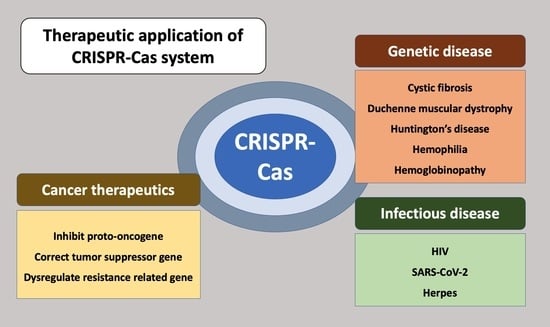
Therapeutic Applications of the CRISPR-Cas System
Abstract
:1. Introduction
2. Therapeutic Approach for Genetic Disease
2.1. Cystic Fibrosis
2.2. Duchenne Muscular Dystrophy
2.3. Huntington’s Disease
2.4. Hemophilia
2.5. Hemoglobinopathy
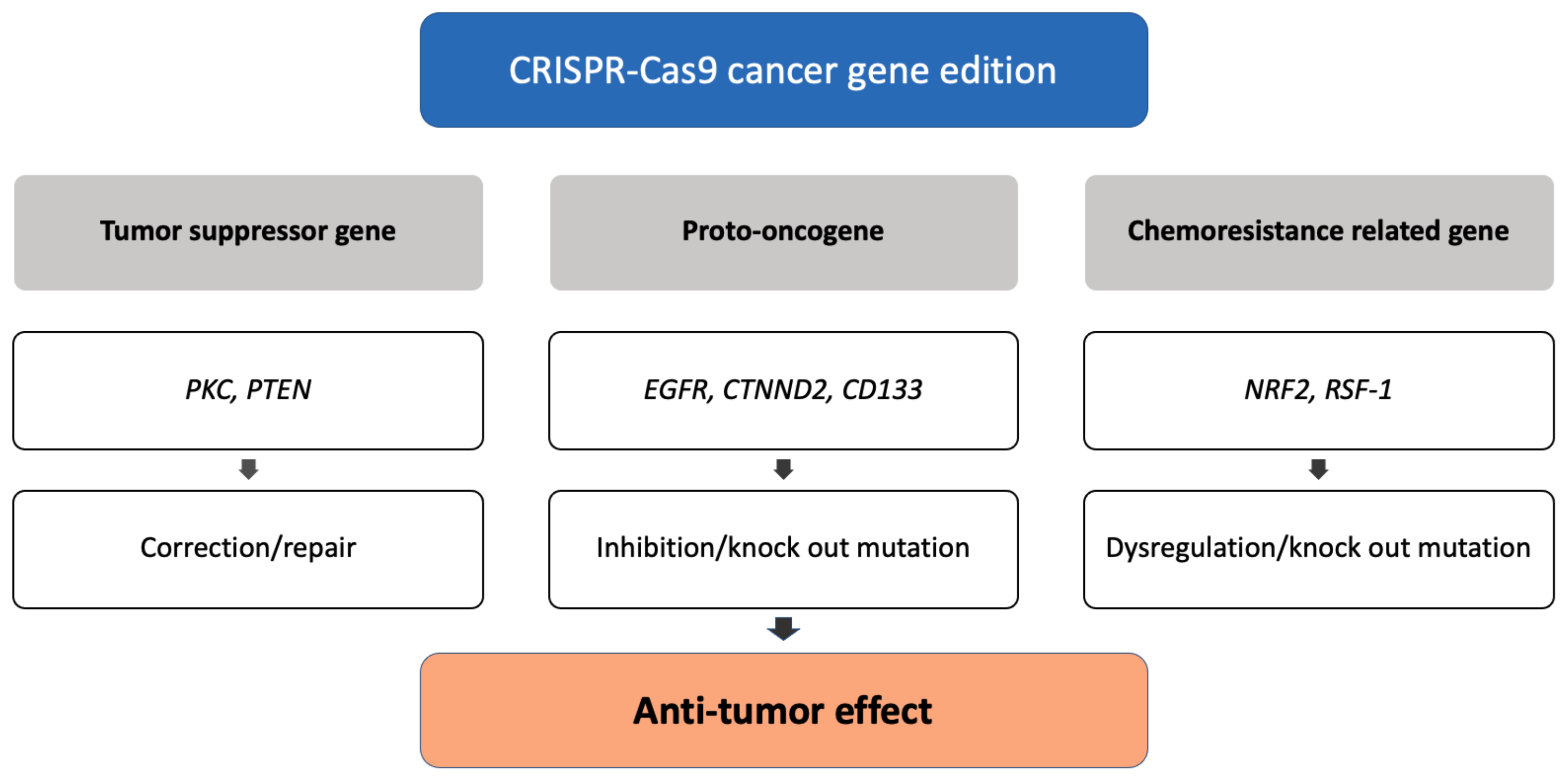
3. Cancer Therapeutics
3.1. Genome Editing of Cancer Cells
3.1.1. Correction of Tumor Suppressor Genes
3.1.2. Inhibition of Proto-Oncogenes
3.1.3. Dysregulation of Chemoresistance-Related Genes
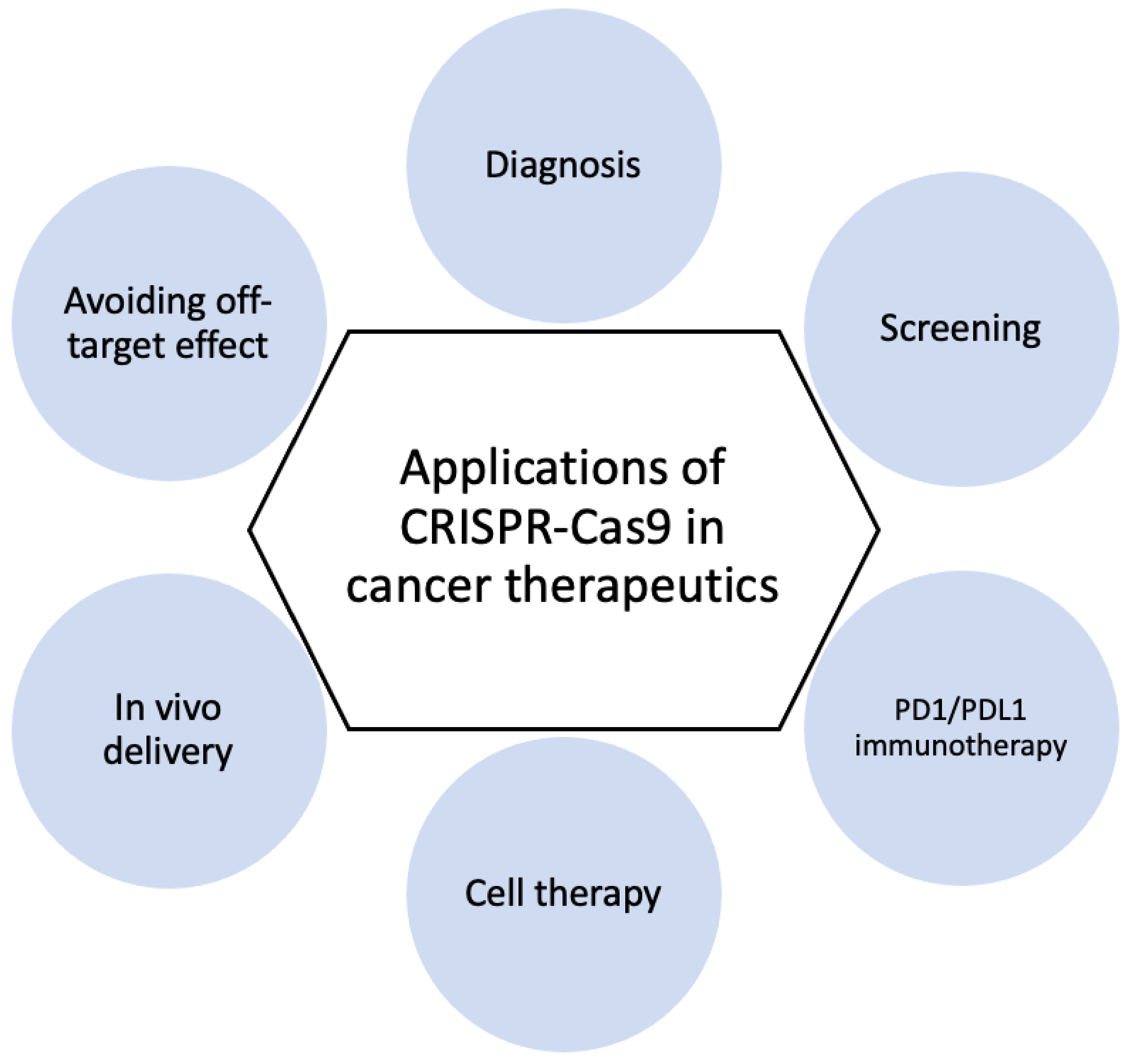
3.2. Cancer Diagnosis and Therapy
3.2.1. CRISPR-Based Diagnostics
3.2.2. CRISPR Screening
3.2.3. PD1/PDL1 Immunotherapy
3.2.4. Cell Therapy
3.2.5. In Vivo Delivery
3.2.6. Avoiding Off-Target Effect
4. Therapeutic Approaches for Infectious Disease
- The modification of receptors for viral entry: Interactions between viral proteins and cell membrane receptors allow the virus to enter the host cell. In addition to interfering with viral tropism, CRISPR-Cas-induced editing of receptor genes can prevent virus–receptor binding and limit virus entry and spread. Modifying these receptors, which also aid in viral genome replication and packaging, can impede viral multiplication [105].
- The segmentation of host viral factors: For replication and propagation, the virus is primarily dependent on host proteins. Some of the genes that encode proteins essential to viruses can be silenced using the CRISPR-Cas-induced knock out, preventing viral replication [106].
- The induction of host transcriptional restriction factor: These factors are restricted by the coupling of inactive Cas9 and viral RNA, which blocks replication and leads to a reduction in viral RNA [107].
- The excision and deletion of integrated viral genome: Viral genes may be excised using CRISPR-Cas in cases where viruses integrate their DNA into the host genome via the deletion and inactivation of genes [108].

4.1. HIV
- (1)
- C-X-C chemokine receptor type 4 (CXCR4) and C-C chemokine receptor type 5 (CCR5) are co-receptors required for HIV-1 entry into CD4+ T cells. Therefore, HIV infection can be prevented by using CRISPR-Cas9 to create a defect in this receptor, and the disruption of genes encoding these co-receptors showed no obvious cytotoxic effects on cell viability, as well as a significant protective effect against HIV-1 infection when compared to unmodified cells [111].
- (2)
- To treat latent HIV infection, proviral DNA integrated in the host genome must be inactivated. CRISPR-Cas9 targeting of the long terminal region (LTR) enables this. The results of gene editing of the HIV-1 LTR U3 region revealed that Cas9/gRNAs completely excised a 9709 bp fragment of integrated proviral DNA spanning from 5 to 3 LTRs, resulting in viral gene expression inactivation and virus replication restriction in HIV-1 latently infected cells. CRISPR-Cas9-targeted proviral DNA has also been shown to prevent new HIV infections [112]. HIV-1 RNA editing with CRISPR-Cas13 is another effective treatment for HIV eradication. The findings suggest that the CRISPR-Cas13 system can effectively inhibit HIV-1 in primary CD4+ T-cells and reduce HIV-1 reactivation in latently infected cells [113].
- (3)
- The activation of restriction factor expression in host cells could be an alternate method for preventing HIV-1 replication. Restriction factors found recently, including the human silencing hub (HUSH) and NONO, are expected to effectively suppress HIV replication through reactivation using the CRIPSR-Cas9 [114,115].
4.2. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)
4.3. Herpes Viruses
5. Limitations and Challenges
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Javed, M.R.; Sadaf, M.; Ahmed, T.; Jamil, A.; Nawaz, M.; Abbas, H.; Ijaz, A. CRISPR-Cas system: History and prospects as a genome editing tool in microorganisms. Curr. Microbiol. 2018, 75, 1675–1683. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Mohanraju, P.; Makarova, K.S.; Zetsche, B.; Zhang, F.; Koonin, E.V.; Van der Oost, J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 2016, 353, aad5147. [Google Scholar] [CrossRef]
- Jore, M.M.; Lundgren, M.; Van Duijn, E.; Bultema, J.B.; Westra, E.R.; Waghmare, S.P.; Wiedenheft, B.; Pul, Ü.; Wurm, R.; Wagner, R.; et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011, 18, 529–536. [Google Scholar] [CrossRef]
- Hayes, R.P.; Xiao, Y.; Ding, F.; Van Erp, P.B.; Rajashankar, K.; Bailey, S.; Wiedenheft, B.; Ke, A. Structural basis for promiscuous PAM recognition in type I–E Cascade from E. coli. Nature 2016, 530, 499–503. [Google Scholar] [CrossRef]
- Wiedenheft, B.; Lander, G.C.; Zhou, K.; Jore, M.M.; Brouns, S.J.; van der Oost, J.; Doudna, J.A.; Nogales, E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 2011, 477, 486–489. [Google Scholar] [CrossRef]
- Nam, K.H.; Haitjema, C.; Liu, X.; Ding, F.; Wang, H.; DeLisa, M.P.; Ke, A. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype IC/Dvulg CRISPR-Cas system. Structure 2012, 20, 1574–1584. [Google Scholar] [CrossRef]
- Riesenberg, S.; Helmbrecht, N.; Kanis, P.; Maricic, T.; Pääbo, S. Improved gRNA secondary structures allow editing of target sites resistant to CRISPR-Cas9 cleavage. Nat. Commun. 2022, 13, 489. [Google Scholar] [CrossRef]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [Green Version]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef]
- Schindele, P.; Wolter, F.; Puchta, H. Transforming plant biology and breeding with CRISPR/Cas9, Cas12 and Cas13. FEBS Lett. 2018, 592, 1954–1967. [Google Scholar] [CrossRef]
- Khan, F.A.; Pandupuspitasari, N.S.; Chun-Jie, H.; Ao, Z.; Jamal, M.; Zohaib, A.; Khan, F.A.; Hakim, M.R.; ShuJun, Z. CRISPR/Cas9 therapeutics: A cure for cancer and other genetic diseases. Oncotarget 2016, 7, 52541. [Google Scholar] [CrossRef]
- Hall, J.G. Genomic imprinting: Review and relevance to human diseases. Am. J. Hum. Genet. 1990, 46, 857. [Google Scholar]
- Ghasemi, N.; Razavi, S.; Nikzad, E. Multiple sclerosis: Pathogenesis, symptoms, diagnoses and cell-based therapy. Cell J. 2017, 19, 1. [Google Scholar]
- Men, K.; Duan, X.; He, Z.; Yang, Y.; Yao, S.; Wei, Y. CRISPR/Cas9-mediated correction of human genetic disease. Sci. China Life Sci. 2017, 60, 447–457. [Google Scholar] [CrossRef]
- Wu, Y.; Liang, D.; Wang, Y.; Bai, M.; Tang, W.; Bao, S.; Yan, Z.; Li, D.; Li, J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 2013, 13, 659–662. [Google Scholar] [CrossRef]
- Musunuru, K. The hope and hype of CRISPR-Cas9 genome editing: A review. JAMA Cardiol. 2017, 2, 914–919. [Google Scholar] [CrossRef]
- Wojtal, D.; Kemaladewi, D.U.; Malam, Z.; Abdullah, S.; Wong, T.W.; Hyatt, E.; Baghestani, Z.; Pereira, S.; Stavropoulos, J.; Mouly, V.; et al. Spell checking nature: Versatility of CRISPR/Cas9 for developing treatments for inherited disorders. Am. J. Hum. Genet. 2016, 98, 90–101. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020, 5, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Hopkin, K. Understanding Cystic Fibrosis; University Press of Mississippi: Jackson, MS, USA, 2010. [Google Scholar]
- Firth, A.L.; Menon, T.; Parker, G.S.; Qualls, S.J.; Lewis, B.M.; Ke, E.; Dargitz, C.T.; Wright, R.; Khanna, A.; Gage, F.H.; et al. Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Rep. 2015, 12, 1385–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marangi, M.; Pistritto, G. Innovative therapeutic strategies for cystic fibrosis: Moving forward to CRISPR technique. Front. Pharmacol. 2018, 9, 396. [Google Scholar] [CrossRef]
- Alton, E.W.; Armstrong, D.K.; Ashby, D.; Bayfield, K.J.; Bilton, D.; Bloomfield, E.V.; Boyd, A.C.; Brand, J.; Buchan, R.; Calcedo, R.; et al. Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: A randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2015, 3, 684–691. [Google Scholar] [CrossRef]
- Graham, C.; Hart, S. CRISPR/Cas9 gene editing therapies for cystic fibrosis. Expert Opin. Biol. Ther. 2021, 21, 767–780. [Google Scholar] [CrossRef]
- Duan, D.; Goemans, N.; Takeda, S.; Mercuri, E.; Aartsma-Rus, A. Duchenne muscular dystrophy. Nat. Rev. Dis. Prim. 2021, 7, 13. [Google Scholar] [CrossRef]
- Petrof, B.J. The molecular basis of activity-induced muscle injury in Duchenne muscular dystrophy. Mol. Cell. Biochem. 1998, 179, 111–124. [Google Scholar] [CrossRef]
- Lu, Q.L.; Yokota, T.; Takeda, S.; Garcia, L.; Muntoni, F.; Partridge, T. The status of exon skipping as a therapeutic approach to duchenne muscular dystrophy. Mol. Ther. 2011, 19, 9–15. [Google Scholar] [CrossRef]
- Amoasii, L.; Hildyard, J.C.; Li, H.; Sanchez-Ortiz, E.; Mireault, A.; Caballero, D.; Harron, R.; Stathopoulou, T.R.; Massey, C.; Shelton, J.M.; et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 2018, 362, 86–91. [Google Scholar] [CrossRef]
- Min, Y.L.; Li, H.; Rodriguez-Caycedo, C.; Mireault, A.A.; Huang, J.; Shelton, J.M.; McAnally, J.R.; Amoasii, L.; Mammen, P.P.; Bassel-Duby, R.; et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 2019, 5, eaav4324. [Google Scholar] [CrossRef]
- White, J.K.; Auerbach, W.; Duyao, M.P.; Vonsattel, J.P.; Gusella, J.F.; Joyner, A.L.; MacDonald, M.E. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat. Genet. 1997, 17, 404–410. [Google Scholar] [CrossRef]
- Ekman, F.K.; Ojala, D.S.; Adil, M.M.; Lopez, P.A.; Schaffer, D.V.; Gaj, T. CRISPR-Cas9-mediated genome editing increases lifespan and improves motor deficits in a Huntington’s disease mouse model. Mol.-Ther.-Nucleic Acids 2019, 17, 829–839. [Google Scholar] [CrossRef] [Green Version]
- Peyvandi, F.; Jayandharan, G.; Chandy, M.; Srivastava, A.; Nakaya, S.; Johnson, M.; Thompson, A.; Goodeve, A.; Garagiola, I.; Lavoretano, S.; et al. Genetic diagnosis of haemophilia and other inherited bleeding disorders. Haemophilia 2006, 12, 82–89. [Google Scholar] [CrossRef]
- Ohmori, T.; Nagao, Y.; Mizukami, H.; Sakata, A.; Muramatsu, S.i.; Ozawa, K.; Tominaga, S.i.; Hanazono, Y.; Nishimura, S.; Nureki, O.; et al. CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci. Rep. 2017, 7, 4159. [Google Scholar] [CrossRef]
- Stephens, C.J.; Lauron, E.J.; Kashentseva, E.; Lu, Z.H.; Yokoyama, W.M.; Curiel, D.T. Long-term correction of hemophilia B using adenoviral delivery of CRISPR/Cas9. J. Control. Release 2019, 298, 128–141. [Google Scholar] [CrossRef]
- Ohmori, T. Advances in gene therapy for hemophilia: Basis, current status, and future perspectives. Int. J. Hematol. 2020, 111, 31–41. [Google Scholar] [CrossRef]
- Sonati, M.d.F.; Costa, F.F. The genetics of blood disorders: Hereditary hemoglobinopathies. J. Pediatr. 2008, 84, S40–S51. [Google Scholar] [CrossRef]
- Moreno, A.D. Gene Therapy Approaches to Promote Fetal Hemoglobin Production for the Treatment of β-Hemoglobinopathies. Ph.D. Thesis, Eberhard Karls Universität Tübingen, Tübingen, Germany, 2021. [Google Scholar]
- Khosravi, M.A.; Abbasalipour, M.; Concordet, J.P.; Vom Berg, J.; Zeinali, S.; Arashkia, A.; Azadmanesh, K.; Buch, T.; Karimipoor, M. Targeted deletion of BCL11A gene by CRISPR-Cas9 system for fetal hemoglobin reactivation: A promising approach for gene therapy of beta thalassemia disease. Eur. J. Pharmacol. 2019, 854, 398–405. [Google Scholar] [CrossRef]
- Koninckx, P.R.; Ussia, A.; Adamyan, L.; Wattiez, A.; Gomel, V.; Martin, D.C. Pathogenesis of endometriosis: The genetic/epigenetic theory. Fertil. Steril. 2019, 111, 327–340. [Google Scholar] [CrossRef]
- Chira, S.; Gulei, D.; Hajitou, A.; Zimta, A.A.; Cordelier, P.; Berindan-Neagoe, I. CRISPR/Cas9: Transcending the reality of genome editing. Mol.-Ther.-Nucleic Acids 2017, 7, 211–222. [Google Scholar] [CrossRef]
- Kwok, M.; Wu, C.J. Clonal evolution of high-risk chronic lymphocytic leukemia: A contemporary perspective. Front. Oncol. 2021, 11, 790004. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrowicz, R.; Taciak, B.; Król, M. Drug delivery systems improving chemical and physical properties of anticancer drugs currently investigated for treatment of solid tumors. J. Physiol. Pharmacol. 2017, 68, 165–174. [Google Scholar]
- Liu, J.; Sareddy, G.R.; Zhou, M.; Viswanadhapalli, S.; Li, X.; Lai, Z.; Tekmal, R.R.; Brenner, A.; Vadlamudi, R.K. Differential effects of estrogen receptor β isoforms on glioblastoma progression. Cancer Res. 2018, 78, 3176–3189. [Google Scholar] [CrossRef] [Green Version]
- Maurya, A.K.; Vinayak, M. Anticarcinogenic action of quercetin by downregulation of phosphatidylinositol 3-kinase (PI3K) and protein kinase C (PKC) via induction of p53 in hepatocellular carcinoma (HepG2) cell line. Mol. Biol. Rep. 2015, 42, 1419–1429. [Google Scholar] [CrossRef]
- Isakov, N. Protein kinase C (PKC) isoforms in cancer, tumor promotion and tumor suppression. Semin. Cancer Biol. 2018, 48, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Antal, C.E.; Hudson, A.M.; Kang, E.; Zanca, C.; Wirth, C.; Stephenson, N.L.; Trotter, E.W.; Gallegos, L.L.; Miller, C.J.; Furnari, F.B.; et al. Cancer-associated protein kinase C mutations reveal kinase’s role as tumor suppressor. Cell 2015, 160, 489–502. [Google Scholar] [CrossRef]
- Moses, C.; Nugent, F.; Waryah, C.B.; Garcia-Bloj, B.; Harvey, A.R.; Blancafort, P. Activating PTEN tumor suppressor expression with the CRISPR/dCas9 system. Mol.-Ther.-Nucleic Acids 2019, 14, 287–300. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef]
- Koo, T.; Yoon, A.R.; Cho, H.Y.; Bae, S.; Yun, C.O.; Kim, J.S. Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Res. 2017, 45, 7897–7908. [Google Scholar] [CrossRef]
- Huang, F.; Chen, J.; Wang, Z.; Lan, R.; Fu, L.; Zhang, L. δ-Catenin promotes tumorigenesis and metastasis of lung adenocarcinoma. Oncol. Rep. 2018, 39, 809–817. [Google Scholar] [CrossRef]
- Jing, F.; Kim, H.J.; Kim, C.H.; Kim, Y.J.; Lee, J.H.; Kim, H.R. Colon cancer stem cell markers CD44 and CD133 in patients with colorectal cancer and synchronous hepatic metastases. Int. J. Oncol. 2015, 46, 1582–1588. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cho, M.Y.; Lee, S.; Jang, M.; Park, J.; Park, R. CRISPR-Cas9 mediated CD133 knockout inhibits colon cancer invasion through reduced epithelial-mesenchymal transition. PLoS ONE 2019, 14, e0220860. [Google Scholar] [CrossRef]
- Singh, A.; Boldin-Adamsky, S.; Thimmulappa, R.K.; Rath, S.K.; Ashush, H.; Coulter, J.; Blackford, A.; Goodman, S.N.; Bunz, F.; Watson, W.H.; et al. RNAi-mediated silencing of nuclear factor erythroid-2–related factor 2 gene expression in non–small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008, 68, 7975–7984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bialk, P.; Wang, Y.; Banas, K.; Kmiec, E.B. Functional gene knockout of NRF2 increases chemosensitivity of human lung cancer A549 cells in vitro and in a xenograft mouse model. Mol.-Ther.-Oncolytics 2018, 11, 75–89. [Google Scholar] [CrossRef]
- Chen, X.; Sun, X.; Guan, J.; Gai, J.; Xing, J.; Fu, L.; Shen, F.; Chen, K.; Li, W.; Han, L.; et al. Rsf-1 influences the sensitivity of non-small cell lung cancer to paclitaxel by regulating NF-κB pathway and its downstream proteins. Cell. Physiol. Biochem. 2017, 44, 2322–2336. [Google Scholar] [CrossRef] [PubMed]
- Rahman, N. Mainstreaming genetic testing of cancer predisposition genes. Clin. Med. 2014, 14, 436. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Burd, E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef]
- Michaud, D.S.; Langevin, S.M.; Eliot, M.; Nelson, H.H.; Pawlita, M.; McClean, M.D.; Kelsey, K.T. High-risk HPV types and head and neck cancer. Int. J. Cancer 2014, 135, 1653–1661. [Google Scholar] [CrossRef]
- Geng, Y.; Pertsinidis, A. Simple and versatile imaging of genomic loci in live mammalian cells and early pre-implantation embryos using CAS-LiveFISH. Sci. Rep. 2021, 11, 12220. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Zhang, F. High-throughput functional genomics using CRISPR–Cas9. Nat. Rev. Genet. 2015, 16, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Dersh, D.; Phelan, J.D.; Gumina, M.E.; Wang, B.; Arbuckle, J.H.; Holly, J.; Kishton, R.J.; Markowitz, T.E.; Seedhom, M.O.; Fridlyand, N.; et al. Genome-wide screens identify lineage-and tumor specific-genes modulating MHC-I and MHC-II immunosurveillance in human lymphomas. Immunity 2020, 54, 116–131.e10. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.S.; Zhang, W.; Wang, X.; Jiang, P.; Traugh, N.; Li, Z.; Meyer, C.; Stewig, B.; Xie, Y.; Bu, X.; et al. Therapeutically increasing MHC-I expression potentiates immune checkpoint blockade. Cancer Discov. 2021, 11, 1524–1541. [Google Scholar] [CrossRef]
- Wang, X.; Tokheim, C.; Gu, S.S.; Wang, B.; Tang, Q.; Li, Y.; Traugh, N.; Zeng, Z.; Zhang, Y.; Li, Z.; et al. In vivo CRISPR screens identify the E3 ligase Cop1 as a modulator of macrophage infiltration and cancer immunotherapy target. Cell 2021, 184, 5357–5374. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef]
- Tzelepis, K.; Koike-Yusa, H.; De Braekeleer, E.; Li, Y.; Metzakopian, E.; Dovey, O.M.; Mupo, A.; Grinkevich, V.; Li, M.; Mazan, M.; et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 2016, 17, 1193–1205. [Google Scholar] [CrossRef]
- Blank, C.; Mackensen, A. Contribution of the PD-L1/PD-1 pathway to T-cell exhaustion: An update on implications for chronic infections and tumor evasion. Cancer Immunol. Immunother. 2007, 56, 739–745. [Google Scholar] [CrossRef]
- Dong, H.; Strome, S.E.; Salomao, D.R.; Tamura, H.; Hirano, F.; Flies, D.B.; Roche, P.C.; Lu, J.; Zhu, G.; Tamada, K.; et al. Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 2002, 8, 793–800. [Google Scholar] [CrossRef]
- Su, S.; Hu, B.; Shao, J.; Shen, B.; Du, J.; Du, Y.; Zhou, J.; Yu, L.; Zhang, L.; Chen, F.; et al. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 2016, 6, 20070. [Google Scholar] [CrossRef]
- Cyranoski, D. CRISPR gene-editing tested in a person for the first time. Nature 2016, 539, 479. [Google Scholar] [CrossRef] [PubMed]
- Cyranoski, D. Chinese scientists to pioneer first human CRISPR trial. Nature 2016, 535, 476–477. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Li, J. CRISPR-Cas9 therapeutics in cancer: Promising strategies and present challenges. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2016, 1866, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Rafiq, S.; Hackett, C.S.; Brentjens, R.J. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol. 2020, 17, 147–167. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.j.; Yin, E.T.S.; Hu, Y.x.; Huang, H. Combination of CRISPR/Cas9 System and CAR-T Cell Therapy: A New Era for Refractory and Relapsed Hematological Malignancies. Curr. Med. Sci. 2021, 41, 420–430. [Google Scholar] [CrossRef]
- Hong, M.; Clubb, J.D.; Chen, Y.Y. Engineering CAR-T cells for next-generation cancer therapy. Cancer Cell 2020, 38, 473–488. [Google Scholar] [CrossRef]
- Sharma, G.; Sharma, A.R.; Bhattacharya, M.; Lee, S.S.; Chakraborty, C. CRISPR-Cas9: A preclinical and clinical perspective for the treatment of human diseases. Mol. Ther. 2021, 29, 571–586. [Google Scholar] [CrossRef]
- Zhao, Y.; Moon, E.; Carpenito, C.; Paulos, C.M.; Liu, X.; Brennan, A.L.; Chew, A.; Carroll, R.G.; Scholler, J.; Levine, B.L.; et al. Multiple Injections of Electroporated Autologous T Cells Expressing a Chimeric Antigen Receptor Mediate Regression of Human Disseminated TumorAutologous RNA CAR T Cells Mediate Tumor Regression. Cancer Res. 2010, 70, 9053–9061. [Google Scholar] [CrossRef]
- Ren, J.; Liu, X.; Fang, C.; Jiang, S.; June, C.H.; Zhao, Y. Multiplex Genome Editing to Generate Universal CAR T Cells Resistant to PD1 InhibitionMultiplex Genome Editing to Generate Universal CAR T Cells. Clin. Cancer Res. 2017, 23, 2255–2266. [Google Scholar] [CrossRef]
- Eyquem, J.; Mansilla-Soto, J.; Giavridis, T.; van der Stegen, S.J.; Hamieh, M.; Cunanan, K.M.; Odak, A.; Gönen, M.; Sadelain, M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 2017, 543, 113–117. [Google Scholar] [CrossRef]
- Liang, X.; Potter, J.; Kumar, S.; Zou, Y.; Quintanilla, R.; Sridharan, M.; Carte, J.; Chen, W.; Roark, N.; Ranganathan, S.; et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015, 208, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.A. State-of-the-art gene-based therapies: The road ahead. Nat. Rev. Genet. 2011, 12, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Naso, M.F.; Tomkowicz, B.; Perry, W.L.; Strohl, W.R. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 2017, 31, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Kalaydina, R.V.; Bajwa, K.; Qorri, B.; Decarlo, A.; Szewczuk, M.R. Recent advances in “smart” delivery systems for extended drug release in cancer therapy. Int. J. Nanomed. 2018, 13, 4727. [Google Scholar] [CrossRef] [PubMed]
- Palmerston Mendes, L.; Pan, J.; Torchilin, V.P. Dendrimers as nanocarriers for nucleic acid and drug delivery in cancer therapy. Molecules 2017, 22, 1401. [Google Scholar] [CrossRef]
- Chen, F.; Alphonse, M.; Liu, Q. Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1609. [Google Scholar] [CrossRef]
- Xu, X.; Liu, C.; Wang, Y.; Koivisto, O.; Zhou, J.; Shu, Y.; Zhang, H. Nanotechnology-based delivery of CRISPR/Cas9 for cancer treatment. Adv. Drug Deliv. Rev. 2021, 176, 113891. [Google Scholar] [CrossRef]
- Song, R.; Murphy, M.; Li, C.; Ting, K.; Soo, C.; Zheng, Z. Current development of biodegradable polymeric materials for biomedical applications. Drug Des. Dev. Ther. 2018, 12, 3117. [Google Scholar] [CrossRef]
- Basha, M. Nanotechnology as a promising strategy for anticancer drug delivery. Curr. Drug Deliv. 2018, 15, 497–509. [Google Scholar] [CrossRef]
- Battaglia, L.; Gallarate, M. Lipid nanoparticles: State of the art, new preparation methods and challenges in drug delivery. Expert Opin. Drug Deliv. 2012, 9, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Iturrioz-Rodríguez, N.; Correa-Duarte, M.A.; Fanarraga, M.L. Controlled drug delivery systems for cancer based on mesoporous silica nanoparticles. Int. J. Nanomed. 2019, 14, 3389. [Google Scholar] [CrossRef] [PubMed]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Choi, K.S.; Warther, D.; Huffman, K.; Landeros, S.; Freeman, W.R.; Sailor, M.J.; Cheng, L. A sustained dual drug delivery system for proliferative vitreoretinopathy. Drug Deliv. 2020, 27, 1461–1473. [Google Scholar] [CrossRef]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.R.J.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef]
- Shen, C.C.; Hsu, M.N.; Chang, C.W.; Lin, M.W.; Hwu, J.R.; Tu, Y.; Hu, Y.C. Synthetic switch to minimize CRISPR off-target effects by self-restricting Cas9 transcription and translation. Nucleic Acids Res. 2019, 47, e13. [Google Scholar] [CrossRef]
- Gonçalves, B.C.; Lopes Barbosa, M.G.; Silva Olak, A.P.; Belebecha Terezo, N.; Nishi, L.; Watanabe, M.A.; Marinello, P.; Zendrini Rechenchoski, D.; Dejato Rocha, S.P.; Faccin-Galhardi, L.C. Antiviral therapies: Advances and perspectives. Fundam. Clin. Pharmacol. 2021, 35, 305–320. [Google Scholar] [CrossRef]
- Traylen, C.M.; Patel, H.R.; Fondaw, W.; Mahatme, S.; Williams, J.F.; Walker, L.R.; Dyson, O.F.; Arce, S.; Akula, S.M. Virus reactivation: A panoramic view in human infections. Future Virol. 2011, 6, 451–463. [Google Scholar] [CrossRef]
- Virgin, H.W.; Wherry, E.J.; Ahmed, R. Redefining chronic viral infection. Cell 2009, 138, 30–50. [Google Scholar] [CrossRef] [PubMed]
- Schiffer, J.T.; Aubert, M.; Weber, N.D.; Mintzer, E.; Stone, D.; Jerome, K.R. Targeted DNA mutagenesis for the cure of chronic viral infections. J. Virol. 2012, 86, 8920–8936. [Google Scholar] [CrossRef] [PubMed]
- McDougall, W.M.; Perreira, J.M.; Reynolds, E.C.; Brass, A.L. CRISPR genetic screens to discover host–virus interactions. Curr. Opin. Virol. 2018, 29, 87–100. [Google Scholar] [CrossRef]
- Nuñez, J.K.; Bai, L.; Harrington, L.B.; Hinder, T.L.; Doudna, J.A. CRISPR immunological memory requires a host factor for specificity. Mol. Cell 2016, 62, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Bogerd, H.P.; Kornepati, A.V.; Marshall, J.B.; Kennedy, E.M.; Cullen, B.R. Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proc. Natl. Acad. Sci. USA 2015, 112, E7249–E7256. [Google Scholar] [CrossRef]
- Li, H.; Sheng, C.; Wang, S.; Yang, L.; Liang, Y.; Huang, Y.; Liu, H.; Li, P.; Yang, C.; Yang, X.; et al. Removal of integrated hepatitis B virus DNA using CRISPR-Cas9. Front. Cell. Infect. Microbiol. 2017, 7, 91. [Google Scholar] [CrossRef]
- Fanales-Belasio, E.; Raimondo, M.; Suligoi, B.; Buttò, S. HIV virology and pathogenetic mechanisms of infection: A brief overview. Ann. Dell’istituto Super. Sanita 2010, 46, 5–14. [Google Scholar] [CrossRef]
- Lengauer, T.; Sing, T. Bioinformatics-assisted anti-HIV therapy. Nat. Rev. Microbiol. 2006, 4, 790–797. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, S.; Jin, X.; Wang, Q.; Yang, K.; Li, C.; Xiao, Q.; Hou, P.; Liu, S.; Wu, S.; et al. Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4+ T cells from HIV-1 infection. Cell Biosci. 2017, 7, 47. [Google Scholar] [CrossRef]
- Hu, W.; Kaminski, R.; Yang, F.; Zhang, Y.; Cosentino, L.; Li, F.; Luo, B.; Alvarez-Carbonell, D.; Garcia-Mesa, Y.; Karn, J.; et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11461–11466. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.; Wilson, H.; Jayakumar, S.; Kulkarni, V.; Kulkarni, S. Efficient inhibition of HIV using CRISPR/Cas13d nuclease system. Viruses 2021, 13, 1850. [Google Scholar] [CrossRef] [PubMed]
- Chougui, G.; Munir-Matloob, S.; Matkovic, R.; Martin, M.M.; Morel, M.; Lahouassa, H.; Leduc, M.; Ramirez, B.C.; Etienne, L.; Margottin-Goguet, F. HIV-2/SIV viral protein X counteracts HUSH repressor complex. Nat. Microbiol. 2018, 3, 891–897. [Google Scholar] [CrossRef]
- Lahaye, X.; Gentili, M.; Silvin, A.; Conrad, C.; Picard, L.; Jouve, M.; Zueva, E.; Maurin, M.; Nadalin, F.; Knott, G.J.; et al. NONO detects the nuclear HIV capsid to promote cGAS-mediated innate immune activation. Cell 2018, 175, 488–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Feng, F.; Hu, G.; Wang, Y.; Yu, Y.; Zhu, Y.; Xu, W.; Cai, X.; Sun, Z.; Han, W.; et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021, 12, 961. [Google Scholar] [CrossRef] [PubMed]
- Javalkote, V.S.; Kancharla, N.; Bhadra, B.; Shukla, M.; Soni, B.; Sapre, A.; Goodin, M.; Bandyopadhyay, A.; Dasgupta, S. CRISPR-based assays for rapid detection of SARS-CoV-2. Methods 2020, 203, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R.; et al. Development of CRISPR as a prophylactic strategy to combat novel coronavirus and influenza. bioRxiv 2020. [Google Scholar] [CrossRef]
- Tanaka, P.; Santos, J.; Oliveira, E.; Miglioli, N.; Assis, A.; Monteleone-Cassiano, A.; Ribeiro, V.; Duarte, M.; Machado, M.; Mascarenhas, R.; et al. A Crispr-Cas9 system designed to introduce point mutations into the human ACE2 gene to weaken the interaction of the ACE2 receptor with the SARS-CoV-2 S protein. Preprints 2020. [Google Scholar] [CrossRef]
- Van Diemen, F.R.; Kruse, E.M.; Hooykaas, M.J.; Bruggeling, C.E.; Schürch, A.C.; van Ham, P.M.; Imhof, S.M.; Nijhuis, M.; Wiertz, E.J.; Lebbink, R.J. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016, 12, e1005701. [Google Scholar] [CrossRef]
- Gergen, J.; Coulon, F.; Creneguy, A.; Elain-Duret, N.; Gutierrez, A.; Pinkenburg, O.; Verhoeyen, E.; Anegon, I.; Nguyen, T.H.; Halary, F.A.; et al. Multiplex CRISPR/Cas9 system impairs HCMV replication by excising an essential viral gene. PLoS ONE 2018, 13, e0192602. [Google Scholar] [CrossRef]
- Karpov, D.; Karpov, V.; Klimova, R.; Demidova, N.; Kushch, A. A plasmid-expressed CRISPR/Cas9 system suppresses replication of HSV type I in a vero cell culture. Mol. Biol. 2019, 53, 70–78. [Google Scholar] [CrossRef]
- Yin, D.; Ling, S.; Wang, D.; Dai, Y.; Jiang, H.; Zhou, X.; Paludan, S.R.; Hong, J.; Cai, Y. Targeting herpes simplex virus with CRISPR–Cas9 cures herpetic stromal keratitis in mice. Nat. Biotechnol. 2021, 39, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Q.; Joung, J.K. Defining and improving the genome-wide specificities of CRISPR–Cas9 nucleases. Nat. Rev. Genet. 2016, 17, 300–312. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wang, H.; Wei, Y.; Jiang, Z.; Tang, Y.; Chen, Y.; Xu, H. GuidePro: A multi-source ensemble predictor for prioritizing sgRNAs in CRISPR/Cas9 protein knockouts. Bioinformatics 2021, 37, 134–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Qin, C.; An, C.; Zheng, X.; Wen, S.; Chen, W.; Liu, X.; Lv, Z.; Yang, P.; Xu, W.; et al. Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol. Cancer 2021, 20, 126. [Google Scholar] [CrossRef]
- Zhuo, C.; Zhang, J.; Lee, J.H.; Jiao, J.; Cheng, D.; Liu, L.; Kim, H.W.; Tao, Y.; Li, M. Spatiotemporal control of CRISPR/Cas9 gene editing. Signal Transduct. Target. Ther. 2021, 6, 238. [Google Scholar] [CrossRef]
- Wei, T.; Cheng, Q.; Farbiak, L.; Anderson, D.G.; Langer, R.; Siegwart, D.J. Delivery of tissue-targeted scalpels: Opportunities and challenges for in vivo CRISPR/Cas-based genome editing. ACS Nano 2020, 14, 9243–9262. [Google Scholar] [CrossRef]
- Mehta, A.; Merkel, O.M. Immunogenicity of Cas9 protein. J. Pharm. Sci. 2020, 109, 62–67. [Google Scholar] [CrossRef]
- Sullivan, N.T.; Allen, A.G.; Atkins, A.J.; Chung, C.H.; Dampier, W.; Nonnemacher, M.R.; Wigdahl, B. Designing safer CRISPR/Cas9 therapeutics for HIV: Defining factors that regulate and technologies used to detect off-target editing. Front. Microbiol. 2020, 11, 1872. [Google Scholar] [CrossRef]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef]
- Aquino-Jarquin, G. Current advances in overcoming obstacles of CRISPR/Cas9 off-target genome editing. Mol. Genet. Metab. 2021, 134, 77–86. [Google Scholar] [CrossRef]
- Simeonov, D.R.; Brandt, A.J.; Chan, A.Y.; Cortez, J.T.; Li, Z.; Woo, J.M.; Lee, Y.; Carvalho, C.; Indart, A.C.; Roth, T.L.; et al. A large CRISPR-induced bystander mutation causes immune dysregulation. Commun. Biol. 2019, 2, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| NCT No. | Disease Type | Disease | Target | Intervention | Phase | Country |
|---|---|---|---|---|---|---|
| NCT03655678 | genetic disease | -thalassemia | disruption of the erythroid | ex vivo-modified hematopietic stem cell | I/II | USA |
| NCT04208529 | genetic disease | -thalassemia | disruption of the erythroid | ex vivo-modified hematopietic stem cell | I/II | USA |
| NCT03745287 | genetic disease | sickle cell disease | disruption of the erythroid | ex vivo-modified hematopietic stem cell | I/II | USA |
| NCT04925206 | genetic disease | -thalassemia | disruption of the erythroid | ex vivo-modified hematopietic stem cell | I | China |
| NCT04774536 | genetic disease | Sickle cell disease | disruption of the erythroid | ex vivo-modified hematopietic stem cell | I/II | USA |
| NCT03872479 | genetic disease | Congenital Amaurosis | eliminate CEP290 mutation | gene editing product | I | USA |
| NCT04601051 | genetic disease | Amyloidosis | disruption of the amyloid | Gene edit product in nanoparticle | I | UK, Swden |
| NCT04637763 | cancer | B-cell lymphoma | creation of CD19-directed T cell | CAR-T cell to CD19 | I | USA |
| NCT04035434 | cancer | B-cell lymphoma | creation of CD19-directed T cell | CAR-T cell to CD19 | I | USA |
| NCT05066165 | cancer | Acute Myeloid Leukemia | create CD19-directed T cell | CAR-T cell to WT1 | I | USA |
| NCT02793856 | cancer | Non small cell lung cancer | PD-1 knock out | CAR-T cell with PD-1 knock out | I | China |
| NCT04842812 | cancer | solid tumor | PD-1 knock out | CAR-T cell with PD-1 knock out | I | China |
| NCT04990557 | Infectious disease | COVID-19 | PD1 and ACE2 knockout | ex vivo-modified T cell | I/II | not specified |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, K.; Song, Y.; Kim, I.; Kim, T.-J. Therapeutic Applications of the CRISPR-Cas System. Bioengineering 2022, 9, 477. https://doi.org/10.3390/bioengineering9090477
Kang K, Song Y, Kim I, Kim T-J. Therapeutic Applications of the CRISPR-Cas System. Bioengineering. 2022; 9(9):477. https://doi.org/10.3390/bioengineering9090477
Chicago/Turabian StyleKang, Kyungmin, Youngjae Song, Inho Kim, and Tae-Jung Kim. 2022. "Therapeutic Applications of the CRISPR-Cas System" Bioengineering 9, no. 9: 477. https://doi.org/10.3390/bioengineering9090477