Cervical Net: A Novel Cervical Cancer Classification Using Feature Fusion
Abstract
1. Introduction
2. Review of Study
3. Materials and Methods
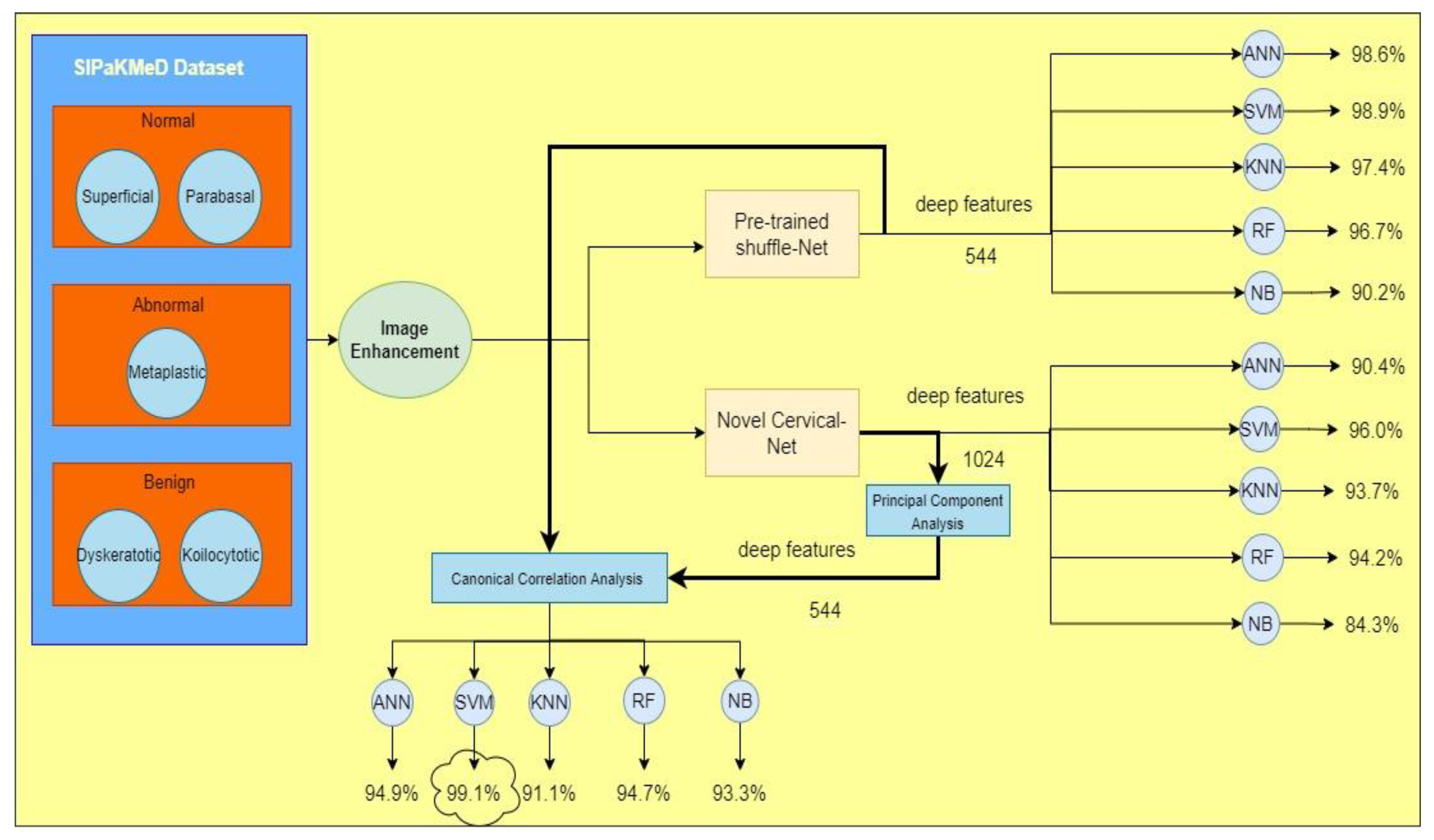
3.1. Image Acquisition
3.2. Image Enhancement
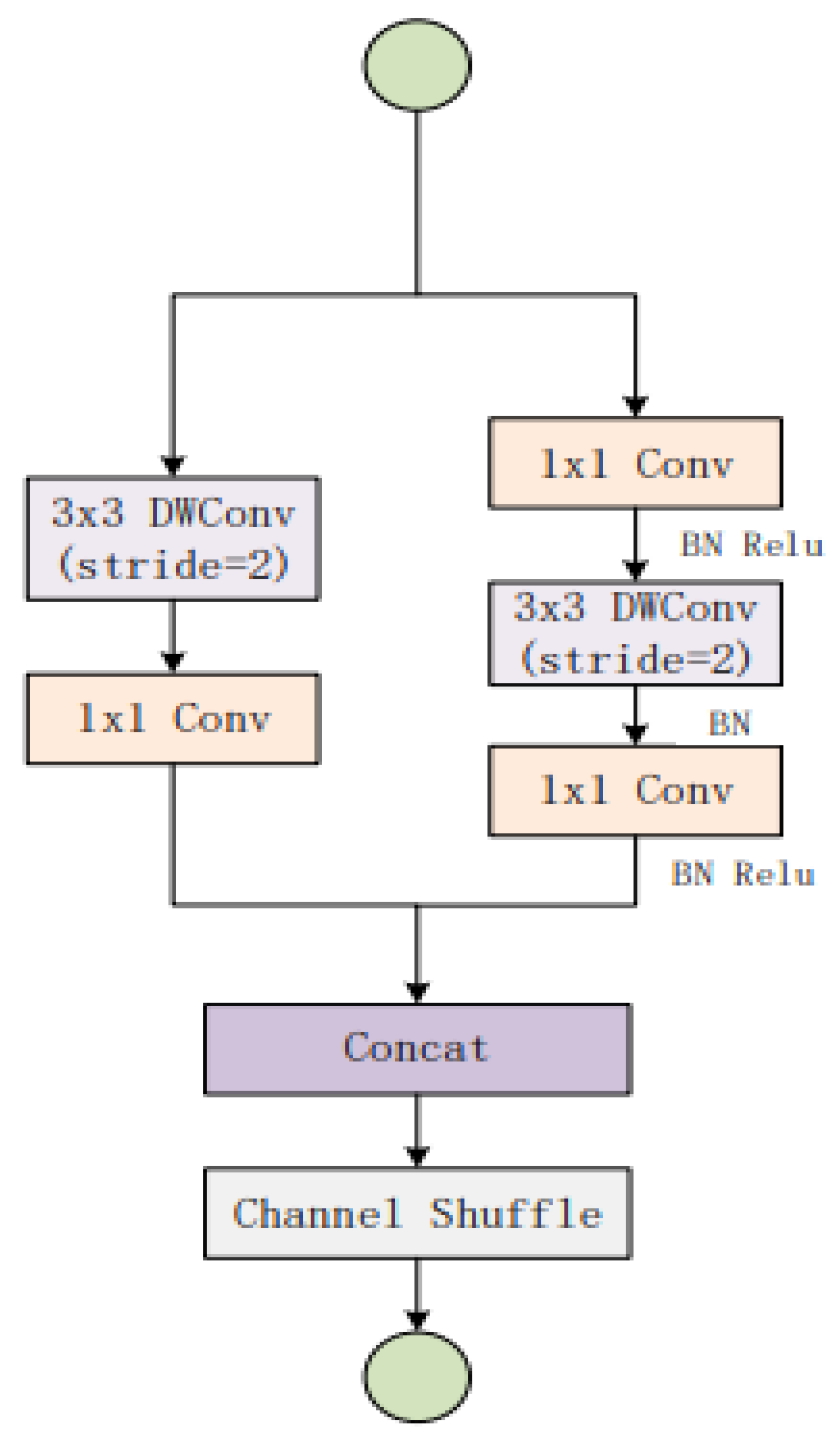
3.3. Cervical Net
3.4. Pre-Trained Shuffle Net
3.5. Deep Features Extraction
3.6. Feature Selection
3.7. Feature Fusion
3.8. Machine Learning Classifiers
3.8.1. Support Vector Machine (SVM)
3.8.2. Artificial Neural Networks (ANN)
3.8.3. Naive Bayes
3.8.4. k-Nearest Neighbour (KNN)
3.8.5. Random Forest (RF)
4. Results and Discussion
4.1. Shuffle Net Features
4.2. Novel Cervical Net Features
4.3. Feature Fusion (CCA)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Cancer Regional Profile 2020; International Agency for Research on Cancer: Lyon, France, 2020; pp. 1–2. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Mustafa, W.A.; Halim, A.; Nasrudin, M.W.; Rahman, K.S.A. Cervical cancer situation in Malaysia: A systematic literature review. Biocell 2022, 46, 367–381. [Google Scholar] [CrossRef]
- Nahrawi, N.; Mustafa, W.A.; Kanafiah, S.N.A.M. Knowledge of Human Papillomavirus ( HPV ) and Cervical Cancer among Malaysia Residents: A Review. Sains Malays. 2020, 49, 1687–1695. [Google Scholar] [CrossRef]
- William, W.; Ware, A.; Basaza-Ejiri, A.H.; Obungoloch, J. A pap-smear analysis tool (PAT) for detection of cervical cancer from pap-smear images. Biomed. Eng. Online 2019, 18, 16. [Google Scholar] [CrossRef]
- Nkwabong, E.; Badjan, I.L.B.; Sando, Z. Pap smear accuracy for the diagnosis of cervical precancerous lesions. Trop. Doct. 2019, 49, 34–39. [Google Scholar] [CrossRef]
- Mustafa, W.A.; Halim, A.; Jamlos, M.A.; Idrus, Z.S.S. A Review: Pap Smear Analysis Based on Image Processing Approach. J. Phys. Conf. Ser. 2020, 1529, 022080. [Google Scholar] [CrossRef]
- Mustafa, W.A.; Halim, A.; Rahman, K.S.A. A Narrative Review: Classification of Pap Smear Cell Image for Cervical Cancer Diagnosis. Oncologie 2020, 22, 53–63. [Google Scholar] [CrossRef]
- Varalakshmi, P.; Lakshmi, A.A.; Swetha, R.; Rahema, M.A. A Comparative Analysis of Machine and Deep Learning Models for Cervical Cancer Classification. In Proceedings of the 2021 International Conference on System, Computation, Automation and Networking (ICSCAN), Puducherry, India, 30–31 July 2021. [Google Scholar] [CrossRef]
- Mbaga, A.H.; ZhiJun, P. Pap Smear Images Classification for Early Detection of Cervical Cancer. Int. J. Comput. Appl. 2015, 118, 10–16. [Google Scholar] [CrossRef]
- Win, K.P.; Kitjaidure, Y.; Hamamoto, K.; Aung, T.M. Computer-assisted screening for cervical cancer using digital image processing of pap smear images. Appl. Sci. 2020, 10, 1800. [Google Scholar] [CrossRef]
- Plissiti, M.E.; Dimitrakopoulos, P.; Sfikas, G.; Nikou, C.; Krikoni, O.; Charchanti, A. Sipakmed: A New Dataset for Feature and Image Based Classification of Normal and Pathological Cervical Cells in Pap Smear Images. In Proceedings of the International Conference on Image Processing, ICIP, Athens, Greece, 7–10 October 2018; pp. 3144–3148. [Google Scholar] [CrossRef]
- Basak, H.; Kundu, R.; Chakraborty, S.; Das, N. Cervical Cytology Classification Using PCA and GWO Enhanced Deep Features Selection. SN Comput. Sci. 2021, 2, 369. [Google Scholar] [CrossRef]
- Park, Y.R.; Kim, Y.J.; Ju, W.; Nam, K.; Kim, S.; Kim, K.G. Comparison of machine and deep learning for the classification of cervical cancer based on cervicography images. Sci. Rep. 2021, 11, 16143. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Arora, A.; Bhan, A. Classification of cervical cancer using Deep Learning Algorithm. In Proceedings of the 5th International Conference on Intelligent Computing and Control Systems, ICICCS 2021, Madurai, India, 6–8 May 2021; pp. 1210–1218. [Google Scholar] [CrossRef]
- AlMubarak, H.A.; Stanley, J.; Guo, P.; Long, R.; Antani, S.; Thoma, G.; Zuna, R.; Frazier, S.; Stoecker, W. A hybrid deep learning and handcrafted feature approach for cervical cancer digital histology image classification. Int. J. Healthc. Inf. Syst. Inform. 2019, 14, 66–87. [Google Scholar] [CrossRef]
- Alyafeai, Z.; Ghouti, L. A fully-automated deep learning pipeline for cervical cancer classification. Expert Syst. Appl. 2020, 141, 112951. [Google Scholar] [CrossRef]
- Alquran, H.; Mustafa, W.A.; Qasmieh, I.A.; Yacob, Y.M.; Alsalatie, M.; Al-Issa, Y.; Alqudah, A.M. Cervical Cancer Classification Using Combined Machine Learning and Deep Learning Approach. Comput. Mater. Contin. 2022, 72, 5117–5134. [Google Scholar] [CrossRef]
- Dhawan, S.; Singh, K.; Arora, M. Cervix image classification for prognosis of cervical cancer using deep neural network with transfer learning. EAI Endorsed Trans. Pervasive Health Technol. 2021, 7, e5. [Google Scholar] [CrossRef]
- Huang, P.; Tan, X.; Chen, C.; Lv, X.; Li, Y. AF-SENet: Classification of cancer in cervical tissue pathological images based on fusing deep convolution features. Sensors 2021, 21, 122. [Google Scholar] [CrossRef]
- Mulmule, P.V.; Kanphade, R.D. Supervised classification approach for cervical cancer detection using Pap smear images. Int. J. Med. Eng. Inform. 2021, 1, 1. [Google Scholar] [CrossRef]
- Nikookar, E.; Naderi, E.; Rahnavard, A. Cervical cancer prediction by merging features of different colposcopic images and using ensemble classifier. J. Med. Signals Sens. 2021, 11, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Yaman, O.; Tuncer, T. Exemplar pyramid deep feature extraction based cervical cancer image classification model using pap-smear images. Biomed. Signal Process. Control 2022, 73, 103428. [Google Scholar] [CrossRef]
- Coppola, F.; Faggioni, L.; Gabelloni, M.; De Vietro, F.; Mendola, V.; Cattabriga, A.; Cocozza, M.A.; Vara, G.; Piccinino, A.; Lo Monaco, S.; et al. Human, All Too Human? An All-Around Appraisal of the ‘Artificial Intelligence Revolution’ in Medical Imaging. Front. Psychol. 2021, 12, 710982. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lei, T.; Cui, R.; Zhang, B.; Meng, H.; Nandi, A.K. Medical image segmentation using deep learning: A survey. IET Image Process. 2022, 16, 1243–1267. [Google Scholar] [CrossRef]
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine learning for medical imaging. Radiographics 2017, 37, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, W.A.; Sam, S.; Jamlos, M.A.; Khairunizam, W. Effect of different filtering techniques on medical and document image. Lect. Notes Electr. Eng. 2021, 666, 727–736. [Google Scholar] [CrossRef]
- Alqudah, A.; Alqudah, A.M.; Alquran, H.; Al-zoubi, H.R.; Al-qodah, M.; Al-khassaweneh, M.A. Recognition of handwritten arabic and hindi numerals using convolutional neural networks. Appl. Sci. 2021, 11, 1573. [Google Scholar] [CrossRef]
- Alsharif, R.; Al-Issa, Y.; Alqudah, A.M.; Qasmieh, I.A.; Mustafa, W.A.; Alquran, H. Pneumonianet: Automated detection and classification of pediatric pneumonia using chest X-ray images and cnn approach. Electronics 2021, 10, 2949. [Google Scholar] [CrossRef]
- Alawneh, K.; Alquran, H.; Alsalatie, M.; Mustafa, W.A.; Al-Issa, Y.; Alqudah, A.; Badarneh, A. LiverNet: Diagnosis of Liver Tumors in Human CT Images. Appl. Sci. 2022, 12, 5501. [Google Scholar] [CrossRef]
- Liu, H.; Yao, D.; Yang, J.; Li, X. Lightweight convolutional neural network and its application in rolling bearing fault diagnosis under variable working conditions. Sensors 2019, 19, 4827. [Google Scholar] [CrossRef]
- Brownlee, J. A Gentle Introduction to Pooling Layers for Convolutional Neural Networks. Mach. Learn. Mastery 2019, 22, 1–16. [Google Scholar]
- Basak, H.; Kundu, R. Comparative Study of Maturation Profiles of Neural Cells in Different Species with the Help of Computer Vision and Deep Learning. Commun. Comput. Inf. Sci. 2021, 1365, 352–366. [Google Scholar] [CrossRef]
- Basak, H.; Ghosal, S.; Sarkar, M.; Das, M.; Chattopadhyay, S. Monocular Depth Estimation Using Encoder-Decoder Architecture and Transfer Learning from Single RGB Image. In Proceedings of the IEEE 7th Uttar Pradesh Section International Conference on Electrical, Electronics and Computer Engineering (UPCON), Prayagraj, India, 27–29 November 2020. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, L.; Huang, H. Sparse additive discriminant canonical correlation analysis for multiple features fusion. Neurocomputing 2021, 463, 185–197. [Google Scholar] [CrossRef]
- Shi, J.; Chen, C.; Liu, H.; Wang, Y.; Shu, M.; Zhu, Q. Automated Atrial Fibrillation Detection Based on Feature Fusion Using Discriminant Canonical Correlation Analysis. Comput. Math. Methods Med. 2021, 2021, 6691177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Li, Y.; Wang, J.; Wang, Z.; Li, X. Feature fusion for multimodal emotion recognition based on deep canonical correlation analysis. IEEE Signal Process. Lett. 2021, 28, 1898–1902. [Google Scholar] [CrossRef]
- Pisner, D.A.; Schnyer, D.M. Support vector machine. In Machine Learning: Methods and Applications to Brain Disorders; Academic Press: Cambridge, MA, USA, 2019; pp. 101–121. [Google Scholar] [CrossRef]
- Alquran, H.; Qasmieh, I.A.; Alqudah, A.M.; Alhammouri, S.; Alawneh, E.; Abughazaleh, A.; Hasayen, F. The melanoma skin cancer detection and classification using support vector machine. In Proceedings of the 2017 IEEE Jordan Conference on Applied Electrical Engineering and Computing Technologies, AEECT 2017, Aqaba, Jordan, 11–13 October 2017; pp. 1–5. [Google Scholar] [CrossRef]
- Haykin, S. Neural Networks and Learning Machines, 3rd ed.; Prentice Hall: Hoboken, NJ, USA, 2009. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Chen, S.; Webb, G.I.; Liu, L.; Ma, X. A novel selective naïve Bayes algorithm. Knowl.-Based Syst. 2020, 192, 105361. [Google Scholar] [CrossRef]
- Fix, E.; Hodges, J.L. Discriminatory Analysis. Nonparametric Discrimination: Consistency Properties. Int. Stat. Rev. Rev. Int. Stat. 1989, 57, 238. [Google Scholar] [CrossRef]
- Alquran, H.; Alsleti, M.; Alsharif, R.; Qasmieh, I.A.; Alqudah, A.M.; Harun, N.H.B. Employing texture features of chest x-ray images and machine learning in covid-19 detection and classification. Mendel 2021, 27, 9–17. [Google Scholar] [CrossRef]
- Sun, G.; Li, S.; Cao, Y.; Lang, F. Cervical cancer diagnosis based on random forest. Int. J. Perform. Eng. 2017, 13, 446–457. [Google Scholar] [CrossRef]


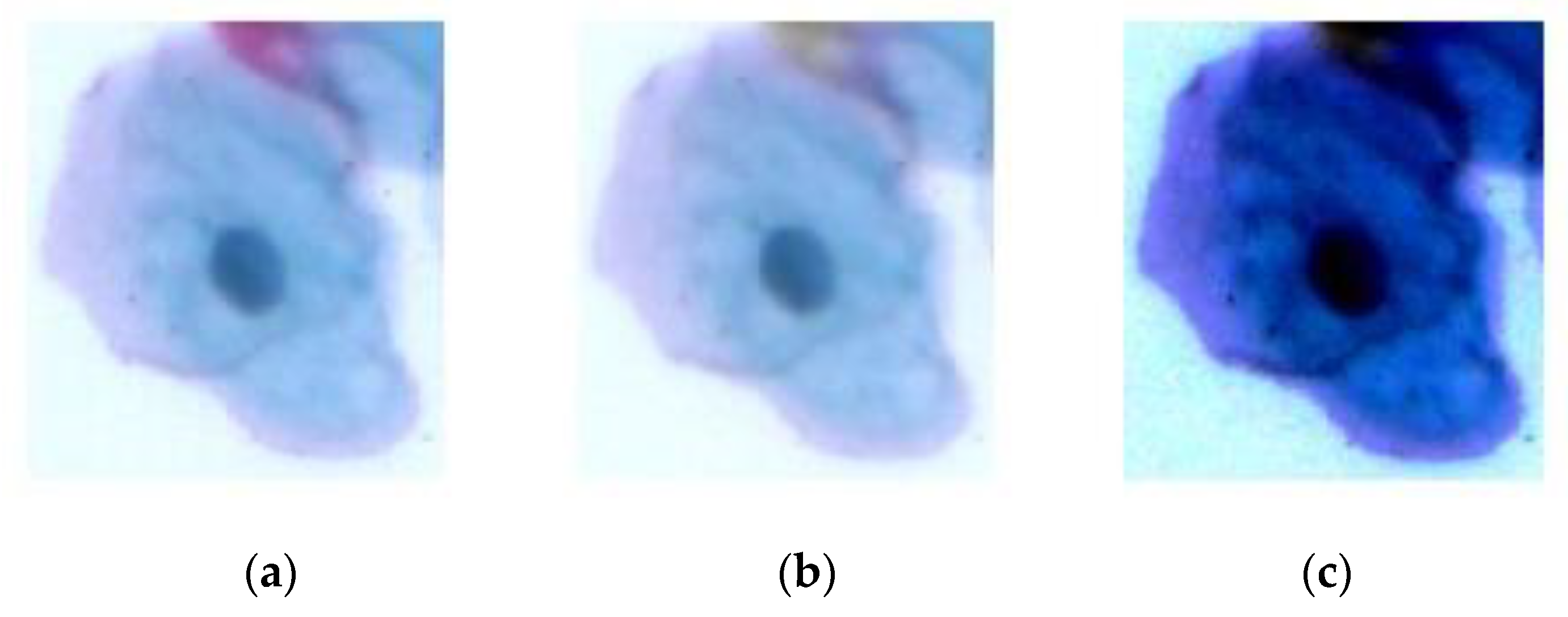




| Class | Number of Images | Number of Cells |
|---|---|---|
| Normal Class | ||
| 1. Superficial–Intermediate Cells | 126 | 831 |
| 2. Parabasal Cells | 108 | 787 |
| Benign Cell | ||
| 3. Metaplastic Cells | 271 | 793 |
| Abnormal Cells | ||
| 4. Dyskeratotic Cells | 223 | 813 |
| 5. Koilocytotic Cells | 238 | 825 |
| Total | 966 | 4049 |
| Layer | Information |
|---|---|
| Input Layer | Size: 224 × 224 × 3 |
| conv1 | Number of Filters: 64 |
| Kernel Size: 7 × 7 | |
| Stride: 2 × 2 | |
| Padding: 0 | |
| Activation Layer | ReLU |
| Pooling Layer | Type: Average Pooling |
| Kernel size: 3 × 3 | |
| Stride: 2 × 2 | |
| Padding: 0 | |
| Grouped Convolutional Layer | Number of Groups: 2 |
| Number of Filters: 94 | |
| Kernel Size: 5 × 5 | |
| Padding: 2 × 2 × 2 × 2 | |
| Activation Layer | ReLU |
| Pooling Layer | Type: Average Pooling |
| Kernel Size: 3 × 3 | |
| Stride: 2 × 2 | |
| Padding: 0 | |
| Convolutional Layer | Number of Filters: 128 |
| Kernel Size: 3 × 3 | |
| Padding: (1 × 1 ×1 × 1) | |
| Activation Layer | ReLU |
| Grouped Convolutional Layer | Number of Groups: 2 |
| Number of Filters: 192 | |
| Kernel Size: 3 × 3 | |
| Padding: (1 × 1 × 1 × 1) | |
| Activation Layer | ReLU |
| Grouped Convolutional Layer | Number of Groups: 2 |
| Number of Filters: 128 | |
| Kernel Size: 3 × 3 | |
| Padding: (1 × 1 × 1 × 1) | |
| Activation Layer | ReLU |
| Pooling Layer | Type: Global Average Pooling |
| Fully connected Layer | 5 neurons |
| Softmax Layer | |
| Classification Layer |
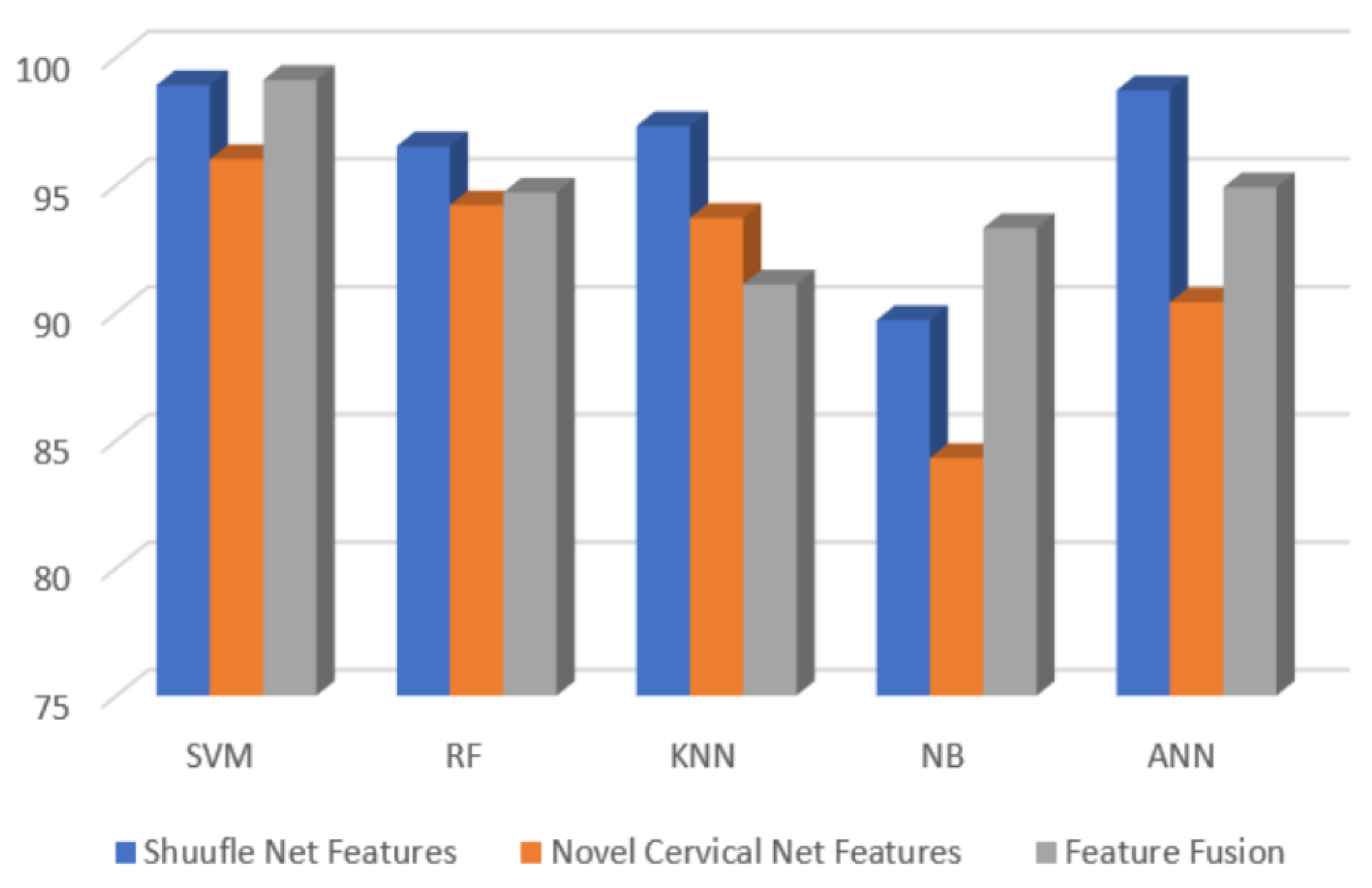
| Shuffle Net | Cervical Net | Feature Fusion (CCA) | |
|---|---|---|---|
| SVM | 98.90% | 96.00% | 99.10% |
| RF | 96.70% | 94.20% | 94.70% |
| KNN | 97.40% | 93.70% | 91.10% |
| Naïve Bayes | 90.20% | 84.30% | 93.30% |
| ANN | 98.60% | 90.40% | 94.90% |
| Study | Method | Dataset | Classes | Accuracy |
|---|---|---|---|---|
| Mbaga et al. [11] | SVM | Herlev dataset | 7 classes | 92.96% |
| Win et al. [12] | SVM, KNN, boosted trees, bagged trees, and major voting | SIPaKMeD dataset | 2 classes 5 classes | 98.27% 94.09% |
| Plissiti et al. [13] | MLP and SVM | SIPaKMeD dataset | 5 classes | 95.35% |
| Basak et al. [14] | feature selection and DL | SIPaKMeD dataset | 5 classes | 97.87% |
| Park et al. [15] | ResNet-50 and SVM | Cervicography images | 2 classes | 82.00% |
| Tripathi et al. [16] | ResNet-152 | SIPaKMeD dataset | 5 classes | 94.89% |
| Al Mubarak et al. [17] | Fusion based and CNN | 4 classes | 80.72% | |
| Alquran et al. [19] | DL and cascading SVM | Herlev dataset | 7 classes | Up to 92% |
| Dhawan et al. [20] | InceptionV3 | Kaggle dataset | 3 classes | 96.10% |
| Huang et al. [21] | ResNet-50V2 and DenseNet-121 | Tissue biopsy image dataset | 4 classes | 95.33% |
| Mulmule and Kanphade [22] | MLP with three kernels and SVM | Benchmark database | 97.14% | |
| Nikookar et al. [23] | Artificial intelligence | Digital colposcopy dataset | 2 classes | 96% for sensitivity and 94% for specificity |
| Yaman and 155 Tuncer [24] | SVM | SIPaKMeD Mendeley | 2 classes | 98.26% 99.47% |
| This study | Cervical Net and feature fusion with ML classifiers | SIPaKMeD | 5 classes | 99.1% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alquran, H.; Alsalatie, M.; Mustafa, W.A.; Abdi, R.A.; Ismail, A.R. Cervical Net: A Novel Cervical Cancer Classification Using Feature Fusion. Bioengineering 2022, 9, 578. https://doi.org/10.3390/bioengineering9100578
Alquran H, Alsalatie M, Mustafa WA, Abdi RA, Ismail AR. Cervical Net: A Novel Cervical Cancer Classification Using Feature Fusion. Bioengineering. 2022; 9(10):578. https://doi.org/10.3390/bioengineering9100578
Chicago/Turabian StyleAlquran, Hiam, Mohammed Alsalatie, Wan Azani Mustafa, Rabah Al Abdi, and Ahmad Rasdan Ismail. 2022. "Cervical Net: A Novel Cervical Cancer Classification Using Feature Fusion" Bioengineering 9, no. 10: 578. https://doi.org/10.3390/bioengineering9100578
APA StyleAlquran, H., Alsalatie, M., Mustafa, W. A., Abdi, R. A., & Ismail, A. R. (2022). Cervical Net: A Novel Cervical Cancer Classification Using Feature Fusion. Bioengineering, 9(10), 578. https://doi.org/10.3390/bioengineering9100578







