPCR | CC16S-F | CGTAGTTCGGATTGCAGGC | [105] | |
| CC16S-R | GTGTTGCCGACTCTCGTG | |||||
| CC16S-Probe | QUASAR670CGGAATTGCTAGTAATCGCBHQ-2 | |||||
| 5 | Alicyclobacillus | PCR–RFLP Restriction endonucleases: BsuR I, Hinf I, Msp I, Rsa I | P1 | CGGGATCCAGAGTTTGATCCTGCGTCAGAACGAACGCT | [106] | |
| P2 | CGGGATCCTAGGGCTACCTTGTTACGACTTCACCCC | |||||
| 6 | Alicyclobacillus acidoterrestris | PCR-RFLP Restriction endonuclease: HhaI | P1 | GCGGCGTGCCTAATACATGC | [104] | |
| P4 | ATCTACGCATTTCACCGCTAC | |||||
| 7 | Alicyclobacillus | PCR-RFLP Restriction endonucleases: BsuRI, Hin6I, HphI | vdc fr | CTGTTGGCTCAATGGCGGCTGAGCGAT | [107] | |
| vdc rev | TTATCAGCGGTTTATCCGCGGTGGAACAGTC | |||||
| vdc1 fr | AACGACGCAGGTGTGGAAAC | |||||
| vdc1 rev | AGCGTGGGCAAGTTGTCATGTG | |||||
| vdc K | TTGGCAACGGAGAAGTGGGAG | |||||
| and vdc S | AATCACGCGCTGATGATGGG | |||||
| Bur 5 | GCCGACGTGATGCTCAARGAGCGCA | |||||
| Bur 6 | GTSGCRTCGAGAATCATCTTGTG | |||||
| Gru3 | CGYGACGTDCACTAYTCBCACTA | |||||
| Gru4 | GCCCANACYTCCATCTCRCCRAA | |||||
| Gru5 | CGCGACGTACACTATTCGCACTA | |||||
| Gru6 | GCCCAAACCTCCATCTCACCAAA | |||||
| 8 | Alicyclobacillus acidoterrestris | qPCR | vdcCF1 | TAYGAAATGGCMGGTGC | [108] | |
| vdcCR1 | GGAAGGTTGAAYGGATC | |||||
| 9 | Alicyclobacillus | qPCR | F | ATGCGTAGATATGTGGAGGA | [109] | |
| R | CAGGCGGAGTGCTTATTG | |||||
| 10 | Yeast | PCR-RFLP Restriction endonucleases: CfoI, HaeIII, HinfI | ITS1 | TCCGTAGGTGAACCTGCGG | [111] | |
| ITS4 | TCCTCCGCTTATTGATATGC | |||||
| 11 | Zygosaccharomyces bailii, Zygosaccharomyces rouxii, Candida krusei, Rhodotorula glutinis, Saccharomyces cerevisiae | qPCR | ITS3 | GCATCGATGAAGAACGCAGC | [112] | |
| ITS4 | TCCTCCGCTTATTGATATGC | |||||
| CS Fwd | GCATATGGTGGTTATGAGAGG | |||||
| CS Rev | AGCAGAAACATTACCACCTTC | |||||
| 12 | Trypanosoma cruzi | qPCR | 32F | TTTGGGAGGGGCGTTCA | [116] | |
| 148R | ATATTACACCAACCCCAATCGAA | |||||
| probe71 | FAMCATCTCACCCGTACATT3NFQ | |||||
| 1 | Streptococcus uberis | LAMP | Su sodA F3 | TGGCGTTATTATCTGATGTGT | [124] | |
| Su sodA B3 | AGAYCCAAAACGTCCCGT | |||||
| Su sodA FIP | ATGGTTAAGATGTCCGCCTCCCATCAATTCCAGAAGATATTCGT | |||||
| Su sodA BIP | TTCACCTGAGAAAACAGAAATCACTTCTTTAAATGCATCAAAAGAACC | |||||
| Su sodA Bloop | CGGAAGTAGCTTCTGCTATTGAT | |||||
| Su sodA FIP | DIG-ATGGTTAAGATGTCCGCCTCCCATCAATTCCAGAAGATATTCGT | |||||
| Milk beverages | Su sodA BIP | Biotin-TTCACCTGAGAAAACAGAAATCACTTCTTTAAATGCATCAAAAGAACC | ||||
| 2 | Streptococcus aureus | multiplex PCR | Sau 327/SAU1 | GGACGACATTAGACGAATCA | [130] | |
| Sau 1645/SAU2 | CGGGCACCTATTTTCTATCT | [131] | ||||
| STAUR4 | ACGGAGTTACAAAGGACGAC | [129] | ||||
| STAUR6 | AGCTCAGCCTTAACGAGTAC | |||||
| 3 | Streptococcus agalactiae | Sag 432/SAGA1 | CGTTGGTAGGAGTGGAAAAT | [130] | ||
| Sag 1018/SAGA2 | CTGCTCCGAAGAGAAAGCCT | [131] | ||||
| SIP3/SIP3 | TGAAAATGCAGGGCTCCAACCTCA | |||||
| SIP4/SIP4 | GATCTGGCATTGCATTCCAAGTAT | |||||
| 4 | Escherichia coli | Eco 2083/Ecoli1 | GCTTGACACTGAACATTGAG | [130] | ||
| Eco 2745/Ecoli2 | GCACTTATCTCTTCCGCATT | [131] | ||||
| FOP | CCGTTAAAGTGCC | |||||
| BOP | GCTTTACGTGCCGCC | |||||
| 5 | Cronobacter sakazakii | QLAMP | FIP | TGCTGCTCAACCGCCGATTTCTCCCCCCCCCCCCACCACCAAAGACA | [133] | |
| BIP | GATGAACGAGCTGCTGGCCGTCGATAATTTTGCCGA | |||||
| FLP | CACCTCGGAGGAGACC | |||||
| BLP | CTGCTGGAGAACCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nesterova, E.; Morozova, P.; Gladkikh, M.; Kazemzadeh, S.; Syromyatnikov, M. Molecular Methods for Detecting Microorganisms in Beverages. Beverages 2024, 10, 46. https://doi.org/10.3390/beverages10020046
Nesterova E, Morozova P, Gladkikh M, Kazemzadeh S, Syromyatnikov M. Molecular Methods for Detecting Microorganisms in Beverages. Beverages. 2024; 10(2):46. https://doi.org/10.3390/beverages10020046
Chicago/Turabian StyleNesterova, Ekaterina, Polina Morozova, Mariya Gladkikh, Shima Kazemzadeh, and Mikhail Syromyatnikov. 2024. "Molecular Methods for Detecting Microorganisms in Beverages" Beverages 10, no. 2: 46. https://doi.org/10.3390/beverages10020046
APA StyleNesterova, E., Morozova, P., Gladkikh, M., Kazemzadeh, S., & Syromyatnikov, M. (2024). Molecular Methods for Detecting Microorganisms in Beverages. Beverages, 10(2), 46. https://doi.org/10.3390/beverages10020046





