A Multimodality Myocardial Perfusion Phantom: Initial Quantitative Imaging Results
Abstract
:1. Introduction
2. Materials and Methods
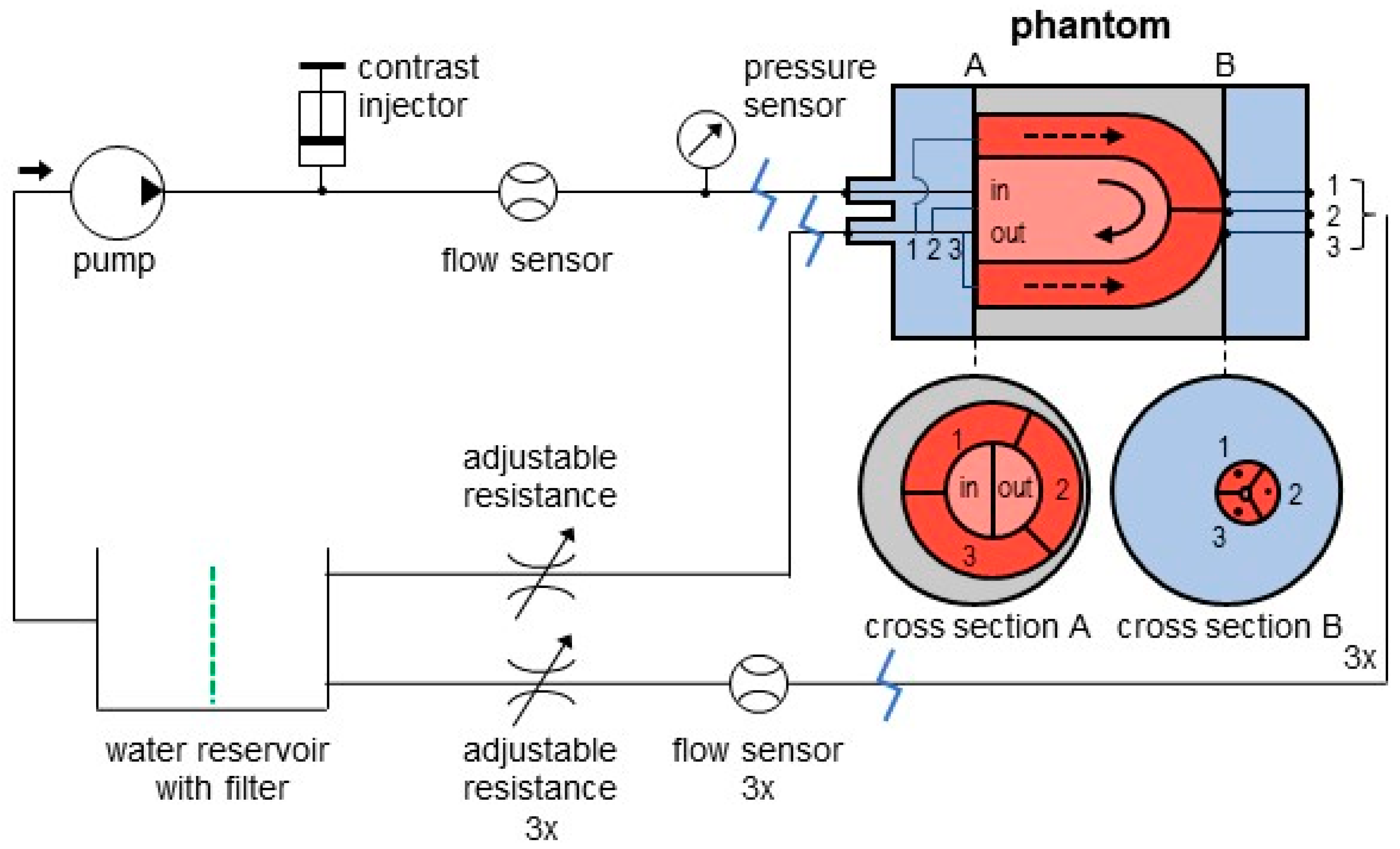
2.1. Phantom Setup
- A previously adapted myocardial perfusion phantom (cylindrical cardiac insert);
- A commercial anthropomorphic thorax phantom;
- An in-house built flow circuit.
2.2. Flow Circuit Adjustments
2.3. Tissue Mimicking Adjustments
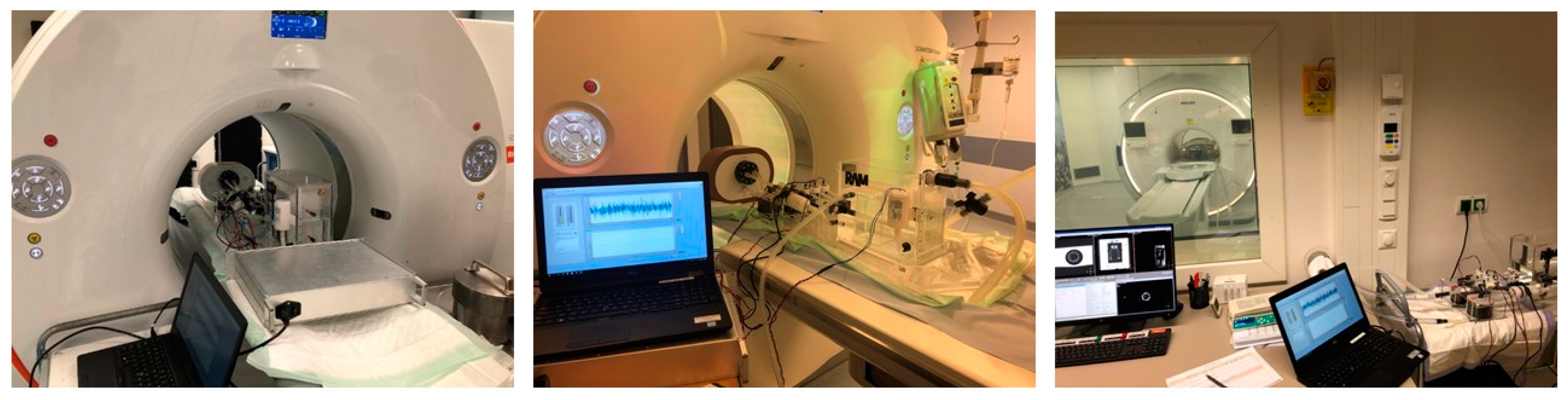
2.4. Phantom Measurements
2.5. Dynamic Myocardial Perfusion Imaging
2.5.1. PET/CT Imaging
2.5.2. CT Imaging
2.5.3. MR Imaging
2.6. Perfusion Analysis
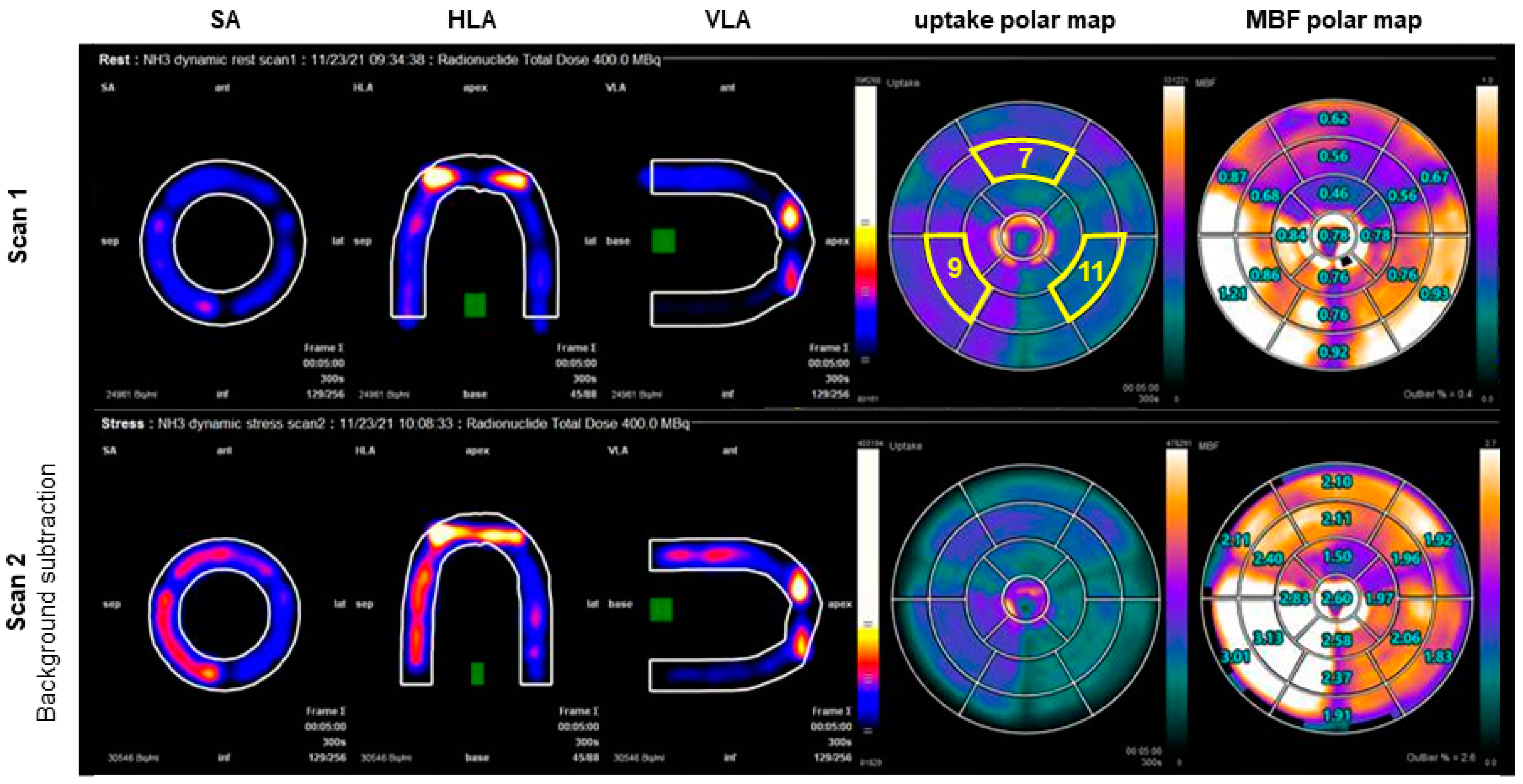
2.6.1. PET/CT Analysis
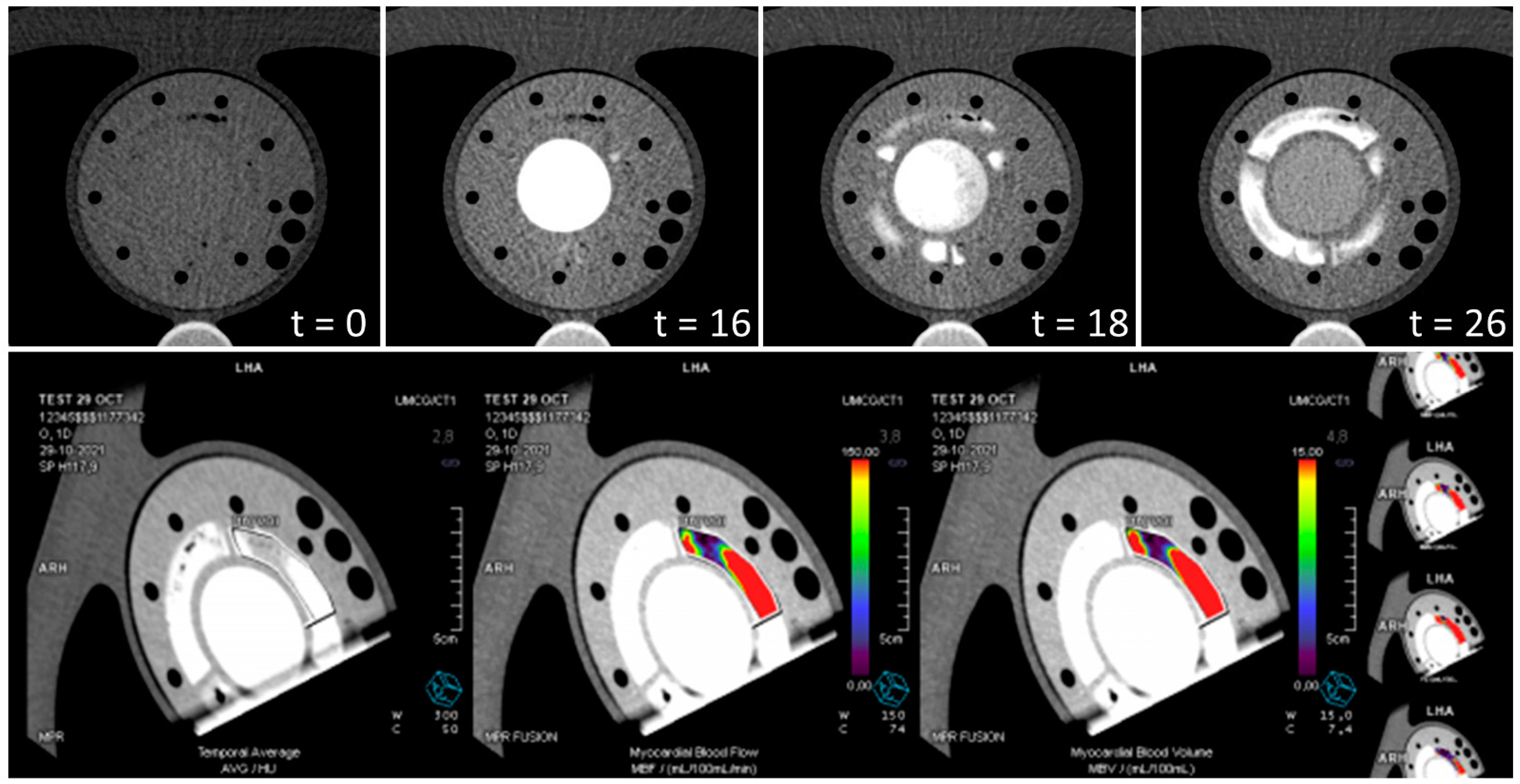
2.6.2. CT Analysis
2.6.3. MR Analysis
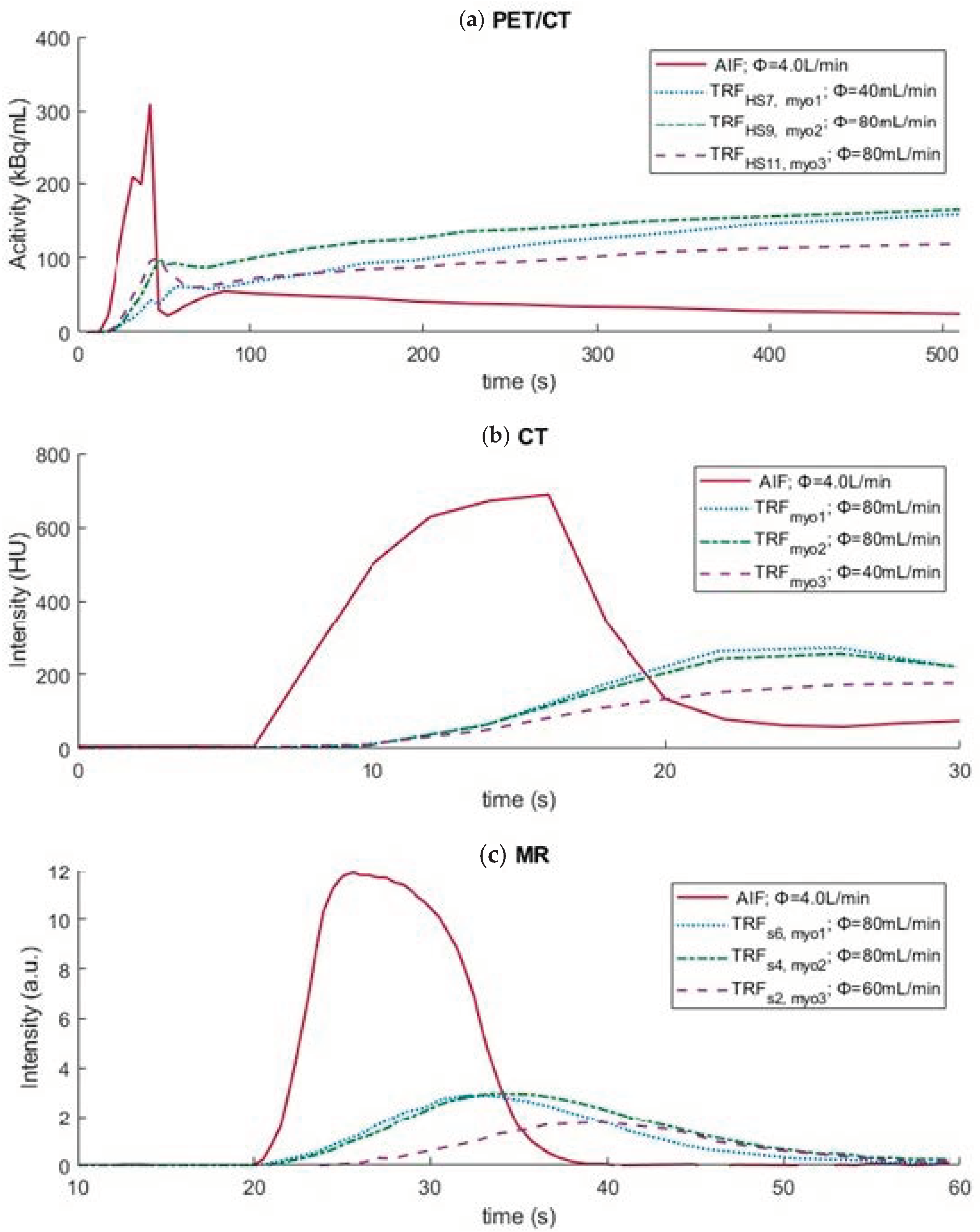
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Slomka, P.J.; Miller, R.J.H.; Hu, L.H.; Germano, G.; Berman, D.S. Solid-State Detector SPECT Myocardial Perfusion Imaging. J. Nucl. Med. 2019, 60, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Hyafil, F.; Gimelli, A.; Slart, R.; Georgoulias, P.; Rischpler, C.; Lubberink, M.; Sciagra, R.; Bucerius, J.; Agostini, D.; Verberne, H.J.; et al. EANM procedural guidelines for myocardial perfusion scintigraphy using cardiac-centered gamma cameras. Eur. J. Hybrid Imaging 2019, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Sciagra, R.; Lubberink, M.; Hyafil, F.; Saraste, A.; Slart, R.; Agostini, D.; Nappi, C.; Georgoulias, P.; Bucerius, J.; Rischpler, C.; et al. EANM procedural guidelines for PET/CT quantitative myocardial perfusion imaging. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1040–1069. [Google Scholar] [CrossRef] [PubMed]
- Bindschadler, M.; Modgil, D.; Branch, K.R.; La Riviere, P.J.; Alessio, A.M. Comparison of blood flow models and acquisitions for quantitative myocardial perfusion estimation from dynamic CT. Phys. Med. Biol. 2014, 59, 1533–1556. [Google Scholar] [CrossRef]
- Pelgrim, G.J.; Handayani, A.; Dijkstra, H.; Prakken, N.H.; Slart, R.H.; Oudkerk, M.; Van Ooijen, P.M.; Vliegenthart, R.; Sijens, P.E. Quantitative Myocardial Perfusion with Dynamic Contrast-Enhanced Imaging in MRI and CT: Theoretical Models and Current Implementation. Biomed Res. Int. 2016, 2016, 1734190. [Google Scholar] [CrossRef] [PubMed]
- Carson, R.E. Tracer Kinetic Modeling in PET. In Positron Emission Tomography: Basic Sciences; Bailey, D.L., Townsend, D.W., Valk, P.E., Maisey, M.N., Eds.; Springer: London, UK, 2005; pp. 127–159. [Google Scholar] [CrossRef]
- Juarez-Orozco, L.E.; Tio, R.A.; Alexanderson, E.; Dweck, M.; Vliegenthart, R.; El Moumni, M.; Prakken, N.; Gonzalez-Godinez, I.; Slart, R. Quantitative myocardial perfusion evaluation with positron emission tomography and the risk of cardiovascular events in patients with coronary artery disease: A systematic review of prognostic studies. Eur. Heart J. Cardiovasc. Imaging 2018, 19, 1179–1187. [Google Scholar] [CrossRef]
- Ziadi, M.C. Myocardial flow reserve (MFR) with positron emission tomography (PET)/computed tomography (CT): Clinical impact in diagnosis and prognosis. Cardiovasc. Diagn. Ther. 2017, 7, 206–218. [Google Scholar] [CrossRef]
- Murthy, V.L.; Bateman, T.M.; Beanlands, R.S.; Berman, D.S.; Borges-Neto, S.; Chareonthaitawee, P.; Cerqueira, M.D.; deKemp, R.A.; DePuey, E.G.; Dilsizian, V.; et al. Clinical Quantification of Myocardial Blood Flow Using PET: Joint Position Paper of the SNMMI Cardiovascular Council and the ASNC. J. Nucl. Med. 2018, 59, 273–293. [Google Scholar] [CrossRef]
- Piekarski, E.; Manrique, A.; Rouzet, F.; Le Guludec, D. Current Status of Myocardial Perfusion Imaging with New SPECT/CT Cameras. Semin. Nucl. Med. 2020, 50, 219–226. [Google Scholar] [CrossRef]
- Nieman, K.; Balla, S. Dynamic CT myocardial perfusion imaging. J. Cardiovasc. Comput. Tomogr. 2020, 14, 303–306. [Google Scholar] [CrossRef]
- Jerosch-Herold, M.; Seethamraju, R.T.; Swingen, C.M.; Wilke, N.M.; Stillman, A.E. Analysis of myocardial perfusion MRI. J. Magn. Reson. Imaging 2004, 19, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Everaars, H.; van Diemen, P.A.; Bom, M.J.; Schumacher, S.P.; de Winter, R.W.; van de Ven, P.M.; Raijmakers, P.G.; Lammertsma, A.A.; Hofman, M.B.M.; van der Geest, R.J.; et al. Comparison between quantitative cardiac magnetic resonance perfusion imaging and [15O]H2O positron emission tomography. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1688–1697. [Google Scholar] [CrossRef] [PubMed]
- Vogel, R.; Indermuhle, A.; Reinhardt, J.; Meier, P.; Siegrist, P.T.; Namdar, M.; Kaufmann, P.A.; Seiler, C. The quantification of absolute myocardial perfusion in humans by contrast echocardiography: Algorithm and validation. J. Am. Coll. Cardiol. 2005, 45, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Bulte, C.S.; Slikkerveer, J.; Meijer, R.I.; Gort, D.; Kamp, O.; Loer, S.A.; de Marchi, S.F.; Vogel, R.; Boer, C.; Bouwman, R.A. Contrast-enhanced ultrasound for myocardial perfusion imaging. Anesth. Analg. 2012, 114, 938–945. [Google Scholar] [CrossRef]
- Kamphuis, M.E.; Kuipers, H.; Verschoor, J.; van Hespen, J.C.G.; Greuter, M.J.W.; Slart, R.; Slump, C.H. Development of a dynamic myocardial perfusion phantom model for tracer kinetic measurements. EJNMMI Phys. 2022, 9, 31. [Google Scholar] [CrossRef]
- Zito, F.; De Bernardi, E.; Soffientini, C.; Canzi, C.; Casati, R.; Gerundini, P.; Baselli, G. The use of zeolites to generate PET phantoms for the validation of quantification strategies in oncology. Med. Phys. 2012, 39, 5353–5361. [Google Scholar] [CrossRef]
- Chai, J.W.; Chen, J.H.; Kao, Y.H.; Liao, J.R.; Chen, C.C.; Lee, S.K.; Chu, W.C. Spoiled gradient-echo as an arterial spin tagging technique for quick evaluation of local perfusion. J. Magn. Reson. Imaging 2002, 16, 51–59. [Google Scholar] [CrossRef]
- Potdevin, T.C.; Fowlkes, J.B.; Moskalik, A.P.; Carson, P.L. Analysis of refill curve shape in ultrasound contrast agent studies. Med. Phys. 2004, 31, 623–632. [Google Scholar] [CrossRef]
- Ohno, N.; Miyati, T.; Chigusa, T.; Usui, H.; Ishida, S.; Hiramatsu, Y.; Kobayashi, S.; Gabata, T.; Alperin, N. Technical Note: Development of a cranial phantom for assessing perfusion, diffusion, and biomechanics. Med. Phys. 2017, 44, 1646–1654. [Google Scholar] [CrossRef]
- Cho, G.Y.; Kim, S.; Jensen, J.H.; Storey, P.; Sodickson, D.K.; Sigmund, E.E. A versatile flow phantom for intravoxel incoherent motion MRI. Magn. Reson. Med. 2012, 67, 1710–1720. [Google Scholar] [CrossRef]
- Hutchins, G.D.; Schwaiger, M.; Rosenspire, K.C.; Krivokapich, J.; Schelbert, H.; Kuhl, D.E. Noninvasive quantification of regional blood flow in the human heart using N-13 ammonia and dynamic positron emission tomographic imaging. J. Am. Coll. Cardiol. 1990, 15, 1032–1042. [Google Scholar] [CrossRef]
- Cerqueira, M.D.; Weissman, N.J.; Dilsizian, V.; Jacobs, A.K.; Kaul, S.; Laskey, W.K.; Pennell, D.J.; Rumberger, J.A.; Ryan, T.; Verani, M.S.; et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation 2002, 105, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rienmuller, R.; Baumgartner, C.; Kern, R.; Harb, S.; Aigner, R.; Fueger, G.; Weihs, W. Quantitative determination of left ventricular myocardial perfusion with electron beam computerized tomography. Herz 1997, 22, 63–71. [Google Scholar] [CrossRef]
- Groves, A.M.; Goh, V.; Rajasekharan, S.; Kayani, I.; Endozo, R.; Dickson, J.C.; Menezes, L.J.; Shastry, M.; Habib, S.B.; Ell, P.J.; et al. CT coronary angiography: Quantitative assessment of myocardial perfusion using test bolus data-initial experience. Eur. Radiol. 2008, 18, 2155–2163. [Google Scholar] [CrossRef] [PubMed]
- Kamphuis, M.E.; de Vries, G.J.; Kuipers, H.; Saaltink, M.; Verschoor, J.; Greuter, M.J.W.; Slart, R.; Slump, C.H. Development of a dedicated 3D printed myocardial perfusion phantom: Proof-of-concept in dynamic SPECT. Med. Biol. Eng. Comput. 2022, 60, 1541–1550. [Google Scholar] [CrossRef]
- Mabrouk, R.; Dubeau, F.; Bentabet, L. Dynamic cardiac PET imaging: Extraction of time-activity curves using ICA and a generalized Gaussian distribution model. IEEE Trans. Biomed. Eng. 2013, 60, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.H.; Kim, Y.H. CT myocardial perfusion imaging: Current status and future perspectives. Int. J. Cardiovasc. Imaging 2017, 33, 1009–1020. [Google Scholar] [CrossRef]
- Coelho-Filho, O.R.; Rickers, C.; Kwong, R.Y.; Jerosch-Herold, M. MR myocardial perfusion imaging. Radiology 2013, 266, 701–715. [Google Scholar] [CrossRef]
- Aramendia-Vidaurreta, V.; Echeverria-Chasco, R.; Vidorreta, M.; Bastarrika, G.; Fernandez-Seara, M.A. Quantification of Myocardial Perfusion With Vasodilation Using Arterial Spin Labeling at 1.5T. J. Magn. Reson. Imaging 2021, 53, 777–788. [Google Scholar] [CrossRef]


| Study Variables | PET/CT | CT | MR |
|---|---|---|---|
| imaging settings | |||
| scan duration | 8.5 min | 30 s | 60 s |
| (radiolabeled) contrast medium | 400 MBq [13N]ammonia | 12.6 g iomeprol | 7.5 mmol gadolinium |
| Vtracer/Vcontrast | 10 | 60 | 15 |
| vtracer/vcontrast (mL/s) | 1.0 | 6.0 | 3.0 |
| Vendflush (mL) | 30 | 32 | 20 |
| vendflush (mL/s) | 5.0 | 6.0 | 3.0 |
| phantom settings | |||
| 29.0 g zeolite | sponge | 7.0 g activated carbon 14.0 g silica granulates |
| 14.5 g zeolite 13.7 g silica granulates | sponge | 7.0 g activated carbon 14.0 g silica granulates |
| 7.3 g zeolite 12.8 g activated carbon | sponge | 7.0 g activated carbon 14.0 g silica granulates |
| ΦLVC (L/min) | 4.0 | 4.0 | 4.0 |
| Φmyo1–3 (mL/min) | [40 80] | [40 80 120] | [60 80] |
| Imaging Modality | Myocardial Region | Φ (mL/g/min) | MBF (mL/g/min) | TTP (s) |
|---|---|---|---|---|
| PET/CT | myo1 (HS7) | 0.8 | 0.6 | - |
| myo2 (HS9) | 1.6 | 0.9 | - | |
| myo3 (HS11) | 1.6 | 0.8 | - | |
| CT | myo1 myo2 myo3 | 1.6 0.8 1.6 | 4.5 2.8 4.3 | - - - |
| MR | myo1 (s6) myo2 (s4) myo3 (s2) | 1.6 1.6 0.8 | - - - | 32.0 32.0 38.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamphuis, M.E.; Kuipers, H.; Liefers, H.R.; van Es, J.; Simonis, F.F.J.; Greuter, M.J.W.; Slump, C.H.; Slart, R.H.J.A. A Multimodality Myocardial Perfusion Phantom: Initial Quantitative Imaging Results. Bioengineering 2022, 9, 436. https://doi.org/10.3390/bioengineering9090436
Kamphuis ME, Kuipers H, Liefers HR, van Es J, Simonis FFJ, Greuter MJW, Slump CH, Slart RHJA. A Multimodality Myocardial Perfusion Phantom: Initial Quantitative Imaging Results. Bioengineering. 2022; 9(9):436. https://doi.org/10.3390/bioengineering9090436
Chicago/Turabian StyleKamphuis, Marije E., Henny Kuipers, H. Remco Liefers, Jan van Es, Frank F. J. Simonis, Marcel J. W. Greuter, Cornelis H. Slump, and Riemer H. J. A. Slart. 2022. "A Multimodality Myocardial Perfusion Phantom: Initial Quantitative Imaging Results" Bioengineering 9, no. 9: 436. https://doi.org/10.3390/bioengineering9090436
APA StyleKamphuis, M. E., Kuipers, H., Liefers, H. R., van Es, J., Simonis, F. F. J., Greuter, M. J. W., Slump, C. H., & Slart, R. H. J. A. (2022). A Multimodality Myocardial Perfusion Phantom: Initial Quantitative Imaging Results. Bioengineering, 9(9), 436. https://doi.org/10.3390/bioengineering9090436







