NDG-CAM: Nuclei Detection in Histopathology Images with Semantic Segmentation Networks and Grad-CAM
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
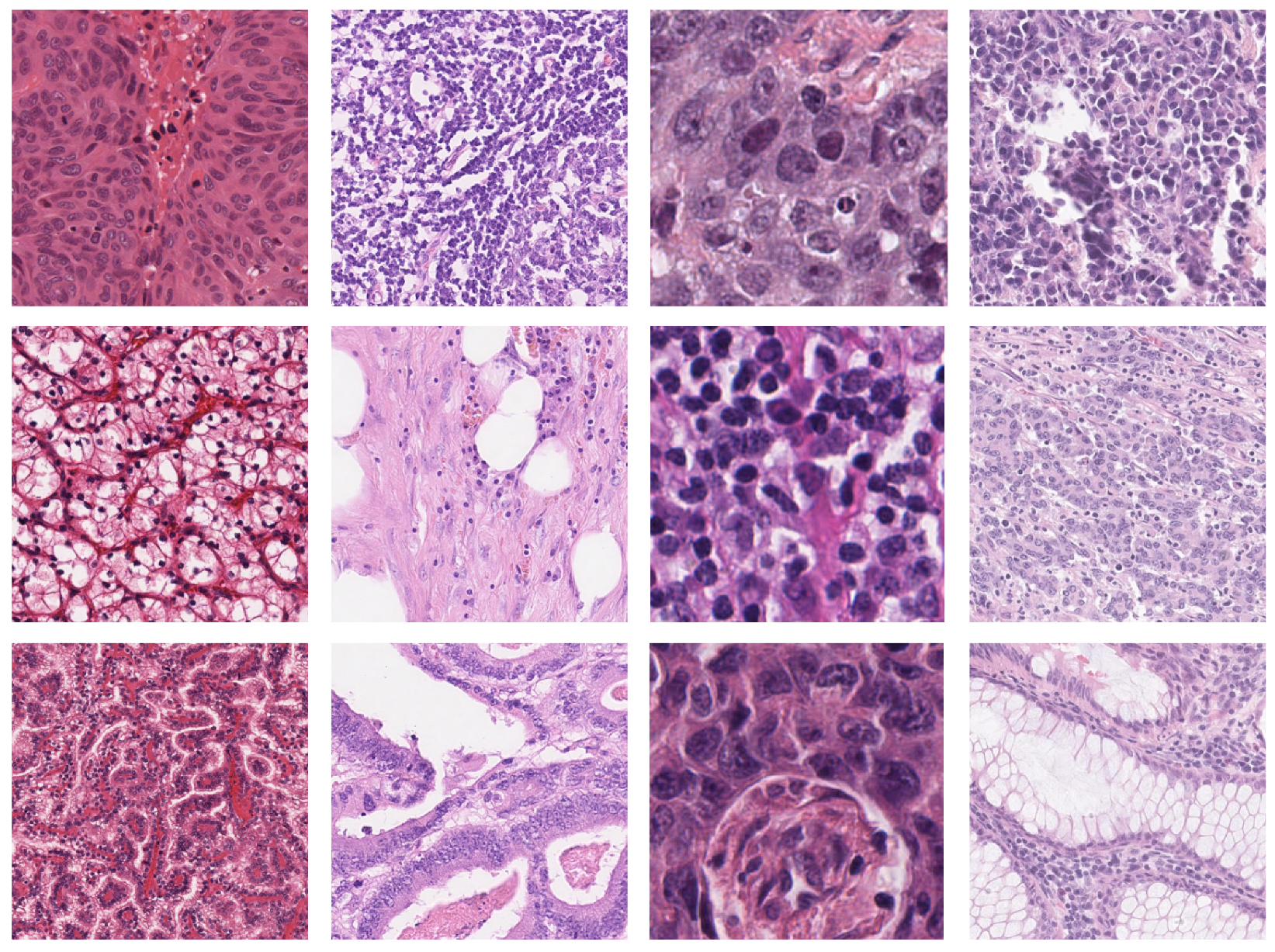
- MoNuSeg [1,18,19]. The cell nucleus segmentation dataset used in this work is publicly accessible from the 2018 Data Science Bowl challenge [20]. The dataset contains a large number of segmented nuclei images and includes different cell types; there are 30 training H&E images containing 21,623 hand-annotated nuclear boundaries from the breast, kidney, prostate, liver, colon, bladder, and stomach. Moreover, there are also 14 H&E test images containing 7000 nuclear boundary annotations from the breast, kidney, prostate, colon, bladder, lung, and brain. All images, each of size 1000 × 1000, were captured at 40× magnification. The nuclear contour annotations are provided through XML files.
- CRCHistoPhenotypes: Labeled Cell Nuclei Data [14,21]. This publicly available dataset contains 100 H&E-stained histology images of colon cell nuclei obtained from WSI of 10 patients with a magnification factor of 20×. Tiles have a size of 500 × 500. Nuclear annotations are provided through the coordinates of the centroids in .mat format, resulting in a total of 29,756 annotated nuclei for detection purposes.
- NuCLS [22]. The dataset contains over 220,000 labeled nuclei from breast cancer images from TCGA, obtained from 125 patients with breast cancer (1 slide per patient) and captured with a magnification factor of 40×. These nuclei were annotated through the collaborative effort of pathologists, pathology residents, and medical students. Data from both single-rater and multi-rater studies are provided. For single-rater data, there are both pathologist-reviewed and uncorrected annotations. For multi-rater datasets, there are annotations generated with and without suggestions from weak segmentation and classification algorithms. We used only the single-rater dataset, which is already split into train and test sets. The annotations for the single-rater dataset include 59,485 nuclei and 19,680 boundaries, extracted from 1744 H&E image tiles of variable dimensions between 200 and 400 pixels.
- Local dataset from Pathology Department of IRCCS Istituto Tumori Giovanni Paolo II [23]. This consists of 19 H&E image tiles which overall contain more than 6378 nuclei from patients with colorectal cancer. Images have a size of 512 × 512 and were captured at 40× magnification. Annotations have been provided by a biologist with experience in analyzing histopathological data.
2.2. NDG-CAM
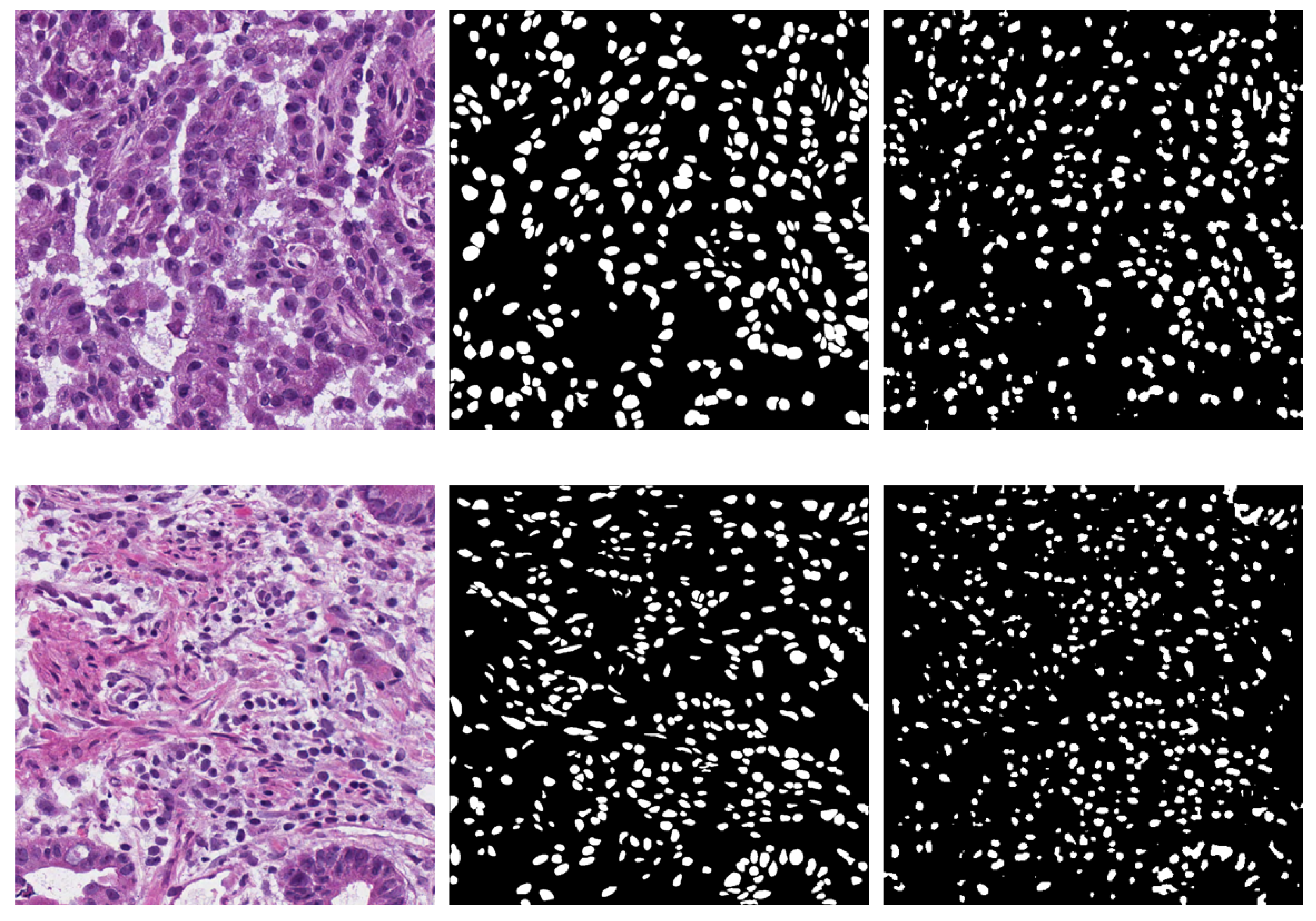
2.2.1. Semantic Segmentation Workflow
- a
- Train on D2 and validation on V1 at 20× resolution.
- b
- Train on T1 and validation on V1 at 20× resolution.
- c
- Train on T1 and validation on V1 at 40× resolution.
2.2.2. Network Architectures
- U-Net [24]. It is a fully convolutional network to perform the semantic segmentation task. The U-Net architecture consists of a series of encoding layers and contractions that are used to extract the context of the image, followed by a sequence of symmetrical decoding layers and expansions to recover the spatial information. In our MATLAB setting, the network is characterized by 58 convolutional layers; the first layer deals with a z-score normalization of the inputs, whereas the last one presents the Dice function as a loss function.
- SegNet [25]. This is another encoder–decoder architecture. In this case, the decoding blocks exploit max pooling indices received from the corresponding contraction block to perform the oversampling, instead of using trainable upsampling layers as transposed convolutions. In our MATLAB setting, this CNN consists of 31 layers with a cross-entropy loss function.
- DeepLab v3+ [26]. This architecture features atrous spatial pyramid pooling (ASPP) and the encoder–decoder paradigm. The first aspect concerns a particular way of combining layers of atrous and depthwise convolution, with which the model captures and concatenates features at different scales. For this network, the backbone is customizable. Three different basic CNN encoders were used: ResNet18, ResNet50, and MobileNet-v2. The DeepLab v3+ has 100 layers, of which the last is a softmax layer that is used to obtain the probabilities that each pixel belongs to the nucleus or background class; in this case, the chosen loss function is the Dice loss.
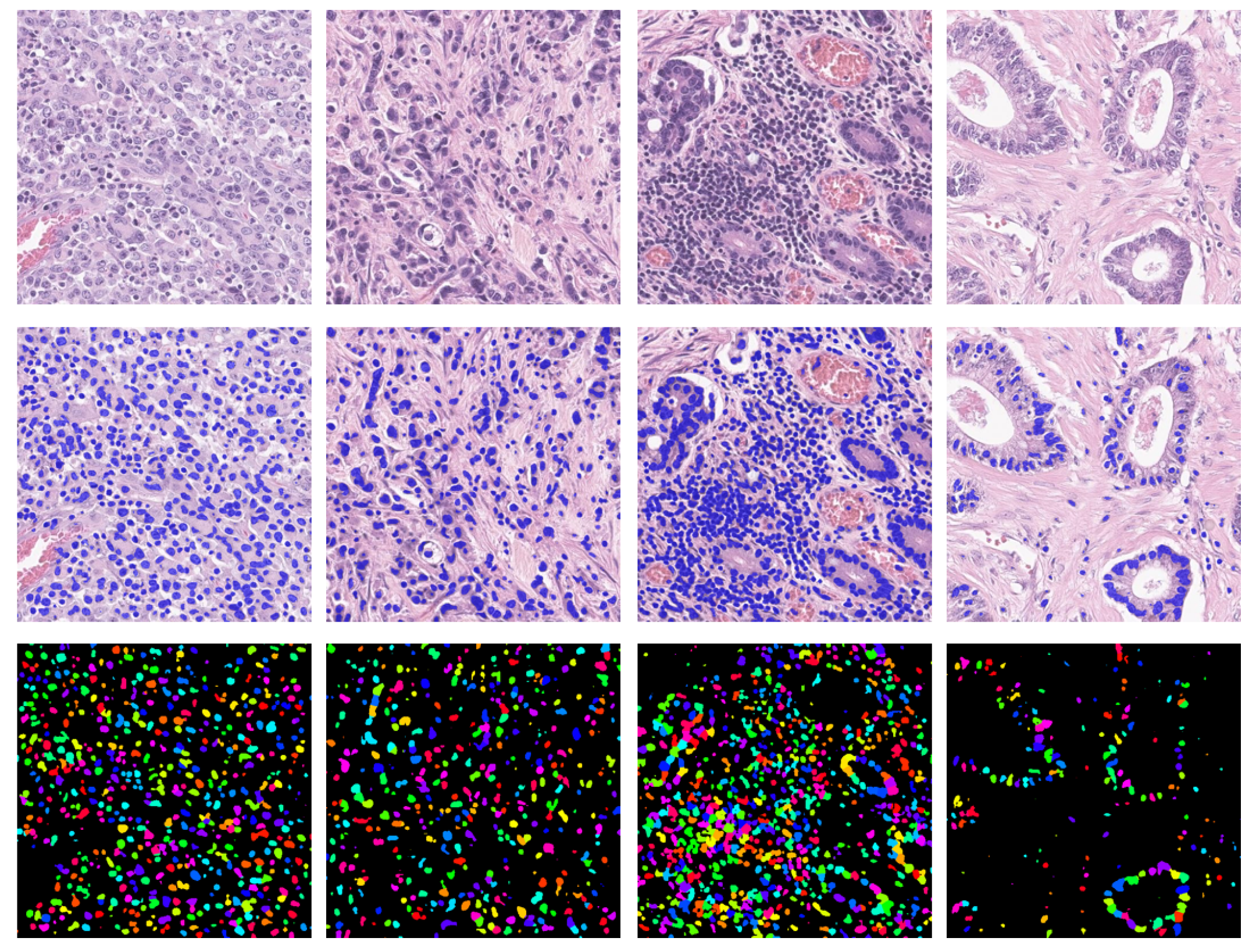
2.2.3. Nuclei Detection with Grad-CAM
2.3. Instance Segmentation
2.4. Implementation Details
2.5. Combined Model
2.6. Evaluation Metrics
| Algorithm 1: Object Detection TP, FP, FN calculation. |
 |
3. Results
4. Discussion
5. Conclusions and Future Works
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, N.; Verma, R.; Sharma, S.; Bhargava, S.; Vahadane, A.; Sethi, A. A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans. Med. Imaging 2017, 36, 1550–1560. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, F.; Borders, D.; Chen, R.J.; McKay, G.N.; Salimian, K.J.; Baras, A.; Durr, N.J. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans. Med. Imaging 2019, 39, 3257–3267. [Google Scholar] [CrossRef] [PubMed]
- Höfener, H.; Homeyer, A.; Weiss, N.; Molin, J.; Lundström, C.F.; Hahn, H.K. Deep learning nuclei detection: A simple approach can deliver state-of-the-art results. Comput. Med. Imaging Graph. 2018, 70, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Alom, Z.; Asari, V.K.; Parwani, A.; Taha, T.M. Microscopic nuclei classification, segmentation, and detection with improved deep convolutional neural networks (DCNN). Diagn. Pathol. 2022, 17, 38. [Google Scholar] [CrossRef]
- Shu, J.; Fu, H.; Qiu, G.; Kaye, P.; Ilyas, M. Segmenting overlapping cell nuclei in digital histopathology images. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5445–5448. [Google Scholar]
- Xu, J.; Xiang, L.; Liu, Q.; Gilmore, H.; Wu, J.; Tang, J.; Madabhushi, A. Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images. IEEE Trans. Med. Imaging 2015, 35, 119–130. [Google Scholar] [CrossRef]
- Sornapudi, S.; Stanley, R.J.; Stoecker, W.V.; Almubarak, H.; Long, R.; Antani, S.; Thoma, G.; Zuna, R.; Frazier, S.R. Deep learning nuclei detection in digitized histology images by superpixels. J. Pathol. Inform. 2018, 9, 5. [Google Scholar] [CrossRef]
- Larson, N.B.; Fridley, B.L. PurBayes: Estimating tumor cellularity and subclonality in next-generation sequencing data. Bioinformatics 2013, 29, 1888–1889. [Google Scholar] [CrossRef]
- Prencipe, B.; Altini, N.; Cascarano, G.D.; Brunetti, A.; Guerriero, A.; Bevilacqua, V. Focal Dice Loss-Based V-Net for Liver Segments Classification. Appl. Sci. 2022, 12, 3247. [Google Scholar] [CrossRef]
- Altini, N.; Brunetti, A.; Napoletano, V.P.; Girardi, F.; Allegretti, E.; Hussain, S.M.; Brunetti, G.; Triggiani, V.; Bevilacqua, V.; Buongiorno, D. A Fusion Biopsy Framework for Prostate Cancer Based on Deformable Superellipses and nnU-Net. Bioengineering 2022, 9, 343. [Google Scholar] [CrossRef]
- Altini, N.; Cascarano, G.D.; Brunetti, A.; Marino, F.; Rocchetti, M.T.; Matino, S.; Venere, U.; Rossini, M.; Pesce, F.; Gesualdo, L.; et al. semantic segmentation framework for glomeruli detection and classification in kidney histological sections. Electronics 2020, 9, 503. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Failmezger, H.; Rueda, O.M.; Ali, H.R.; Gräf, S.; Chin, S.F.; Schwarz, R.F.; Curtis, C.; Dunning, M.J.; Bardwell, H.; et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci. Transl. Med. 2012, 4, 157ra143. [Google Scholar] [CrossRef] [PubMed]
- Kuse, M.; Wang, Y.F.; Kalasannavar, V.; Khan, M.; Rajpoot, N. Local isotropic phase symmetry measure for detection of beta cells and lymphocytes. J. Pathol. Inform. 2011, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Sirinukunwattana, K.; Raza, S.E.A.; Tsang, Y.W.; Snead, D.R.; Cree, I.A.; Rajpoot, N.M. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imaging 2016, 35, 1196–1206. [Google Scholar] [CrossRef]
- Alom, M.Z.; Yakopcic, C.; Hasan, M.; Taha, T.M.; Asari, V.K. Recurrent residual U-Net for medical image segmentation. J. Med. Imaging 2019, 6, 014006. [Google Scholar] [CrossRef]
- Kainz, P.; Urschler, M.; Schulter, S.; Wohlhart, P.; Lepetit, V. You should use regression to detect cells. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 276–283. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- MoNuSeg—Grand Challenge. Available online: https://monuseg.grand-challenge.org/Data/ (accessed on 7 April 2022).
- Kumar, N.; Verma, R.; Anand, D.; Zhou, Y.; Onder, O.F.; Tsougenis, E.; Chen, H.; Heng, P.A.; Li, J.; Hu, Z.; et al. A multi-organ nucleus segmentation challenge. IEEE Trans. Med. Imaging 2019, 39, 1380–1391. [Google Scholar] [CrossRef] [PubMed]
- Caicedo, J.C.; Goodman, A.; Karhohs, K.W.; Cimini, B.A.; Ackerman, J.; Haghighi, M.; Heng, C.; Becker, T.; Doan, M.; McQuin, C.; et al. Nucleus segmentation across imaging experiments: The 2018 Data Science Bowl. Nat. Methods 2019, 16, 1247–1253. [Google Scholar] [CrossRef]
- CRCHistoPhenotypes—Labeled Cell Nuclei Data, Tissue Image Analytics (TIA) Centre, Warwick. Available online: https://warwick.ac.uk/fac/cross_fac/tia/data/crchistolabelednucleihe (accessed on 7 April 2022).
- Amgad, M.; Elfandy, H.; Hussein, H.; Atteya, L.A.; Elsebaie, M.A.; Abo Elnasr, L.S.; Sakr, R.A.; Salem, H.S.; Ismail, A.F.; Saad, A.M.; et al. Structured crowdsourcing enables convolutional segmentation of histology images. Bioinformatics 2019, 35, 3461–3467. [Google Scholar] [CrossRef]
- Altini, N.; Marvulli, T.M.; Caputo, M.; Mattioli, E.; Prencipe, B.; Cascarano, G.D.; Brunetti, A.; Tommasi, S.; Bevilacqua, V.; Summa, S.D.; et al. Multi-class Tissue Classification in Colorectal Cancer with Handcrafted and Deep Features. In Proceedings of the International Conference on Intelligent Computing, Shenzhen, China, 12–15 August 2021; pp. 512–525. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European conference on computer vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Altini, N.; Prencipe, B.; Cascarano, G.D.; Brunetti, A.; Brunetti, G.; Triggiani, V.; Carnimeo, L.; Marino, F.; Guerriero, A.; Villani, L.; et al. Liver, kidney and spleen segmentation from CT scans and MRI with deep learning: A survey. Neurocomputing 2022, 490, 30–53. [Google Scholar] [CrossRef]
- Altini, N.; Prencipe, B.; Brunetti, A.; Brunetti, G.; Triggiani, V.; Carnimeo, L.; Marino, F.; Guerriero, A.; Villani, L.; Scardapane, A.; et al. A Tversky loss-based convolutional neural network for liver vessels segmentation. In Proceedings of the International Conference on Intelligent Computing, Bari, Italy, 2–5 October 2020; pp. 342–354. [Google Scholar]
- Bevilacqua, V.; Altini, N.; Prencipe, B.; Brunetti, A.; Villani, L.; Sacco, A.; Morelli, C.; Ciaccia, M.; Scardapane, A. Lung Segmentation and Characterization in COVID-19 Patients for Assessing Pulmonary Thromboembolism: An Approach Based on Deep Learning and Radiomics. Electronics 2021, 10, 2475. [Google Scholar] [CrossRef]
- Tjoa, E.; Guan, C. A survey on explainable artificial intelligence (xai): Toward medical xai. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 4793–4813. [Google Scholar] [CrossRef] [PubMed]
- Gunning, D.; Stefik, M.; Choi, J.; Miller, T.; Stumpf, S.; Yang, G.Z. XAI—Explainable artificial intelligence. Sci. Robot. 2019, 4, eaay7120. [Google Scholar] [CrossRef] [PubMed]
- Antoniadi, A.M.; Du, Y.; Guendouz, Y.; Wei, L.; Mazo, C.; Becker, B.A.; Mooney, C. Current challenges and future opportunities for XAI in machine learning-based clinical decision support systems: A systematic review. Appl. Sci. 2021, 11, 5088. [Google Scholar] [CrossRef]
- Hussain, S.M.; Buongiorno, D.; Altini, N.; Berloco, F.; Prencipe, B.; Moschetta, M.; Bevilacqua, V.; Brunetti, A. Shape-Based Breast Lesion Classification Using Digital Tomosynthesis Images: The Role of Explainable Artificial Intelligence. Appl. Sci. 2022, 12, 6230. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Vinogradova, K.; Dibrov, A.; Myers, G. Towards interpretable semantic segmentation via gradient-weighted class activation mapping (student abstract). In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 13943–13944. [Google Scholar]
- Du, L.; Zhang, R.; Wang, X. Overview of two-stage object detection algorithms. J. Phys. Conf. Ser. 2020, 1544, 012033. [Google Scholar] [CrossRef]
- Wu, Y.; Kirillov, A.; Massa, F.; Lo, W.Y.; Girshick, R. Detectron2. 2019. Available online: https://github.com/facebookresearch/detectron2 (accessed on 7 September 2022).
- Altini, N.; Cascarano, G.D.; Brunetti, A.; De Feudis, I.; Buongiorno, D.; Rossini, M.; Pesce, F.; Gesualdo, L.; Bevilacqua, V. A deep learning instance segmentation approach for global glomerulosclerosis assessment in donor kidney biopsies. Electronics 2020, 9, 1768. [Google Scholar] [CrossRef]





| Dataset | Publication Year | Organs | Resolution | Number of H&E images | Number of Nuclei | Size (pixels) | Annotations Format |
|---|---|---|---|---|---|---|---|
| MoNuSeg—Train (T1) [1] | 2017 | breast, kidney, prostate, liver, colon, bladder, stomach | 40× | 30 | 21,623 | 1000 × 1000 | Nuclei Contours |
| MoNuSeg—Test (V1) [1] | breast, kidney, prostate, colon, bladder, lung, brain | 14 | 7000 | ||||
| CRCHistoPhenotypes (D2) [14] | 2016 | colon | 20× | 100 | 29,756 | 500 × 500 | Nuclei Centroids |
| NuCLS (D3) [22] | 2019 | breast | 40× | 1744 | 59,485 | 200–400 per side | Nuclei Contours or Bounding Boxes |
| Local (V4) | 2022 | colon | 40× | 19 | 6378 | 512 × 512 | Nuclei Centroids |
| Network | Metric | Experiment (a) | Experiment (b) | Experiment (c) |
|---|---|---|---|---|
| U-Net | DICE PRECISION RECALL | 66.74 ± 3.44 57.13 ± 8.15 83.56 ± 10.61 | 65.71 ± 8.57 52.69 ± 11.96 91.65 ± 6.57 | 60.74 ± 11.65 45.43 ± 11.77 96.46 ± 2.44 |
| SegNet | DICE PRECISION RECALL | 56.44 ± 9.31 67.09 ± 8.01 52.60 ± 16.20 | 65.05 ± 6.32 58.93 ± 14.23 81.35 ± 17.69 | 62.02 ± 12.28 51.67 ± 14.96 85.05 ± 13.24 |
| DeepLab v3+ [ResNet18] | DICE PRECISION RECALL | 52.21 ± 11.99 76.78 ± 6.60 41.76 ± 13.55 | 74.23 ± 4.85 76.42 ± 8.69 74.25 ± 11.23 | 72.17 ± 8.03 62.76 ± 11.78 87.17 ± 5.64 |
| DeepLab v3+ [ResNet50] | DICE PRECISION RECALL | 57.87 ± 6.88 59.70 ± 6.35 57.10 ± 10.43 | 61.68 ± 8.75 63.69 ± 7.51 60.71 ± 11.94 | 65.98 ± 7.84 54.14 ± 13.81 90.95 ± 10.02 |
| DeepLab v3+ [mobilenetv2] | DICE PRECISION RECALL | 56.64 ± 6.60 66.49 ± 5.56 50.66 ± 10.50 | 73.01 ± 7.56 73.50 ± 11.76 75.07 ± 10.38 | 66.31 ± 13.80 57.52 ± 16.31 85.35 ± 9.43 |
| Method | Precision | Recall | Dice |
|---|---|---|---|
| CRImage [12] | 0.657 | 0.461 | 0.542 |
| CNN [12] | 0.783 | 0.804 | 0.793 |
| SSAE [6] | 0.617 | 0.644 | 0.630 |
| LIPSyM [13] | 0.725 | 0.517 | 0.604 |
| SC-CNN (M = 1) [14] | 0.758 | 0.827 | 0.791 |
| SC-CNN (M = 2) [14] | 0.781 | 0.823 | 0.802 |
| UD-Net [4] | 0.822 | 0.842 | 0.828 |
| NDG-CAM (V1) | 0.833 | 0.815 | 0.824 |
| NDG-CAM (V4) | 0.992 | 0.841 | 0.910 |
| Mask R-CNN (V1) | 0.867 | 0.888 | 0.878 |
| Mask R-CNN (V4) | 0.989 | 0.403 | 0.573 |
| Combined (V1) | 0.838 | 0.934 | 0.884 |
| Combined (V4) | 0.986 | 0.850 | 0.914 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altini, N.; Brunetti, A.; Puro, E.; Taccogna, M.G.; Saponaro, C.; Zito, F.A.; De Summa, S.; Bevilacqua, V. NDG-CAM: Nuclei Detection in Histopathology Images with Semantic Segmentation Networks and Grad-CAM. Bioengineering 2022, 9, 475. https://doi.org/10.3390/bioengineering9090475
Altini N, Brunetti A, Puro E, Taccogna MG, Saponaro C, Zito FA, De Summa S, Bevilacqua V. NDG-CAM: Nuclei Detection in Histopathology Images with Semantic Segmentation Networks and Grad-CAM. Bioengineering. 2022; 9(9):475. https://doi.org/10.3390/bioengineering9090475
Chicago/Turabian StyleAltini, Nicola, Antonio Brunetti, Emilia Puro, Maria Giovanna Taccogna, Concetta Saponaro, Francesco Alfredo Zito, Simona De Summa, and Vitoantonio Bevilacqua. 2022. "NDG-CAM: Nuclei Detection in Histopathology Images with Semantic Segmentation Networks and Grad-CAM" Bioengineering 9, no. 9: 475. https://doi.org/10.3390/bioengineering9090475
APA StyleAltini, N., Brunetti, A., Puro, E., Taccogna, M. G., Saponaro, C., Zito, F. A., De Summa, S., & Bevilacqua, V. (2022). NDG-CAM: Nuclei Detection in Histopathology Images with Semantic Segmentation Networks and Grad-CAM. Bioengineering, 9(9), 475. https://doi.org/10.3390/bioengineering9090475