Function of the Long Noncoding RNAs in Hepatocellular Carcinoma: Classification, Molecular Mechanisms, and Significant Therapeutic Potentials
Abstract
:1. Introduction
2. Characteristics and Classification of RNAs
3. Characteristics and Functions of lncRNAs
4. Cancer-Associated lncRNAs
5. LncRNAs in HCC
5.1. Regulation and Modification of Chromatin by lncRNAs
5.2. Transcriptional Regulation and Activation
5.3. Interaction with mRNAs
5.4. Sponge of MicroRNAs
5.5. Protein Binding and/or Modification
5.6. Other Mechanisms and Pathways of lncRNAs in HCC
6. Importance of Gene Expression Regulation in HCC Progression
7. LncRNAs as Diagnostic and Therapeutic Markers in HCC
7.1. LncRNAs as a Potential Biomarker of HCC
7.2. LncRNAs as Promising Therapeutic Potentials for HCC
8. Future Prospects and Conclusions
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HCC | hepatocellular carcinoma |
| LncRNAs | long noncoding RNAs |
| snRNAs | small nuclear RNAs |
| ncRNAs | noncoding RNAs |
| siRNAs | small interfering RNA |
| mRNAs | messenger RNAs |
| tRNAs | transport RNAs |
| rRNAs | ribosomal RNAs |
| ceRNAs | competing endogenous RNAs |
| DNMT1 | DNA methyltransferase1 |
| MAPK6 | lncMAPK6, mitogen-activated protein kinase 6 |
| HULC | highly upregulated in liver cancer |
| LATS1 | long-acting thyroid stimulator 1 |
| HDAC3 | histone deacetylase 3 |
| EMT | epithelial-mesenchymal transformation |
| URHC | upregulated in HCC |
| PTTG3P | pituitary tumor-transforming 3 pseudo gene |
| AFP | alpha-fetoprotein |
| ASOs | antisense oligonucleotides |
| MALAT1 | metastasis-associated in lung adenocarcinoma transcript |
| TINCR | terminal differentiation-induced noncoding RNA |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzmaurice, C.; Akinyemiju, T.F.; Al Lami, F.H.; Alam, T.; Alizadeh-Navaei, R.; Allen, C.; Yonemoto, N. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, a Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018, 4, 1553–1568. [Google Scholar] [PubMed]
- Ferlay, J.; Foucher, E.; Tieulent, J.; Rosso, S.; Coebergh, J.W.; Comber, H.; Forman, D.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur. J. Cancer 2013, 49, 1374–1403. [Google Scholar] [CrossRef] [Green Version]
- Ferlay, J.; Foucher, E.; Tieulent, J.; Rosso, S.; Coebergh, J.W.; Comber, H.; Forman, D.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef] [PubMed]
- Döbrössy, L. Cancer mortality in central-eastern Europe: Facts behind the figures. Lancet Oncol. 2002, 3, 374–381. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef] [Green Version]
- Glassman, M.L.; de Groot, N.; Hochberg, A. Relaxation of imprinting in carcinogenesis. Cancer Genet. Cytogenet. 1996, 89, 69–73. [Google Scholar] [CrossRef]
- Nandwani, A.; Rathore, S.; Datta, M. LncRNAs in cancer: Regulatory and therapeutic implications. Cancer Lett. 2021, 501, 162–171. [Google Scholar] [CrossRef]
- Chen, W.; Sauer, A.M.G.; Chen, M.S., Jr.; Kagawa-Singer, M.; Jemal, A.; Siegel, R.L. Cancer statistics in China. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [Green Version]
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global cancer incidence and mortality rates and trends—An update. Cancer Epidemiol. Biomark. Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef] [Green Version]
- Gotwals, P.; Cameron, S.; Cipolletta, D.; Cremasco, V.; Crystal, A.; Hewes, B.; Mueller, B. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat. Rev. Cancer 2017, 17, 286–301. [Google Scholar] [CrossRef] [PubMed]
- Baudino, A.T. Targeted cancer therapy: The next generation of cancer treatment. Curr. Drug Discov. Technol. 2015, 12, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Da, C.M.; Gong, C.Y.; Nan, W.; Zhou, K.S.; Zuo-Long, W.U.; Zhang, H.H. The role of long non-coding RNA MIAT in cancers. Biomed. Pharmacother. 2020, 129, 110359. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Mester, R.G.; Barzi, A.; Jemal, A. Colorectal cancer statistics. CA Cancer J. Clin. 2017, 67, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Han, X.; Yao, Y.; Li, J.; Li, Z.; Han, X. Role of LncRNAs in the Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 2014. [Google Scholar]
- Han, L.-L.; Lv, Y.; Guo, H.; Ruan, Z.-P.; Nan, K.-J. Implications of biomarkers in human hepatocellular carcinoma pathogenesis and therapy. World J. Gastroenterol. WJG 2014, 20, 10249. [Google Scholar] [CrossRef]
- El–Serag, H.B.; Rudolph, K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, Y. Research Progress on regulating LncRNAs of hepatocellular carcinoma stem cells. OncoTargets Ther. 2021, 14, 917. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; El-Serag, H.B. Hepatocellular carcinoma from epidemiology to prevention: Translating knowledge into practice. J. Clin. Gastroenterol. 2015, 13, 2140–2141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGlynn, K.A.; Petrick, J.L.; London, W.T. Global epidemiology of hepatocellular carcinoma: An emphasis on demographic and regional variability. Clin. Liver Dis. 2015, 19, 223–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledda, C.; Loreto, C.; Zammit, C.; Marconi, A.; Fago, L.; Matera, S.; Constanzo, V.; Fuccio, G.; Palmucci, S.; Ferrante, M.; et al. Non-infective occupational risk factors for hepatocellular carcinoma: A review. Mol. Med. Rep. 2017, 15, 511–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagaraju, G.P.; Dariya, B.; Kasa, P.; Peela, S.; El-Rayes, B.F. Epigenetics in hepatocellular carcinoma. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Wong, L.-S.; Wong, C.-M. Decoding the Roles of Long Noncoding RNAs in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 3137. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Wen, D.Y.; Li, Q.; He, Y.; Yang, H.; Chen, G. Genome-wide analysis of prognostic lncRNAs, miRNAs, and mRNAs forming a competing endogenous RNA network in hepatocellular carcinoma. Cell. Physiol. Biochem. 2018, 48, 1953–1967. [Google Scholar] [CrossRef] [PubMed]
- Longo, D.; Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar]
- Kim, Y.-A.; Park, K.-K.; Lee, S.-J. lncRNAs act as a link between chronic liver disease and hepatocellular carcinoma. Int. J. Mol. Sci. 2020, 21, 2883. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Chen, J.; Zhang, K.; Feng, B.; Wang, R.; Chen, L. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell. Physiol. Biochem. 2015, 36, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, O.; Gnoni, A.; Licchetta, A.; Longo, V.; Calabrese, A.; Argentiero, A.; Delcuratoro, S.; Solimando, A.; Gardini, A.; Silvestris, A. Predictive and prognostic factors in HCC patients treated with sorafenib. Medicina 2019, 55, 707. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Jiang, J.; Xu, Q.; Ni, C.; Yang, L.; Huang, D. A systematic review of long noncoding RNAs in hepatocellular carcinoma: Molecular mechanism and clinical implications. BioMed Res. Int. 2018, 2018, 8126208. [Google Scholar] [CrossRef] [Green Version]
- Ghidini, M.; Braconi, C. Non-coding RNAs in primary liver cancer. Front. Med. 2015, 2, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chu, E.S.H.; Chen, H.Y.; Man, K.; Go, M.Y.Y.; Huang, X.R.; Lan, H.Y.; Sung, J.J.Y.; Yu, J. microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget 2015, 6, 7325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Hu, F.; Hu, X.; Chen, W.; Huang, Y.; Yu, X. Proteomic identification of potential Clonorchis sinensis excretory/secretory products capable of binding and activating human hepatic stellate cells. Parasitol. Res. 2014, 113, 3063–3071. [Google Scholar] [CrossRef] [PubMed]
- Intini, C.; Hodgkinson, T.; Casey, S.M.; Gleeson, J.P.; O’Brien, F.J. Highly Porous Type II Collagen-Containing Scaffolds for Enhanced Cartilage Repair with Reduced Hypertrophic Cartilage Formation. Bioengineering 2022, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Wan, L.Y.; Liang, J.J.; Zhang, Y.Q.; Ai, W.B.; Wu, J.F. The roles of lncRNA in hepatic fibrosis. Cell Biosci. 2018, 8, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piscaglia, F.; Svegliati-Baroni, G.; Barchetti, A.; Pecorelli, A.; Marinelli, S.; Tiribelli, C.; Bellentani, S.; HCC-NAFLD Italian Study Group. Clinical patterns of hepatocellular carcinoma in nonalcoholic fatty liver disease: A multicenter prospective study. Hepatology 2016, 63, 827–838. [Google Scholar] [CrossRef]
- Juhling, F.; Hamdane, N.; Crouchet, E.; Li, S.; El Saghire, H.; Mukherji, A.; Fujiwara, N.; Oudot, M.A.; Thumann, C.; Saviano, A.; et al. Targeting clinical epigenetic reprogramming for chemoprevention of metabolic and viral hepatocellular carcinoma. Gut 2021, 70, 157–169. [Google Scholar] [CrossRef]
- Ogunwobi, O.O.; Harricharran, T.; Huaman, J.; Galuza, A.; Odumuwagun, O.; Tan, Y.; Ma, G.X.; Nguyen, M.T. Mechanisms of hepatocellular carcinoma progression. World J. Gastroenterol. 2019, 25, 2279–2293. [Google Scholar] [CrossRef]
- Okrah, K.; Tarighat, S.; Liu, B.; Koeppen, H.; Wagle, M.C.; Cheng, G.; Sun, C.; Dey, A.; Chang, M.T.; Sumiyoshi, T.; et al. Transcriptomic analysis of hepatocellular carcinoma reveals molecular features of disease progression and tumor immune biology. NPJ Precis. Oncol. 2018, 2, 25. [Google Scholar] [CrossRef] [PubMed]
- Safcak, D.; Drazilova, S.; Gazda, J.; Andrasina, I.; Adamcova-Selcanova, S.; Barila, R.; Mego, M.; Rac, M.; Skladany, L.; Zigrai, M.; et al. Nonalcoholic Fatty Liver Disease-Related Hepatocellular Carcinoma: Clinical Patterns, Outcomes, and Prognostic Factors for Overall Survival—A Retrospective Analysis of a Slovak Cohort. J. Clin. Med. 2021, 10, 3186. [Google Scholar] [CrossRef] [PubMed]
- Geh, D.; Derek, M.; Manas, H.; Reeves, L. Hepatocellular carcinoma in non-alcoholic fatty liver disease—A review of an emerging challenge facing clinicians. Hepatobiliary Surg. Nutr. 2021, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Vesque, A.D.; Decraecker, M.; Blanc, J.-F. Profile of Cabozantinib for the Treatment of Hepatocellular Carcinoma: Patient Selection and Special Considerations. J. Hepatocell. Carcinoma 2020, 7, 91. [Google Scholar] [CrossRef]
- Yang, F.U.; Zhang, L.; Huo, X.S.; Yuan, J.H.; Xu, D.; Yuan, S.X.; Zhu, N.; Zhou, W.P.; Yang, G.S.; Wang, Y.Z.; et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011, 54, 1679–1689. [Google Scholar] [CrossRef]
- Kudo, M. Systemic therapy for hepatocellular carcinoma: 2017 update. Oncology 2017, 93 (Suppl. S1), 135–146. [Google Scholar] [CrossRef]
- Li, D.; Liu, X.; Zhou, J.; Hu, J.; Zhang, D.; Liu, J.; Qiao, Y.; Zhan, Q. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology 2017, 65, 1612–1627. [Google Scholar] [CrossRef] [Green Version]
- Ghanem, I.; Riveiro, M.E.; Paradis, V.; Faivre, S.; de Parga, P.M.V.; Raymond, E. Insights on the CXCL12-CXCR4 axis in hepatocellular carcinoma carcinogenesis. Am. J. Transl. Res. 2014, 6, 340. [Google Scholar]
- Villanueva, A.; Minguez, B.; Forner, A.; Reig, M.; Llovet, J.M. Hepatocellular carcinoma: Novel molecular approaches for diagnosis, prognosis, and therapy. Annu. Rev. Med. 2010, 61, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Dramani Maman, S.T.; Zhu, X.; Liu, X.; Bongolo, C.C.; Liang, C.; Tu, J. Clinical value and potential mechanisms of LINC00221 in hepatocellular carcinoma based on integrated analysis. Epigenomics 2021, 13, 299–317. [Google Scholar] [CrossRef]
- Hu, X.; Chen, R.; Wei, Q.; Xu, X. The Landscape of Alpha Fetoprotein in Hepatocellular Carcinoma: Where Are We? Int. J. Biol. Sci. 2022, 18, 536–551. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Su, H. Long noncoding RNAs act as novel biomarkers for hepatocellular carcinoma: Progress and prospects. Biomed. Res. Int. 2017, 2017, 6049480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.-L.; Su, T.-H.; Chen, P.-J.; Chen, Y.-R. Acute-phase serum amyloid A for early detection of hepatocellular carcinoma in cirrhotic patients with low AFP level. Sci. Rep. 2022, 12, 5799. [Google Scholar] [CrossRef]
- Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017, 18, 655. [Google Scholar] [CrossRef] [PubMed]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.-M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The landscape of circular RNA in cancer. Cell 2019, 176, 869–881.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruber, A.J.; Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 2019, 20, 599–614. [Google Scholar] [CrossRef] [PubMed]
- Dvinge, H.; Guenthoer, J.; Porter, P.L.; Bradley, R.K. RNA components of the spliceosome regulate tissue-and cancer-specific alternative splicing. Genome Res. 2019, 29, 1591–1604. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Rodriguez, D.H.; Pashneh-Tala, S.; Bains, A.K.; Moorehead, R.D.; Kassos, N.; Kelly, A.L.; Paterson, T.E.; Orozco-Diaz, C.A.; Gill, A.A.; Asencio, I.O. Demonstrating the Potential of Using Bio-Based Sustainable Polyester Blends for Bone Tissue Engineering Applications. Bioengineering 2022, 9, 163. [Google Scholar] [CrossRef]
- Ning, J.; Sun, K.; Fan, X.; Jia, K.; Wang, X.; Ma, C.; Wei, L. Necroptosis-related lncRNAs and Hepatocellular Carcinoma Undoubtedly Secret. Res. Sq. 2022, 25. [Google Scholar] [CrossRef]
- Amaral, P.P.; Leonardi, T.; Han, N.; Vire, E.; Gascoigne, D.K.; Arias-Carrasco, R.; Büscher, M.; Pandolfini, L.; Zhang, A.; Pluchino, S.; et al. Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Fernandez-Ruiz, I. A new role for lncRNAs in atherosclerosis. Nat. Rev. Cardiol. 2018, 15, 195. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.-M.; Tsang, F.H.-C.; Ng, I.O.-L. Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, S. LncRNA-ZFAS1 contributes to colon cancer progression through the miR-150-5p/VEGFA axis. Ann. Oncol. 2018, 29, v52–v53. [Google Scholar] [CrossRef]
- Kirchhoff, T.; Simpson, D.; Hekal, T.; Ferguson, R.; Kazlow, E.; Moran, U.; Lee, Y.; Izsak, A.; Wilson, M.; Shapiro, R.; et al. Discovery of novel germline genetic biomarkers of melanoma recurrence impacting exonic and long non-coding RNA (lncRNA) transcripts. Ann. Oncol. 2018, 29, viii463. [Google Scholar] [CrossRef]
- Zhang, J.; Han, X.; Jiang, L.; Han, Z.; Wang, Z. LncRNA CERNA2 is an independent predictor for clinical prognosis and is related to tumor development in gastric cancer. Int. J. Clin. Exp. Pathol. 2018, 11, 5783. [Google Scholar] [PubMed]
- Lei, G.L.; Niu, Y.; Cheng, S.J.; Li, Y.Y.; Bai, Z.F.; Yu, L.X.; Hong, Z.X.; Liu, H.; Liu, H.H.; Yan, J.; et al. Upregulation of long noncoding RNA W42 promotes tumor development by binding with DBN1 in hepatocellular carcinoma. World J. Gastroenterol. 2021, 27, 2586. [Google Scholar] [CrossRef]
- Li, Y.; Guo, D.; Lu, G.; Chowdhury, A.T.M.M.; Zhang, D.; Ren, M.; Chen, Y.; Wang, R.; He, S. LncRNA SNAI3-AS1 promotes PEG10-mediated proliferation and metastasis via decoying of miR-27a-3p and miR-34a-5p in hepatocellular carcinoma. Cell Death Dis. 2020, 11, 685. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Hu, W.; Alvarez-Dominguez, J.R.; Lodish, H.F. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012, 13, 971–983. [Google Scholar] [CrossRef] [Green Version]
- Garcia, L.; Zambalde, E.; Mathias, C.; Barazetti, J.; Gradia, D.; Oliveira, J. lncRNAs in Hallmarks of Cancer and Clinical Applications. In Non-Coding RNAs; IntechOpen: London, UK, 2019. [Google Scholar]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniue, K.; Akimitsu, N. The functions and unique features of lncrnas in cancer development and tumorigenesis. Int. J. Mol. Sci. 2021, 22, 632. [Google Scholar] [CrossRef] [PubMed]
- Parasramka, M.A.; Maji, S.; Matsuda, A.; Yan, I.K.; Patel, T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol. Ther. 2016, 161, 67–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Z.; Shen, J.; Zhang, L.; Li, M.; Hu, W.; Cho, C. Therapeutic targeting of noncoding RNAs in hepatocellular carcinoma: Recent progress and future prospects. Oncol. Lett. 2018, 15, 3395–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhang, Y.; Ding, X.; Li, W. Identification of lncRNA/circRNA-miRNA-mRNA ceRNA Network as Biomarkers for Hepatocellular Carcinoma. Front. Genet. 2022, 538. [Google Scholar] [CrossRef] [PubMed]
- Han, T.-S.; Hur, K.; Cho, H.-S.; Ban, H.S. Epigenetic associations between lncRNA/circRNA and miRNA in hepatocellular Carcinoma. Cancers 2020, 12, 2622. [Google Scholar] [CrossRef]
- Meng, H.; Niu, R.; Huang, C.; Li, J. Circular RNA as a Novel Biomarker and Therapeutic Target for HCC. Cells 2022, 11, 1948. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Glaser, S.; Francis, H.; Alpini, G. Concise review: Functional roles and therapeutic potentials of long non-coding RNAs in cholangiopathies. Front. Med. 2020, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef] [Green Version]
- Kitabayashi, J.; Shirasaki, T.; Shimakami, T.; Nishiyama, T.; Welsch, C.; Funaki, M.; Murai, K.; Sumiyadorj, A.; Takatori, H.; Kitamura, K.; et al. Upregulation of the long non-coding RNA HULC by hepatitis C virus and its regulation of viral replication. J. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Wang, D.; Chen, F.; Zeng, T.; Tang, Q.; Chen, B.; Chen, L.; Dong, Y.; Li, X. Comprehensive biological function analysis of lncRNAs in hepatocellular carcinoma. Genes Dis. 2020, 8, 157–167. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Morales, D.R.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta Rev. Cancer 2015, 1856, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Lin, Z.; Dong, P.; Lu, Z.; Gao, S.; Chen, X.; Wu, C.; Yu, F. Activation of hepatic stellate cells is suppressed by microRNA-150. Int. J. Mol. Med. 2013, 32, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Dong, P.; Gao, S.; Wang, N.; Yu, F. High expression of serum miR-17-5p associated with poor prognosis in patients with hepatocellular Carcinoma. Hepatogastroenterology 2013, 60, 549–552. [Google Scholar] [PubMed]
- Zheng, J.; Wu, C.; Lin, Z.; Guo, Y.; Shi, L.; Dong, P.; Lu, Z.; Gao, S.; Liao, Y.; Chen, B.; et al. Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through micro RNA-mediated control of DNA methylation—A novel mechanism suppressing liver fibrosis. FEBS J. 2014, 281, 88–103. [Google Scholar] [CrossRef]
- Zhou, B.; Yang, H.; Bao, Y.-L.; Yang, S.-M.; Liu, J.; Xiao, Y.-F. Translation of noncoding RNAs and cancer. Cancer Lett. 2021, 497, 89–99. [Google Scholar] [CrossRef]
- Zhang, L.-G.; Zhou, X.-K.; Zhou, R.-J.; Lv, H.-Z.; Li, W.-P. Long non-coding RNA LINC00673 promotes hepatocellular carcinoma progression and metastasis through negatively regulating miR-205. Am. J. Cancer Res. 2017, 7, 2536. [Google Scholar] [PubMed]
- de Oliveira, J.C.; Oliveira, L.C.; Mathias, C.; Pedroso, G.A.; Lemos, D.S.; Salviano-Silva, A.; Jucoski, T.S.; Lobo-Alves, S.C.; Zambalde, E.P.; Cipolla, G.A.; et al. Long non-coding RNAs in cancer: Another layer of complexity. J. Gene Med. 2019, 21, e3065. [Google Scholar] [PubMed] [Green Version]
- ZZhang, T.; Hu, H.; Yan, G.; Wu, T.; Liu, S.; Chen, W.; Ning, Y.; Lu, Z. Long non-coding RNA and breast cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819843889. [Google Scholar]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Meng, X.-M.; Huang, C.; Wu, B.-M.; Zhang, L.; Lv, X.-W.; Li, J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014, 344, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-DiNardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level rulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629. [Google Scholar] [CrossRef] [Green Version]
- Liz, J.; Esteller, M. lncRNAs and microRNAs with a role in cancer development. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 169–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, Z.-S.; Niu, X.-J.; Wang, W.-H. Long non-coding RNAs in hepatocellular carcinoma: Potential roles and clinical implications. World J. Gastroenterol. 2017, 23, 5860. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, A.; Zhou, T.; Tan, C.; Alpini, G.; Glaser, S. Role of Non-Coding RNAs in the Progression of Liver Cancer: Evidence from Experimental Models. Cancers 2019, 11, 1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.-J.; Zheng, J.-J.; Dong, P.-H.; Fan, X.-M. Long non-coding RNAs and hepatocellular carcinoma. Mol. Clin. Oncol. 2015, 3, 13–17. [Google Scholar] [CrossRef] [Green Version]
- Engreitz, J.M.; Ollikainen, N.; Guttman, M. Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 756–770. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Wu, J.; Liangpunsakul, S.; Wang, L. Long non-coding RNA in liver metabolism and disease: Current status. Liver Res. 2017, 1, 163–167. [Google Scholar] [CrossRef]
- Li, C.H.; Chen, Y. Targeting long non-coding RNAs in cancers: Progress and prospects. Int. J. Biochem. Cell Biol. 2013, 45, 1895–1910. [Google Scholar] [CrossRef]
- Wan, Y.; Kertesz, M.; Spitale, R.C.; Segal, E.; Chang, H.Y. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011, 12, 641–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Jayaraj, G.G.; Pandey, S.; Scaria, V.; Maiti, S. Potential G-quadruplexes in the human long non-coding transcriptome. RNA Biol. 2012, 9, 81–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Zhang, G.; Li, J.; Yang, R.; Chen, S.; Wu, S.; Zhang, F.; Bai, Y.; Zhao, H.; Wang, Y.; et al. Long noncoding RNA RGMB-AS1 indicates a poor prognosis and modulates cell proliferation, migration and invasion in lung adenocarcinoma. PLoS ONE 2016, 11, e0150790. [Google Scholar] [CrossRef]
- Wilk, R.; Hu, J.; Blotsky, D.; Krause, H.M. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 2016, 30, 594–609. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Zhao, B.S.; He, C. Nucleic acid modifications in regulation of gene expression. Cell Chem. Biol. 2016, 23, 74–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Sun, J.; Wang, J.; Song, Y.; Gao, P.; Shi, J.; Chen, P.; Wang, Z. Long noncoding RNAs in gastric cancer: Functions and clinical applications. OncoTargets Ther. 2016, 9, 681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Xu, A.; Zhang, J.; He, X.; Pan, Y.; Cheng, G.; Qin, C.; Hua, L.; Wang, Z. Prognostic significance of long non-coding RNA MALAT-1 in various human carcinomas: A meta-analysis. Genet. Mol. Res. 2016, 15, 15017433. [Google Scholar]
- Sun, L.; Guo, Y.; He, P.; Xu, X.; Zhang, X.; Wang, H.; Tang, T.; Zhou, W.; Xu, P.; Xie, P. Genome-wide profiling of long noncoding RNA expression patterns and CeRNA analysis in mouse cortical neurons infected with different strains of borna disease virus. Genes Dis. 2019, 6, 147–158. [Google Scholar] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [PubMed] [Green Version]
- Isin, M.; Dala, Y.N. LncRNAs and neoplasia. Clin. Chim. Acta 2015, 444, 280–288. [Google Scholar]
- Serviss, J.T.; Johnsson, P.; Grandér, D. An emerging role for long non-coding RNAs in cancer metastasis. Front. Genet. 2014, 5, 234. [Google Scholar] [PubMed] [Green Version]
- Harries, L.W. Long non-coding RNAs and human disease. Biochem. Soc. Trans. 2012, 40, 902–906. [Google Scholar] [PubMed]
- Wang, Q.-f.; Wang, Q.-l.; Cao, M.-b. LncRNA PITPNA-AS1 as a potential diagnostic marker and therapeutic target promotes hepatocellular carcinoma progression via modulating miR-448/ROCK1 axis. Front. Med. 2021, 8, 668787. [Google Scholar] [CrossRef]
- Amicone, L.; Citarella, F.; Cicchini, C. Epigenetic regulation in hepatocellular carcinoma requires long noncoding RNAs. Biomed. Res. Int. 2015. [Google Scholar]
- Yari, H.; Jin, L.; Teng, L.; Wang, Y.; Wu, Y.; Liu, G.; Gao, W.; Liang, J.; Xi, Y.; Feng, Y.C.; et al. LncRNA REG1CP promotes tumorigenesis through an enhancer complex to recruit FANCJ helicase for REG3A transcription. Nat. Commun. 2019, 10, 5334. [Google Scholar] [PubMed] [Green Version]
- Yan, X.; Hu, Z.; Feng, Y.; Hu, X.; Yuan, J.; Zhao, S.D.; Zhang, Y.; Yang, L.; Shan, W.; He, Q.; et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 2015, 28, 529–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, T. LncRNA DRAIR is a novel prognostic and diagnostic biomarker for gastric cancer. Mamm. Genome 2021, 32, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.-X.; Wang, J.; Gu, X.-Y.; Huang, C.-H.; Chen, C.; Hong, M.; Chen, Z. TTN-AS1 as a potential diagnostic and prognostic biomarker for multiple cancers. Biomed. Pharmacother. 2021, 135, 111169. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef] [Green Version]
- DiStefano, J.K.; Gerhard, G.S. Long Noncoding RNAs and Human Liver Disease. Annu. Rev. Pathol. 2021, 17, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Yuan, X.-Q.; Zhang, C.-Y.; Peng, J.-Y.; Zhang, Y.-Q.; Pan, X.; Li, G.-C. The emergence of long non-coding RNAs in hepatocellular carcinoma: An update. J. Cancer 2018, 9, 2549. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Chen, J.; Ding, C.; Jin, X.; Jia, Z.; Peng, J. Lnc RNA NR 2F1-AS 1 regulates hepatocellular carcinoma oxaliplatin resistance by targeting ABCC 1 via miR-363. J. Cell. Mol. Med. 2018, 22, 3238–3245. [Google Scholar] [CrossRef]
- El-Ashmawy, N.E.; Hussien, F.Z.; El-Feky, O.A.; Hamouda, S.M.; Al-Ashmawy, G.M. Serum LncRNA-ATB and FAM83H-AS1 as diagnostic/prognostic non-invasive biomarkers for breast cancer. Life Sci. 2020, 259, 118193. [Google Scholar] [CrossRef]
- Maier, I.M.; Maier, A.C. miRNAs and lncRNAs: Potential non-invasive biomarkers for endometriosis. Biomedicines 2021, 9, 1662. [Google Scholar] [CrossRef]
- Herceg, Z.; Hainaut, P. Genetic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis. Mol. Oncol. 2007, 1, 26–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagy, Á.; Munkácsy, G.; Győrffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Menyhárt, O.; Harami-Papp, H.; Sukumar, S.; Schäfer, R.; Magnani, L.; de Barrios, O.; Győrffy, B. Guidelines for the selection of functional assays to evaluate the hallmarks of cancer. Biochim. Biophys. Acta Rev. Cancer 2016, 1866, 300–319. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zitello, E.; Guo, R.; Deng, Y. The function of LncRNAs and their role in the prediction, diagnosis, and prognosis of lung cancer. Clin. Transl. Med. 2021, 11, e367. [Google Scholar] [CrossRef]
- Zhang, R.; Xia, L.Q.; Lu, W.W.; Zhang, J.; Zhu, J.S. LncRNAs and cancer. Oncol. Lett. 2016, 12, 1233–1239. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.; Abbas, M.; Ullah, M.F.; Aziz, M.H.; Beylerli, O.; Alam, M.A.; Syed, M.A.; Uddin, S.; Ahmad, A. Long non-coding RNAs regulated NF-κB signaling in cancer metastasis: Micromanaging by not so small non-coding RNAs. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Tanne, A.; Muniz, L.R.; Puzio-Kuter, A.; Leonova, K.I.; Gudkov, A.V.; Ting, D.T.; Monasson, R.; Cocco, S.; Levine, A.J.; Bhardwaj, N.; et al. Distinguishing the immunostimulatory properties of noncoding RNAs expressed in cancer cells. Proc. Natl. Acad. Sci. USA 2015, 112, 15154–15159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef] [Green Version]
- Pan, L.; Liang, W.; Gu, J.; Zang, X.; Huang, Z.; Shi, H.; Chen, J.; Fu, M.; Zhang, P.; Xiao, X.; et al. Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget 2018, 9, 1915. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Li, J.; Chen, C.; Zhang, R.; Wang, K. Pan-cancer analysis of long non-coding RNA NEAT1 in various cancers. Genes Dis. 2018, 5, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Ren, K.; Wang, M.; Wang, J.; Li, E.; Hou, C.; Su, Y.; Jin, Y.; Zou, Q.; Zhou, P.; et al. Long non-coding RNA RACGAP1P promotes breast cancer invasion and metastasis via miR-345-5p/RACGAP1-mediated mitochondrial fission. Mol. Oncol. 2021, 15, 543. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Lin, F.; Xu, M.; Zhao, X. Long non-coding RNA LINC00665 promotes melanoma cell growth and migration via regulating the miR-224-5p/VMA21 axis. Exp. Dermatol. 2020, 31, 64–73. [Google Scholar] [CrossRef] [PubMed]
- He, R.-Z.; Luo, D.-X.; Mo, Y.-Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019, 6, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.Q.; Zeng, D.; Chen, C.F.; Sun, S.M.; Lu, X.F.; Peng, C.Y.; Lin, H.Y. Long noncoding RNA H19 is a critical oncogenic driver and contributes to epithelial-mesenchymal transition in papillary thyroid Carcinoma. Cancer Manag. Res. 2019, 11, 2059. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Liu, G.; Xu, W.; Liu, H.; Bu, Q.; Sun, D. Long noncoding RNA H19 inhibits cell viability, migration, and invasion via downregulation of IRS-1 in thyroid cancer cells. Technol. Cancer Res. 2017, 16, 1102–1112. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.R.; Yan, L.; Liu, Y.T.; Cao, L.; Guo, Y.H.; Zhang, Y.; Yao, H.; Cai, L.; Shang, H.B.; Rui, W.W.; et al. Inhibition of mTORC1 by lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary tumours. Nat. Commun. 2018, 9, 4624. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Hao, G.; Sun, Y.; Li, L.; Wang, Y. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. OncoTargets Ther. 2018, 11, 8001. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.M.; Xu, S.F.; Zheng, Y.; Wang, P.; Zhang, L.; Shi, S.S.; Wu, T.; Li, Y.; Zhao, J.; Tian, Q.; et al. Long non-coding RNA H19 is responsible for the progression of lung adenocarcinoma by mediating methylation-dependent repression of CDH1 promoter. J. Cell. Mol. Med. 2019, 23, 6411–6428. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Dong, Y.-J. LncRNA SNHG20 promotes migration and invasion of ovarian cancer via modulating the microRNA-148a/ROCK1 axis. J. Ovarian Res. 2021, 14, 168. [Google Scholar] [CrossRef]
- Yan, Q.; Tian, Y.; Hao, F. Downregulation of lncRNA UCA1 inhibits proliferation and invasion of cervical cancer cells through miR-206 expression. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2021. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Li, J.; Li, F.; Li, H.; Bei, S.; Zhang, X.; Yang, Z. Long noncoding RNA VCAN-AS1 contributes to the progression of gastric cancer via regulating p53 expression. J. Cell. Physiol. 2020, 235, 4388–4398. [Google Scholar] [CrossRef]
- Lou, C.; Zhao, J.; Gu, Y.; Li, Q.; Tang, S.; Wu, Y.; Tang, J.; Zhang, C.; Li, Z.; Zhang, Y. LINC01559 accelerates pancreatic cancer cell proliferation and migration through YAP-mediated pathway. J. Cell. Physiol. 2020, 235, 3928–3938. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Duan, Y.; Wang, P. SP1-mediated upregulation of lncRNA SNHG4 functions as a ceRNA for miR-377 to facilitate prostate cancer progression through regulation of ZIC5. J. Cell. Physiol. 2020, 235, 3916–3927. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, F.; Xiong, Y.; Cheng, X.; Qiu, Z.; Song, R. LncRNA TTN-AS1 sponges miR-376a-3p to promote colorectal cancer progression via upregulating KLF15. Life Sci. 2020, 244, 116936. [Google Scholar] [CrossRef] [PubMed]
- Qiao, K.; Ning, S.; Wan, L.; Wu, H.; Wang, Q.; Zhang, X.; Xu, S.; Pang, D. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, T.; Pistoni, M.; Sancisi, V.; Gobbi, G.; Torricelli, F.; Donati, B.; Ribisi, S.; Gugnoni, M.; Ciarrocchi, A. RAIN is a novel Enhancer-associated lncRNA that controls RUNX2 expression and promotes breast and thyroid cancer. Mol. Cancer Res. 2020, 18, 140–152. [Google Scholar] [CrossRef] [Green Version]
- Chang, Q.-Q.; Chen, C.-Y.; Chen, Z.; Chang, S. LncRNA PVT1 promotes proliferation and invasion through enhancing Smad3 expression by sponging miR-140-5p in cervical cancer. Radiol. Oncol. 2019, 53, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Han, Q.; Chen, Y.; Chen, X.; Ma, R.; Chang, Q.; Yin, D. Upregulation of the long non-coding RNA FOXD2-AS1 is correlated with tumor progression and metastasis in papillary thyroid cancer. Am. J. Transl. Res. 2019, 11, 5457. [Google Scholar]
- Ouyang, T.; Zhang, Y.; Tang, S.; Wang, Y. Long non-coding RNA LINC00052 regulates miR-608/EGFR axis to promote progression of head and neck squamous cell Carcinoma. Exp. Mol. Pathol. 2019, 111, 104321. [Google Scholar] [CrossRef]
- Tang, C.; Wang, Y.; Zhang, L.; Wang, J.; Wang, W.; Han, X.; Mu, C.; Gao, D. Identification of novel LncRNA targeting Smad2/PKCα signal pathway to negatively regulate malignant progression of glioblastoma. J. Cell. Physiol. 2020, 235, 3835–3848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Wu, K.; Yang, Y.; Zhu, D.; Zhang, C.; Zhao, S. Long noncoding RNA ADAMTS9-AS2 suppresses the progression of esophageal cancer by mediating CDH3 promoter methylation. Mol. Carcinog. 2020, 59, 32–44. [Google Scholar] [CrossRef]
- Wang, W.; Xia, S.; Zhan, W. The long non-coding RNA ENST00000489676 influences papillary thyroid cancer cell proliferation and invasion through regulating MiR-922. J. Cancer 2019, 10, 5434. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Zhang, J.; Huang, S.; Li, P. lncRNA OSER1-AS1 acts as a ceRNA to promote tumorigenesis in hepatocellular carcinoma by regulating miR-372-3p/Rab23 axis. Biochem. Biophys. Res. Commun. 2020, 521, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Zhang, R.; An, X.; Li, Z.; Fang, C.; Pan, B.; Chen, W.; Xu, G.; Han, W. LncRNA HOXA-AS3 confers cisplatin resistance by interacting with HOXA3 in non-small-cell lung Carcinoma cells. Oncogenesis 2019, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Xu, Z.; Chen, X.; Wang, X.; Zeng, S.; Zhao, Z.; Qian, L.; Li, Z.; Wei, J.; Huo, L.; et al. Novel function of lncRNA ADAMTS9-AS2 in promoting temozolomide resistance in glioblastoma via upregulating the FUS/MDM2 ubiquitination axis. Front. Cell Dev. Biol. 2019, 7, 217. [Google Scholar]
- Yokoyama, Y.; Sakatani, T.; Wada, R.; Ishino, K.; Kudo, M.; Koizumi, M.; Yamada, T.; Yoshida, H.; Naito, Z. In vitro and in vivo studies on the association of long non-coding RNAs H19 and urothelial cancer associated 1 with the susceptibility to 5-fluorouracil in rectal cancer. Int. J. Oncol. 2019, 55, 1361–1371. [Google Scholar] [CrossRef]
- Tamang, S.; Acharya, V.; Roy, D.; Sharma, R.; Aryaa, A.; Sharma, U.; Khandelwal, A.; Prakash, H.; Vasquez, K.M.; Jain, A. SNHG12: An LncRNA as a potential therapeutic target and biomarker for human cancer. Front. Oncol. 2019, 9, 901. [Google Scholar] [CrossRef]
- Avazpour, N.; Hajjari, M.; Birgani, M.T. HOTAIR: A promising long non-coding RNA with potential role in breast invasive carcinoma. Front. Genet. 2017, 8, 170. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Zhou, G.; Wang, H.; Liu, Y.; Chen, B.; Chen, W.; Lin, C.; Wu, S.; Gong, A.; Xu, M. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int. J. Cancer 2020, 146, 2901–2912. [Google Scholar] [CrossRef]
- Li, T.; Xie, J.; Shen, C.; Cheng, D.; Shi, Y.; Wu, Z.; Deng, X.; Chen, H.; Shen, B.; Peng, C.; et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular Carcinoma. Oncogene 2016, 35, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; He, S.; Li, K.; Zhang, K.; Jin, H.; Shi, J.; Cheng, Y.; Wang, L.; Liu, P. LncRNA SNHG8 Promotes Liver Cancer Proliferation and Metastasis by Sponging miR-542-3p and miR-4701-5p. Res. Sq. 2021, 25. [Google Scholar] [CrossRef]
- Wang, F.; Yuan, J.-H.; Wang, S.-B.; Yang, F.; Yuan, S.-X.; Ye, C.; Yang, N.; Zhou, W.-P.; Li, W.-L.; Sun, S.-H. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology 2014, 60, 1278–1290. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Lin, J.; Zhang, J.; Song, Z.; Zheng, D.; Chen, F.; Zhuang, X.; Li, A.; Liu, X. LncRNA SNHG17 contributes to proliferation, migration, and poor prognosis of hepatocellular carcinoma. Can. J. Gastroenterol. Hepatol. 2021; 11, 9990338. [Google Scholar]
- Liu, Y.; Mao, X.; Ma, Z.; Chen, W.; Guo, X.; Yu, L.; Deng, X.; Jiang, F.; Li, T.; Lin, N.; et al. Aberrant regulation of LncRNA TUG1-microRNA-328-3p-SRSF9 mRNA Axis in hepatocellular carcinoma: A promising target for prognosis and therapy. Mol. Cancer 2022, 21, 36. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Cui, J.; Zhong, R.; Sun, G. Role of lncRNA LINC01194 in hepatocellular carcinoma via the miR-655-3p/SMAD family member 5 axis. Bioengineered 2022, 13, 1115–1125. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, G.; Celsa, C.; Giammanco, A.; Spatola, F.; Petta, S. The burden of hepatocellular carcinoma in non-alcoholic fatty liver disease: Screening issue and future perspectives. Int. J. Mol. Sci. 2019, 20, 5613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Q.; Yan, X.; Yang, L.; Liu, Z.; Yuan, Z.; Zhang, Y. lncRNA CYTOR promotes cell proliferation and tumor growth via miR-125b/SEMA4C axis in hepatocellular carcinoma. Oncol. Lett. 2021, 22, 796. [Google Scholar] [CrossRef]
- Fu, C.; Li, J.; Li, P.; Cheng, D. LncRNA DNAJC3-AS1 Promotes Hepatocellular Carcinoma (HCC) Progression via Sponging Premature miR-27b. Cancer Manag. Res. 2021, 13, 8575. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, L.; Chen, T.; Liu, X.; Guo, Y.; Zhu, Q.; Tong, X.; Yang, W.; Xu, Q.; Huang, D.; et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol. Cancer 2019, 18, 28. [Google Scholar] [CrossRef]
- Hu, X.; Li, Q.; Zhang, J. The long noncoding RNA LINC00908 facilitates hepatocellular carcinoma progression via interaction with Sox-4. Cancer Manag. Res. 2019, 11, 8789. [Google Scholar] [CrossRef] [Green Version]
- Dai, W.; Dai, J.L.; Tang, M.H.; Ye, M.S.; Fang, S. lncRNA-SNHG15 accelerates the development of hepatocellular carcinoma by targeting miR-490-3p/histone deacetylase 2 axis. World J. Gastroenterol. 2019, 25, 5789. [Google Scholar] [CrossRef] [PubMed]
- Sui, C.-J.; Zhou, Y.-M.; Shen, W.-F.; Dai, B.-H.; Lu, J.-J.; Zhang, M.-F.; Yang, J.-M. Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J. Mol. Med. 2016, 94, 1281–1296. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.D.; Chen, W.M.; Qi, F.Z.; Xia, R.; Sun, M.; Xu, T.P.; Yin, L.; Zhang, E.B.; De, W.; Shu, Y.Q. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J. Hematol. Oncol. 2015, 8, 50. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.D.; Chen, W.M.; Qi, F.Z.; Sun, M.; Xu, T.P.; Ma, P.; Shu, Y.Q. Long non-coding RNA TUG1 is up-regulated in hepatocellular Carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol. Cancer. 2015, 14, 165. [Google Scholar] [CrossRef] [Green Version]
- Cao, C.; Sun, J.; Zhang, D.; Guo, X.; Xie, L.; Li, X.; Wu, D.; Liu, L. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology 2015, 148, 415–426.e18. [Google Scholar] [CrossRef]
- Chen, L.; Yao, H.; Wang, K.; Liu, X. Long non-coding RNA MALAT1 regulates ZEB1 expression by sponging miR-143-3p and promotes hepatocellular carcinoma progression. J. Cell. Biochem. 2017, 118, 4836–4843. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Liu, S.; Cheng, Y.; Lu, L.; Shi, J.; Xu, G.; Li, N.; Cheng, K.; Wu, M.; Cheng, S.; et al. ICAM-1-related noncoding RNA in cancer stem cells maintains ICAM-1 expression in hepatocellular carcinoma. Clin Cancer Res. 2016, 22, 2041–2050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Xie, J.; Shen, C.; Cheng, D.; Shi, Y.; Wu, Z.; Deng, X.; Chen, H.; Shen, B.; Peng, C.; et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015, 75, 3181–3191. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.-X.; Yang, F.; Yang, Y.; Tao, Q.-F.; Zhang, J.; Huang, G.; Wang, R.-Y.; Yang, S.; Huo, X.-S.; Zhang, L.; et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology 2012, 56, 2231–2241. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, M.; Gross, M.; Goyal, A.; Polycarpou-Schwarz, M.; Miersch, T.; Ernst, A.S.; Leupold, J.; Patil, N.; Warnken, U.; Allgayer, H.; et al. The lncRNA CASC9 and RNA binding protein HNRNPL form a complex and co-regulate genes linked to AKT signaling. Hepatology 2018, 68, 1817–1832. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, J.; Peng, Y.; He, W.; Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 2020, 19, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, L.-J.; Li, Y.; Wang, S.-D.; Wang, X.-S.; Fang, F.; Wang, W.-Y.; Lv, P.; Zhao, D.-H.; Wei, F.; Qi, L. Long noncoding RNA lncCAMTA1 promotes proliferation and cancer stem cell-like properties of liver cancer by inhibiting CAMTA1. Int. J. Mol. Sci. 2016, 17, 1617. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, Q.; Qi, J.; Wang, W.; Zhang, D.; Li, Z.; Qin, C. lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPARγ pathway in hepatocellular carcinoma. Int. J. Oncol. 2018, 53, 551–566. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Z.; Yan, G.X.; Dong, D.S.; Xin, H.; Liu, Z.Y. LncRNA-ATB promotes autophagy by activating Yes-associated protein and inducing autophagy-related protein 5 expression in hepatocellular carcinoma. World J. Gastroenterol. 2019, 25, 5310. [Google Scholar] [CrossRef]
- Pan, W.; Li, W.; Zhao, J.; Huang, Z.; Zhao, J.; Chen, S.; Wang, C.; Xue, Y.; Huang, F.; Fang, Q.; et al. lnc RNA-PDPK 2P promotes hepatocellular carcinoma progression through the PDK 1/AKT/Caspase 3 pathway. Mol. Oncol. 2019, 13, 2246–2258. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Huo, X.; Yang, X.R.; He, J.; Cheng, L.; Wang, N.; Deng, X.; Jin, H.; Wang, N.; Wang, C.; et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer 2017, 16, 136. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Yu, W.; Zhou, Q.; Zhang, J.; Jiang, H.; Hao, D.; Wang, J.; Zhou, Z.; He, C.; Xiao, Z. A novel lncRNA IHS promotes tumor proliferation and metastasis in HCC by regulating the ERK-and AKT/GSK-3β-signaling pathways. Mol. Ther. Nucleic Acids 2019, 16, 707–720. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Xiong, Q.; Li, S.; Yang, X.; Ge, F. Integrated proteomic and transcriptomic analysis reveals long noncoding RNA HOX transcript antisense intergenic RNA (HOTAIR) promotes hepatocellular carcinoma cell proliferation by regulating opioid growth factor receptor (OGFr). Mol. Cell. Proteom. 2018, 17, 146–159. [Google Scholar] [CrossRef] [Green Version]
- Cheng, D.; Deng, J.; Zhang, B.; He, X.; Meng, Z.; Li, G.; Ye, H.; Zheng, S.; Wei, L.; Deng, X.; et al. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine 2018, 36, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Li, J.; Wang, L. Large intervening non-coding RNA HOTAIR is an indicator of poor prognosis and a therapeutic target in human cancers. Int. J. Mol. Sci. 2014, 15, 18985–18999. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Zhang, L.; Zheng, S. Role of the long non-coding RNA HOTAIR in hepatocellular Carcinoma. Oncol. Lett. 2017, 14, 1233–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; An, J.; Wu, M.; Zheng, Q.; Gui, X.; Li, T.; Pu, H.; Lu, D. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget 2015, 6, 27847. [Google Scholar] [CrossRef] [PubMed]
- You, L.-N.; Tai, Q.-W.; Xu, L.; Hao, Y.; Guo, W.-J.; Zhang, Q.; Tong, Q.; Zhang, H.; Huang, W.-K. Exosomal LINC00161 promotes angiogenesis and metastasis via regulating miR-590-3p/ROCK axis in hepatocellular carcinoma. Cancer Gene Ther. 2021, 28, 719–736. [Google Scholar] [CrossRef]
- Lin, Y.; Jian, Z.; Jin, H.; Wei, X.; Zou, X.; Guan, R.; Huang, J. Long non-coding RNA DLGAP1-AS1 facilitates tumorigenesis and epithelial–mesenchymal transition in hepatocellular carcinoma via the feedback loop of miR-26a/b-5p/IL-6/JAK2/STAT3 and Wnt/β-catenin pathway. Cell Death Dis. 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Yi, T.; Wang, T.; Shi, Y.; Peng, X.; Tang, S.; Zhong, L.; Chen, Y.; Li, Y.; He, K.; Wang, M.; et al. Long noncoding RNA 91H overexpression contributes to the growth and metastasis of HCC by epigenetically positively regulating IGF2 expression. Liver Int. 2020, 40, 456–467. [Google Scholar] [CrossRef]
- Teng, F.; Zhang, J.-X.; Chang, Q.-M.; Wu, X.-B.; Tang, W.-G.; Wang, J.-F.; Feng, J.-F.; Zhang, Z.-P.; Hu, Z.-Q. LncRNA MYLK-AS1 facilitates tumor progression and angiogenesis by targeting miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2020, 39, 1–18. [Google Scholar]
- Tian, C.; Abudoureyimu, M.; Lin, X.; Chu, X.; Wang, R. Linc-ROR facilitates progression and angiogenesis of hepatocellular carcinoma by modulating DEPDC1 expression. Cell Death Dis. 2021, 12, 1047. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.; Ma, L. Long noncoding RNA highly upregulated in liver cancer promotes the progression of hepatocellular carcinoma and attenuates the chemosensitivity of oxaliplatin by regulating miR-383-5p/vesicle-associated membrane protein-2 axis. Pharmacol. Res. Perspect. 2021, 9, e00815. [Google Scholar] [CrossRef]
- Qian, H.; Wu, Q.; Wu, J.H.; Tian, X.Y.; Xu, W.; Hao, C.Y. Long noncoding LINC00238 restrains Hepatocellular Carcinoma Malignant Phenotype via Sponging miR-522. Res. Sq. 2021, 21. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, R.; Zhao, J.; Zeng, Z.; Shi, Z.; Lu, Q.; Guo, J.; Li, L.; Yao, Y.; Liu, X.; et al. LncRNA TMEM220-AS1 suppresses hepatocellular carcinoma cell proliferation and invasion by regulating the TMEM220/β-catenin axis. J. Cancer 2021, 12, 6805. [Google Scholar] [CrossRef]
- Sheng, J.-Q.; Wang, M.-R.; Fang, D.; Liu, L.; Huang, W.-J.; Tian, D.-A.; He, X.-X.; Li, P.-Y. LncRNA NBR2 inhibits tumorigenesis by regulating autophagy in hepatocellular carcinoma. Biomed. Pharmacother. 2021, 133, 111023. [Google Scholar] [CrossRef]
- Lei, G.-L.; Fan, H.-X.; Wang, C.; Niu, Y.; Li, T.-L.; Yu, L.-X.; Hong, Z.-X.; Yan, J.; Wang, X.-L.; Zhang, S.-G.; et al. Long non-coding ribonucleic acid W5 inhibits progression and predicts favorable prognosis in hepatocellular carcinoma. World J. Gastroenterol. 2021, 27, 55. [Google Scholar] [CrossRef]
- Pan, W.; Zhang, N.; Liu, W.; Liu, J.; Zhou, L.; Liu, Y.; Yang, M. The long noncoding RNA GAS8-AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8. J. Biol. Chem. 2018, 293, 17154–17165. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Zhou, Y.; Huan, L.; Xu, L.; Shen, M.; Huang, S.; Liang, L. LncRNA MIR22HG inhibits growth, migration and invasion through regulating the miR-10a-5p/NCOR2 axis in hepatocellular carcinoma cells. Cancer Sci. 2019, 110, 973–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.; Tang, Z.; Chen, K.; Liu, Y.; Yu, G.; Chen, Q.; Dang, H.; Chen, F.; Ling, J.; Zhu, L.; et al. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J. Exp. Clin. Cancer Res. 2018, 37, 214. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, Y.; Li, M.; Wang, L. Long noncoding RNA GAS5 inhibits metastasis by targeting miR-182/ANGPTL1 in hepatocellular Carcinoma. Am. J. Cancer Res. 2019, 9, 108. [Google Scholar] [PubMed]
- Yu, Z.; Zhao, H.; Feng, X.; Li, H.; Qiu, C.; Yi, X.; Tang, H.; Zhang, J. Long non-coding RNA FENDRR acts as a miR-423-5p sponge to suppress the Treg-mediated immune escape of hepatocellular Carcinoma cells. Mol. Ther. Nucleic Acids 2019, 17, 516–529. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-G.; Wang, T.; Shi, M.; Zhai, B. Long noncoding RNA EPB41L4A-AS2 inhibits hepatocellular carcinoma development by sponging miR-301a-5p and targeting FOXL1. Exp. Clin. Cancer Res. 2019, 38, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Li, N.; Huang, Z.; Chen, R.; Yi, P.; Kang, R.; Tang, D.; Hu, X.; Fan, X. A novel lncRNA, TCONS_00006195, represses hepatocellular carcinoma progression by inhibiting enzymatic activity of ENO1. Cell Death Dis. 2018, 9, 153. [Google Scholar] [CrossRef]
- Ni, W.; Zhang, Y.; Zhan, Z.; Ye, F.; Liang, Y.; Huang, J.; Chen, K.; Chen, L.; Ding, Y. A novel lncRNA uc. 134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J. Hematol. Oncol. 2017, 10, 91. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Song, C.; Duan, B.; Zhang, X.; Li, D.; Zhu, L.; Gao, H. LncRNA-SVUGP2 suppresses progression of hepatocellular carcinoma. Oncotarget 2017, 8, 97835. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Wang, G.; Liu, Y.; Mei, C.; Yao, Y.; Wu, X.; Chen, X.; Ma, W.; Li, K.; Zhang, Z.; et al. A novel long non-coding RNA RP11-286H15. 1 represses hepatocellular carcinoma progression by promoting ubiquitination of PABPC4. Cancer Lett. 2021, 499, 109–121. [Google Scholar] [CrossRef]
- Huang, J.-F.; Guo, Y.-J.; Zhao, C.-X.; Yuan, S.-X.; Wang, Y.; Tang, G.-N.; Zhou, W.-P.; Sun, S.-H. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology 2013, 57, 1882–1892. [Google Scholar] [CrossRef]
- Zhuang, L.K.; Yang, Y.T.; Ma, X.; Han, B.; Wang, Z.S.; Zhao, Q.Y.; Wu, L.Q.; Qu, Z.Q. MicroRNA-92b promotes hepatocellular Carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016, 7, e2203. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Deng, W.-L.; Zhao, B.-G.; Xu, Y.; Wang, X.-W.; Fang, Y.; Xiao, H.-J. FOXO3-induced lncRNA LOC554202 contributes to hepatocellular carcinoma progression via the miR-485-5p/BSG axis. Cancer Gene Ther. 2021, 29, 326–340. [Google Scholar] [CrossRef]
- Wan, T.; Zheng, J.; Yao, R.; Yang, S.; Zheng, W.; Zhou, P. LncRNA DDX11-AS1 accelerates hepatocellular carcinoma progression via the miR-195-5p/MACC1 pathway. Ann. Hepatol. 2021, 20, 100258. [Google Scholar] [CrossRef]
- Wang, H.; Liang, L.; Dong, Q.; Huan, L.; He, J.; Li, B.; Yang, C.; Jin, H.; Wei, L.; Yu, C.; et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics 2018, 8, 2814. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, H.; Wan, X.; Yang, X.; Zhu, C.; Wang, A.; He, L.; Miao, R.; Chen, S.; Zhao, H. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular Carcinoma. Biomed. Res. Int. 2014, 2014, 780521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, W.Y.; Wei, H.Y.; Li, K.M.; Wang, R.B.; Xu, X.Q.; Feng, R. LINC00511 as a ceRNA promotes cell malignant behaviors and correlates with prognosis of hepatocellular carcinoma patients by modulating miR-195/EYA1 axis. Biomed. Pharmacother. 2020, 121, 109642. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Han, J.; Zhang, J.; Li, G.; Liu, H.; Cui, X.; Xu, Y.; Li, T.; Liu, J.; Wang, C. The long noncoding RNAs PVT1 and uc002mbe. 2 in sera provide a new supplementary method for hepatocellular carcinoma diagnosis. Medicine 2016, 95, e4436. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.-K.; Pang, C.; Yang, Y.; Duan, Q.; Zhang, J.; Liu, W.-C. Serum long noncoding RNA urothelial carcinoma-associated 1: A novel biomarker for diagnosis and prognosis of hepatocellular carcinoma. Int. J. Med. Res. 2018, 46, 348–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Shi, H.; Wang, X.; Wang, B.; Qu, Q.; Geng, H.; Sun, H. Identification of diagnostic long non-coding RNA biomarkers in patients with hepatocellular carcinoma. Mol. Med. Rep. 2019, 20, 1121–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Hu, R.; Pang, J.; Zhang, G.; Yan, W.; Li, Z. Serum long noncoding RNA LRB1 as a potential biomarker for predicting the diagnosis and prognosis of human hepatocellular carcinoma. Oncol. Lett. 2018, 16, 1593–1601. [Google Scholar] [CrossRef] [Green Version]
- Luo, T.; Chen, M.; Zhao, Y.; Wang, D.; Liu, J.; Chen, J.; Luo, H.; Li, L. Macrophage-associated lncRNA ELMO1-AS1: A novel therapeutic target and prognostic biomarker for hepatocellular carcinoma. OncoTargets Ther. 2019, 12, 6203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.-C.; Yang, F.; Yuan, S.-X.; Ma, J.-Z.; Liu, F.; Yuan, J.-H.; Bi, F.-R.; Lin, K.-Y.; Yin, J.-H.; Cao, G.-W.; et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology 2016, 63, 850–863. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, B.; Zhang, M.; Guo, W.; Wu, Z.; Wang, Y.; Jia, L.; Li, S.; Caesar-Johnson, S.J.; Demchok, J.A.; et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell 2018, 33, 706–720.e9. [Google Scholar] [CrossRef] [Green Version]
- Xing, Z.; Lin, A.; Li, C.; Liang, K.; Wang, S.; Liu, Y.; Park, P.K.; Qin, L.; Wei, Y.; Hawke, D.H.; et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 2014, 159, 1110–1125. [Google Scholar] [CrossRef] [Green Version]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef] [Green Version]
- Guccione, E.; Bassi, C.; Casadio, F.; Martinato, F.; Cesaroni, M.; Schuchlautz, H.; Lüscher, B.; Amati, B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 2007, 449, 933–937. [Google Scholar] [CrossRef]
- Chen, X.; Tang, F.-R.; Arfuso, F.; Cai, W.-Q.; Ma, Z.; Yang, J.; Sethi, G. The emerging role of long non-coding RNAs in the metastasis of hepatocellular carcinoma. Biomolecules 2020, 10, 66. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.-X.; Zhang, J.; Xu, Q.-G.; Yang, Y.; Zhou, W.-P. Long noncoding RNA, the methylation of genomic elements and their emerging crosstalk in hepatocellular carcinoma. Cancer Lett. 2016, 379, 239–244. [Google Scholar] [CrossRef]
- Xu, X.; Lou, Y.; Tang, J.; Teng, Y.; Zhang, Z.; Yin, Y.; Zhuo, H.; Tan, Z. The long non-coding RNA Linc-GALH promotes hepatocellular carcinoma metastasis via epigenetically regulating Gankyrin. Cell Death Dis. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Nebel, S.; Lux, M.; Kuth, S.; Bider, F.; Dietrich, W.; Egger, D.; Boccaccini, A.R.; Kasper, C. Alginate Core–Shell Capsules for 3D Cultivation of Adipose-Derived Mesenchymal Stem Cells. Bioengineering 2022, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Xie, Y.; Xu, X.; Yin, Y.; Jiang, R.; Deng, L.; Tan, Z.; Gangarapu, V.; Tang, J.; Sun, B. Bidirectional transcription of Linc00441 and RB1 via H3K27 modification-dependent way promotes hepatocellular carcinoma. Cell Death Dis. 2017, 8, e2675. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Gao, Y.; Yao, L.; Liu, Y.; Huang, L.; Yan, Z.; Zhao, W.; Zhu, P.; Weng, H. LncFZD6 initiates Wnt/β-catenin and liver TIC self-renewal through BRG1-mediated FZD6 transcriptional activation. Oncogene 2018, 37, 3098–3112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, G.X.; Do, B.T.; Webster, D.E.; Khavari, P.A.; Chang, H.Y. Dicer-microRNA-Myc circuit promotes transcription of hundreds of long noncoding RNAs. Nat. Struct. Mol. Biol. 2014, 21, 585–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, D.D.H.; Kessler, C.; E Niehus, S.; Mahnkopf, M.; Koch, A.; Tamura, T. Myc target gene, long intergenic noncoding RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and cell survival by titrating tumor suppressor microRNAs. Oncogene 2018, 37, 75–85. [Google Scholar] [CrossRef]
- Li, W.; Kang, Y. A new Lnc in metastasis: Long noncoding RNA mediates the prometastatic functions of TGF-β. Cancer Cell 2014, 25, 557–559. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.H.; Liu, X.N.; Wang, T.T.; Pan, W.; Tao, Q.F.; Zhou, W.P.; Wang, F.; Sun, S.H. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat. Cell Biol. 2017, 19, 820–832. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zuo, Q.; Hu, B.; Jin, H.; Wang, C.; Cheng, Z.; Deng, X.; Yang, C.; Ruan, H.; Yu, C.; et al. A novel, liver-specific long noncoding RNA LINC01093 suppresses HCC progression by interaction with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett. 2019, 450, 98–109. [Google Scholar] [CrossRef]
- Moore, J.B., IV; Uchida, S. Functional characterization of long noncoding RNAs. Curr. Opin. Cardiol. 2020, 35, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.-H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, S.; Kauppinen, S. Long non-coding RNAs in liver cancer and nonalcoholic steatohepatitis. Non-Coding RNA 2020, 6, 34. [Google Scholar]
- Xie, C.-R.; Wang, F.; Zhang, S.; Wang, F.-Q.; Zheng, S.; Li, Z.; Lv, J.; Qi, H.-Q.; Fang, Q.-L.; Wang, X.-M.; et al. Long noncoding RNA HCAL facilitates the growth and metastasis of hepatocellular carcinoma by acting as a ceRNA of LAPTM4B. Mol. Ther. Nucleic Acids 2017, 9, 440–451. [Google Scholar] [CrossRef] [Green Version]
- Hou, Z.; Xu, X.; Zhou, L.; Fu, X.; Tao, S.; Zhou, J.; Tan, D.; Liu, S. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumor Biol. 2017, 39, 1010428317718135. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Zhou, J.; Sun, Y.; Li, N.; Miao, M.; Jiao, B.; Chen, H. The noncoding RNA HOXD-AS1 is a critical regulator of the metastasis and apoptosis phenotype in human hepatocellular carcinoma. Mol. Cancer 2017, 16, 125. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-P.; Xu, H.-X.; Yu, Y.; He, J.-D.; Wang, Z.; Xu, Y.-J.; Wang, C.-Y.; Zhang, H.-M.; Zhang, R.-X.; Zhang, J.-J.; et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget 2016, 7, 42431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Li, Z.; Liu, L.; Wang, Q.; Li, S.; Chen, D.; Hu, Z.; Yu, T.; Ding, J.; Li, J.; et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology 2018, 67, 171–187. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Li, S.-Y.; Fang, J.-H.; Zhu, Y.; Yang, J.-E. Functional long non-coding RNAs in hepatocellular carcinoma. Cancer Lett. 2020, 500, 281–291. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, J.; Liu, X.; Li, S.; Wang, Q.; Chen, D.; Hu, Z.; Yu, T.; Ding, J.; Li, J.; et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat. Commun. 2018, 9, 1572. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.H.; Yin, C.; Chen, S.J.; Wen, L.Z.; Ding, K.; Lei, S.J.; Liu, J.P.; Wang, J.; Chen, K.X.; Jiang, H.L.; et al. The HNF1α-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1. Mol. Cancer 2018, 17, 63. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Huo, X.-S.; Yuan, S.-X.; Zhang, L.; Zhou, W.-P.; Wang, F.; Sun, S.-H. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell 2013, 49, 1083–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, G.-Z.; Lehwald, N.; Jang, K.Y.; Baek, J.; Xu, B.; Omary, M.B.; Sylvester, K.G. Wnt/β-catenin signaling protects mouse liver against oxidative stress-induced apoptosis through the inhibition of forkhead transcription factor FoxO3. J. Biol. Chem. 2013, 288, 17214–17224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Xu, C.; Liu, Y. Involvement of ERK1/2 signaling in proliferation of eight liver cell types during hepatic regeneration in rats. Genet. Mol. Res. 2013, 12, 665–677. [Google Scholar] [CrossRef]
- Xu, D.; Yang, F.; Yuan, J.-H.; Zhang, L.; Bi, H.-S.; Zhou, C.-C.; Liu, F.; Wang, F.; Sun, S.-H. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/β-Catenin signaling. Hepatology 2013, 58, 739–751. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [Green Version]
- Clevers, H. Wnt/β-catenin signaling in development and disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef] [Green Version]
- Clevers, H.; Nusse, R. Wnt/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef] [Green Version]
- Monga, S.P. β-catenin signaling and roles in liver homeostasis, injury, and tumorigenesis. Gastroenterology 2015, 148, 1294–1310. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Zhu, X.; Qin, F.; Zhang, Y.; Lin, J.; Ding, Y.; Yang, Z.; Shang, Y.; Wang, L.; Zhang, Q.; et al. Linc00210 drives Wnt/β-catenin signaling activation and liver tumor progression through CTNNBIP1-dependent manner. Mol. Cancer 2018, 17, 73. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.-H.; Zhang, J.-B.; Dang, Z.; Li, X.; Zhou, T.; Liu, J.; Wang, D.-S.; Song, W.-J.; Dou, K.-F. Long non-coding RNA URHC regulates cell proliferation and apoptosis via ZAK through the ERK/MAPK signaling pathway in hepatocellular carcinoma. Int. J. Biol. Sci. 2014, 10, 664. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yang, F.U.; Yuan, J.H.; Yuan, S.X.; Zhou, W.P.; Huo, X.S.; Xu, D.; Bi, H.S.; Wang, F.; Sun, S.H. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 2013, 34, 577–586. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Ahn, K.S.; Kim, C.; Siveen, K.S.; Ong, T.H.; Shanmugam, M.K.; Li, F.; Shi, J.; Kumar, A.P.; Wang, L.Z.; et al. Ascochlorin, an isoprenoid antibiotic inhibits growth and invasion of hepatocellular carcinoma by targeting STAT3 signaling cascade through the induction of PIAS3. Mol. Oncol. 2015, 9, 818–833. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, P.; Li, F.; Shanmugam, M.K.; Vali, S.; Abbasi, T.; Kapoor, S.; Ahn, K.S.; Kumar, A.P.; Sethi, G. Honokiol inhibits signal transducer and activator of transcription-3 signaling, proliferation, and survival of hepatocellular carcinoma cells via the protein tyrosine phosphatase SHP-1. J. Cell. Physiol. 2012, 227, 2184–2195. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-L.; Cao, S.-W.; Ou, Q.-S.; Yang, B.; Zheng, S.-H.; Tang, J.; Chen, J.; Hu, Y.-W.; Zheng, L.; Wang, Q. The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol. Cancer 2018, 17, 93. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Y.; Yu, S. Long noncoding RNA AWPPH promotes hepatocellular carcinoma progression through YBX1 and serves as a prognostic biomarker. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1805–1816. [Google Scholar] [CrossRef]
- Tang, J.; Zhuo, H.; Zhang, X.; Jiang, R.; Ji, J.; Deng, L.; Qian, X.; Zhang, F.; Sun, B. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014, 5, e1549. [Google Scholar] [CrossRef] [Green Version]
- Zhi, Y.; Huang, S.; Lina, Z. Suppressor of Cytokine Signaling 6 in cancer development and therapy: Deciphering its emerging and suppressive roles. Cytokine Growth Factor Rev. 2022, 64, 21–32. [Google Scholar]
- Zhang, L.-H.; Ji, J.-F. Molecular profiling of hepatocellular carcinomas by cDNA microarray. World J. Gastroenterol. 2005, 11, 463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aravalli, R.N.; Steer, C.J.; Cressman, E.N. Molecular mechanisms of hepatocellular carcinoma. Hepatology 2008, 48, 2047–2063. [Google Scholar] [CrossRef]
- Ogunwobi, O.O.; Puszyk, W.; Dong, H.-J.; Liu, C. Epigenetic upregulation of HGF and c-Met drives metastasis in hepatocellular carcinoma. PLoS ONE 2013, 8, e63765. [Google Scholar] [CrossRef] [PubMed]
- Long, L.; Xiang, H.; Liu, J.; Zhang, Z.; Sun, L. ZEB1 mediates doxorubicin (Dox) resistance and mesenchymal characteristics of hepatocarcinoma cells. Exp. Mol. Pathol. 2019, 106, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.; Wen, X.; Su, K.; Gou, W. MiR-552 promotes the proliferation, migration and EMT of hepatocellular carcinoma cells by inhibiting AJAP 1 expression. J. Cell Mol. Med. 2019, 23, 1541–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.I.; Lin, N.A.N.; Chen, Z.; Xu, R. Hypoxia-induced secretion of platelet-derived growth factor-BB by hepatocellular carcinoma cells increases activated hepatic stellate cell proliferation, migration and expression of vascular endothelial growth factor-A. Mol. Med. Rep. 2015, 11, 691–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galiè, M.; Sorrentino, C.; Montani, M.; Micossi, L.; Di Carlo, E.; D’Antuono, T.; Calderan, L.; Marzola, P.; Benati, D.; Merigo, F.; et al. Mammary carcinoma provides highly tumourigenic and invasive reactive stromal cells. Carcinogenesis 2005, 26, 1868–1878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez–Gea, V.; Toffanin, S.; Friedman, S.L.; Llovet, J.M. Role of the microenvironment in the pathogenesis and treatment of hepatocellular Carcinoma. Gastroenterology 2013, 144, 512–527. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; E Powers, S.; Zhu, W.; Hannun, Y.A. Substantial contribution of extrinsic risk factors to cancer development. Nature 2016, 529, 43–47. [Google Scholar] [CrossRef]
- Yamashita, T.; Ji, J.; Budhu, A.; Forgues, M.; Yang, W.; Wang, H.Y.; Jia, H.; Ye, Q.; Qin, L.X.; Wauthier, E.; et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009, 136, 1012–1024.e4. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.F.; Ho, D.W.; Ng, M.N.; Lau, C.K.; Yu, W.C.; Ngai, P.; Chu, P.W.; Lam, C.T.; Poon, R.T.; Fan, S.T. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 2008, 13, 153–166. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Chan, K.-W.; Hu, L.; Lee, T.K.-W.; Wo, J.Y.-H.; Ng, I.O.-L.; Zheng, B.-J.; Guan, X.-Y. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007, 132, 2542–2556. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Hao, X.; Yan, M.; Yao, M.; Ge, C.; Gu, J.; Li, J. Cancer stem/progenitor cells are highly enriched in CD133+ CD44+ population in hepatocellular carcinoma. Int. J. Cancer 2010, 126, 2067–2078. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, N.; Ishii, H.; Mimori, K.; Tanaka, F.; Ohkuma, M.; Kim, H.M.; Akita, H.; Takiuchi, D.; Hatano, H.; Nagano, H.; et al. CD13 is a therapeutic target in human liver cancer stem cells. J. Clin. Investig. 2010, 120, 3326–3339. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.-Y.; Hsu, Y.-C.; Ho, H.J.; Chen, Y.-J.; Lee, T.-Y.; Lin, J.-T. Association between ultrasonography screening and mortality in patients with hepatocellular carcinoma: A nationwide cohort study. Gut 2016, 65, 693–701. [Google Scholar] [CrossRef]
- Tzartzeva, K.; Obi, J.; Rich, N.E.; Parikh, N.D.; Marrero, J.A.; Yopp, A.; Waljee, A.K.; Singal, A.G. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: A meta-analysis. Gastroenterology 2018, 154, 1706–1718.e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, T.-S.; Wu, Y.-C.; Tung, S.-Y.; Wei, K.-L.; Hsieh, Y.-Y.; Huang, H.-C.; Chen, W.-M.; Shen, C.-H.; Lu, C.-H.; Wu, C.-S.; et al. Alpha-fetoprotein measurement benefits hepatocellular carcinoma surveillance in patients with cirrhosis. Am. J. Gastroenterol. 2015, 110, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Bisceglie, A.M.; Sterling, R.K.; Chung, R.T.; Everhart, J.E.; Dienstag, J.L.; Bonkovsky, H.L.; Wright, E.C.; Everson, G.T.; Lindsay, K.L.; Lok, A.S.; et al. Serum alpha-fetoprotein levels in patients with advanced hepatitis C: Results from the HALT-C. Trial. J. Hepatol. 2005, 43, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Moon, A.M.; Weiss, N.S.; Beste, L.A.; Su, F.; Ho, S.B.; Jin, G.-Y.; Lowy, E.; Berry, K.; Ioannou, G.N. No association between screening for hepatocellular carcinoma and reduced cancer-related mortality in patients with cirrhosis. Gastroenterology 2018, 155, 1128–1139.e6. [Google Scholar] [CrossRef]
- Xu, C.; Xu, Z.; Zhang, Y.; Evert, M.; Calvisi, D.F.; Chen, X. β-Catenin signaling in hepatocellular carcinoma. J. Clin. Investig. 2022, 132, e154515. [Google Scholar] [CrossRef]
- Li, J.; Yang, X.; Huang, L.; Zhu, X.; Qiu, M.; Yan, J.; Yan, Y.; Wei, S. Treatment Strategy for Post-hepatectomy Recurrent Hepatocellular Carcinoma Within the Milan Criteria: Repeat Resection, Local Ablative Therapy or Transarterial Chemoembolization? Indian J. Surg. 2022, 26, 1–6. [Google Scholar] [CrossRef]
- Kamel, M.M.; Matboli, M.; Sallam, M.; Montasser, I.F.; Saad, A.S.; El-Tawdi, A.H. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl. Res. 2016, 168, 134–145. [Google Scholar] [CrossRef]
- Kodidela, S.; Behera, A.; Reddy, A.B.M. Risk factors and clinical aspects associated with hepatocellular carcinoma: Role of long noncoding RNAs. In Theranostics and Precision Medicine for the Management of Hepatocellular Carcinoma; Elsevier: Amsterdam, The Netherlands, 2022; pp. 341–356. [Google Scholar]
- Luo, P.; Liang, C.; Zhang, X.; Liu, X.; Wang, Y.; Wu, M.; Feng, X.; Tu, J. Identification of long non-coding RNA ZFAS1 as a novel biomarker for diagnosis of HCC. Biosci. Rep. 2018, 31, 38. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhao, J.; Yu, S.; Wang, Z.; He, X.; Su, Y.; Guo, T.; Sheng, H.; Chen, J.; Zheng, Q.; et al. Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin. Chem. 2019, 65, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Chen, Y.; Dong, X.; Wang, X. Serum exosomal long noncoding RNAs ENSG00000258332. 1 and LINC00635 for the diagnosis and prognosis of hepatocellular carcinoma. Cancer Epidemiol. Biomark. Prev. 2018, 27, 710–716. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, R.; Kanda, T.; Yokosuka, O.; Kato, N.; Matsuoka, S.; Moriyama, M. Exosomes and hepatocellular carcinoma: From bench to bedside. Int. J. Mol. Sci. 2019, 20, 1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Ji, Q.; Yang, Y.; Li, Q.; Wang, Z. Exosome: Function and role in cancer metastasis and drug resistance. Technol. Cancer Res. Treat. 2018, 17, 1533033818763450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, W.; Sun, Y.; Liu, L.; Zhou, B.; Wang, S.; Gu, D. Circulating LncRNAs serve as diagnostic markers for hepatocellular carcinoma. Cell. Physiol. Biochem. 2017, 44, 125–132. [Google Scholar] [CrossRef]
- Wu, E.-R.; Hsieh, M.-J.; Chiang, W.-L.; Hsueh, K.-C.; Yang, S.-F.; Su, S.-C. Association of lncRNA CCAT2 and CASC8 gene polymorphisms with hepatocellular carcinoma. Int. J. Environ. Res. 2019, 16, 2833. [Google Scholar] [CrossRef] [Green Version]
- Tonus, C.; Cloquette, K.; Ectors, F.; Piret, J.; Gillet, L.; Antoine, N.; Desmecht, D.; Vanderplasschen, A.; Waroux, O.; Grobet, L. Long term-cultured and cryopreserved primordial germ cells from various chicken breeds retain high proliferative potential and gonadal colonisation competency. Reprod. Fertil. Dev. Rep. 2016, 28, 628–639. [Google Scholar] [CrossRef]
- Hong, D.; Kurzrock, R.; Kim, Y.; Woessner, R.; Younes, A.; Nemunaitis, J.; Fowler, N.; Zhou, T.; Schmidt, J.; Jo, M.; et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 2015, 7, 314ra185. [Google Scholar] [CrossRef] [Green Version]
- Arun, G.; Diermeier, S.; Akerman, M.; Chang, K.-C.; Wilkinson, J.E.; Hearn, S.; Kim, Y.; MacLeod, A.R.; Krainer, A.R.; Norton, L.; et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016, 30, 34–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esposito, R.; Bosch, N.; Lanzós, A.; Polidori, T.; Pulido-Quetglas, C.; Johnson, R. Hacking the cancer genome: Profiling therapeutically actionable long non-coding RNAs using CRISPR-Cas9 screening. Cancer Cell 2019, 35, 545–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, M.; Wang, H.; Hu, X.; Cao, X. lncRNA MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically promote inflammation-related hepatocellular carcinoma progression. Oncoimmunology 2019, 8, e1518628. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Tan, G.; Jiang, X.; Han, P.; Zhai, B.; Dong, X.; Qiao, H.; Jiang, H.; Sun, X. An artificial lncRNA targeting multiple miRNAs overcomes sorafenib resistance in hepatocellular carcinoma cells. Oncotarget 2016, 7, 73257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Ren, F.; Zhang, Y.; Qin, Y.; Shang, J.; Wang, Y.; Wei, P.; Guo, J.; Jia, H.; Zhao, T. Taraxasterol prompted the anti-tumor effect in mice burden hepatocellular carcinoma by regulating T lymphocytes. Cell Death Discov. 2022, 8, 264. [Google Scholar] [CrossRef]
- Su, G.L. AGA Clinical Practice Guideline on Systemic Therapy for Hepatocellular Carcinoma. Gastroenterology 2022, 162, 920–934. [Google Scholar] [CrossRef]
- Park, R.; da Silva, L.L.; Nissaisorakarn, V.; Riano, I.; Williamson, S.; Sun, W.; Saeed, A. Comparison of Efficacy of Systemic Therapies in Advanced Hepatocellular Carcinoma: Updated Systematic Review and Frequentist Network Meta-Analysis of Randomized Controlled Trials. J. Hepatocell Carcinoma 2021, 8, 145–154. [Google Scholar] [CrossRef]
- Marron, T.U.; Fiel, M.I.; Hamon, P.; Fiaschi, N.; Kim, E.; Ward, S.C.; Zhao, Z.; Kim, J.; Kennedy, P.; Gunasekaran, G.; et al. Neoadjuvant cemiplimab for resectable hepatocellular carcinoma: A single-arm, open-label, phase 2 trial. Lancet Gastroenterol. Hepatol. 2022, 7, 219–229. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, W.; Jiang, L.; Chen, Y. Recent advances in systemic therapy for hepatocellular carcinoma. Biomark. Res. 2022, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, T.M.; Leger, A.J.; Pandey, S.K.; MacLeod, A.R.; Nakamori, M.; Cheng, S.H.; Wentworth, B.M.; Bennett, C.F.; Thornton, C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 2012, 488, 111–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Sedano, S.; Allen, R.; Gong, J.; Cho, M.; Sharma, S. Current treatment landscape for advanced hepatocellular carcinoma: Patient outcomes and the impact on quality of life. Cancers 2019, 11, 841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grützner, F.; Kaessmann, H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef]
- Juan, V.; Crain, C.; Wilson, C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000, 28, 1221–1227. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Zhu, Y.; Sun, D.; Zhang, Q. Emerging roles of long non-coding RNAs in the tumor microenvironment. Int. J. Biol. Sci. 2020, 16, 2094. [Google Scholar] [CrossRef]
- Wei, L.; Lee, D.; Law, C.-T.; Zhang, M.S.; Shen, J.; Chin, D.W.-C.; Zhang, A.; Tsang, F.H.-C.; Wong, C.L.-S.; Ng, I.O.-L.; et al. Genome-wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for Sorafenib resistance in HCC. Nat. Commun. 2019, 10, 4681. [Google Scholar] [CrossRef] [Green Version]
- Bester, A.C.; Lee, J.D.; Chavez, A.; Lee, Y.-R.; Nachmani, D.; Vora, S.; Victor, J.; Sauvageau, M.; Monteleone, E.; Rinn, J.L.; et al. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell 2018, 173, 649–664.e20. [Google Scholar] [CrossRef]
- Liu, S.J.; Horlbeck, M.A.; Weissman, J.S.; Lim, D.A. Genome-Scale Perturbation of Long Noncoding RNA Expression Using CRISPR Interference. In Functional Analysis of Long Non-Coding RNAs; Springer: New York, NY, USA, 2021; pp. 323–338. [Google Scholar]
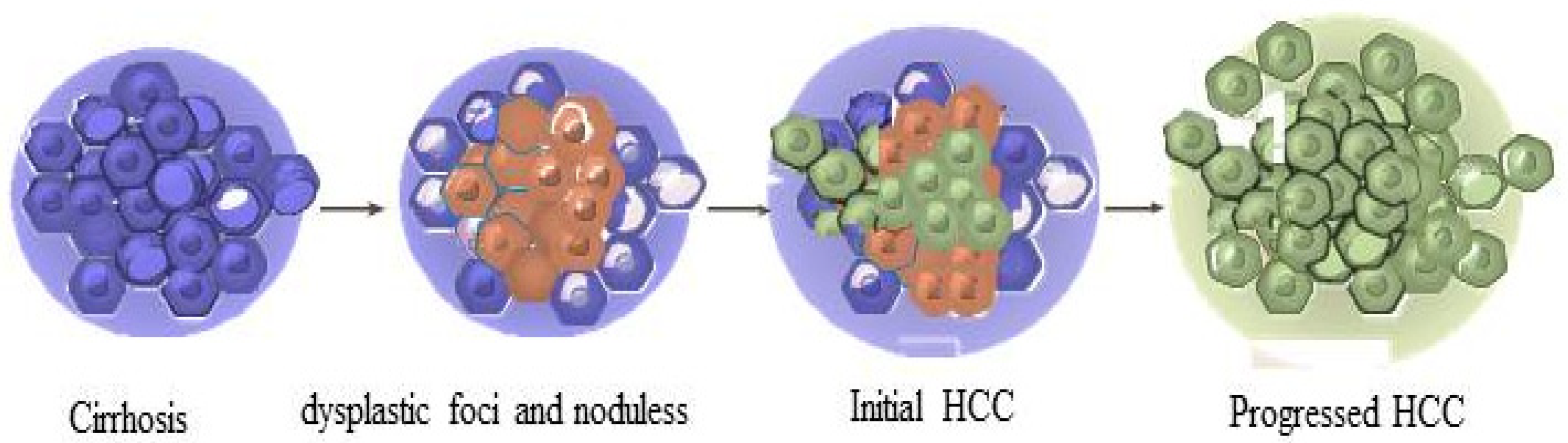
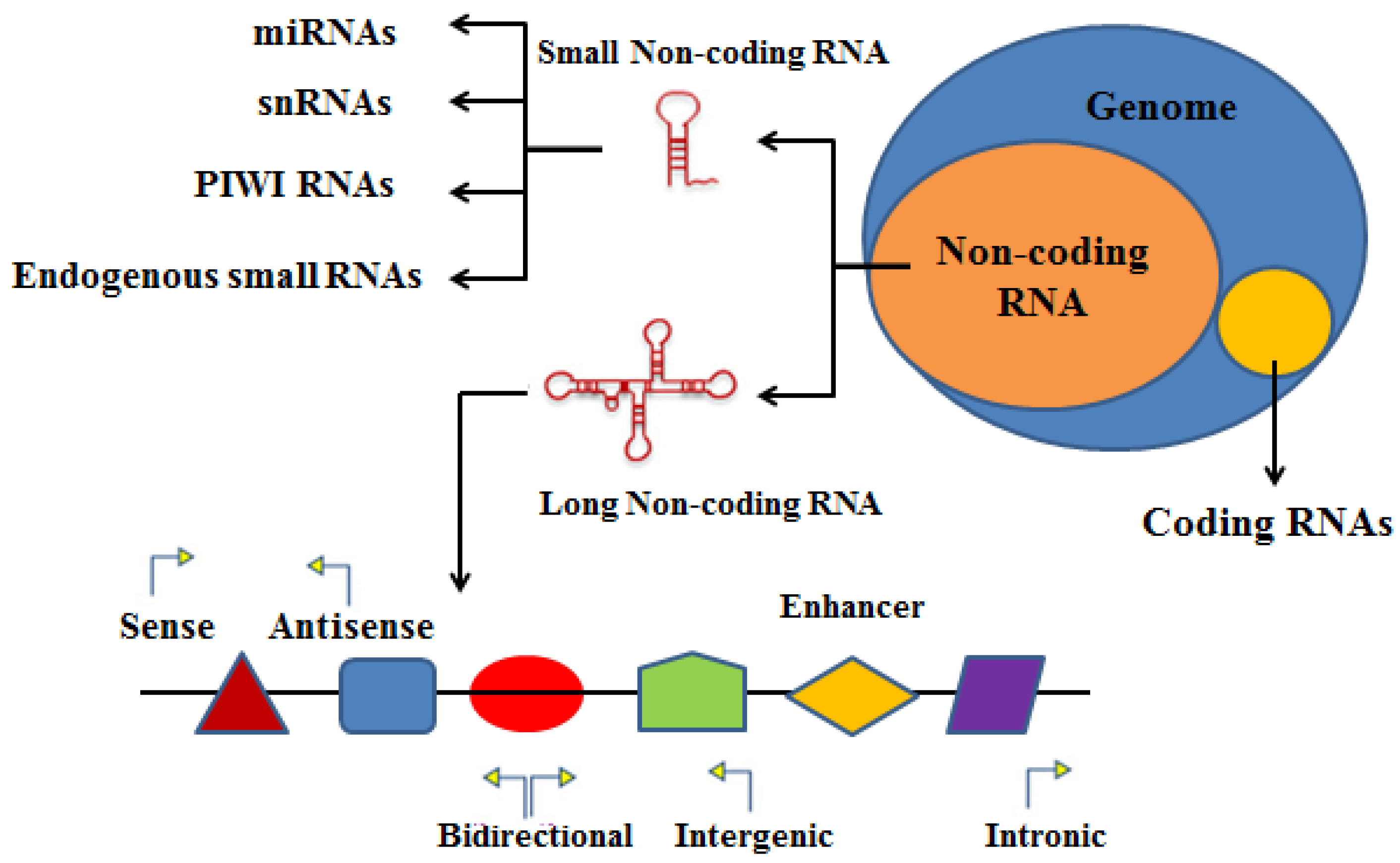
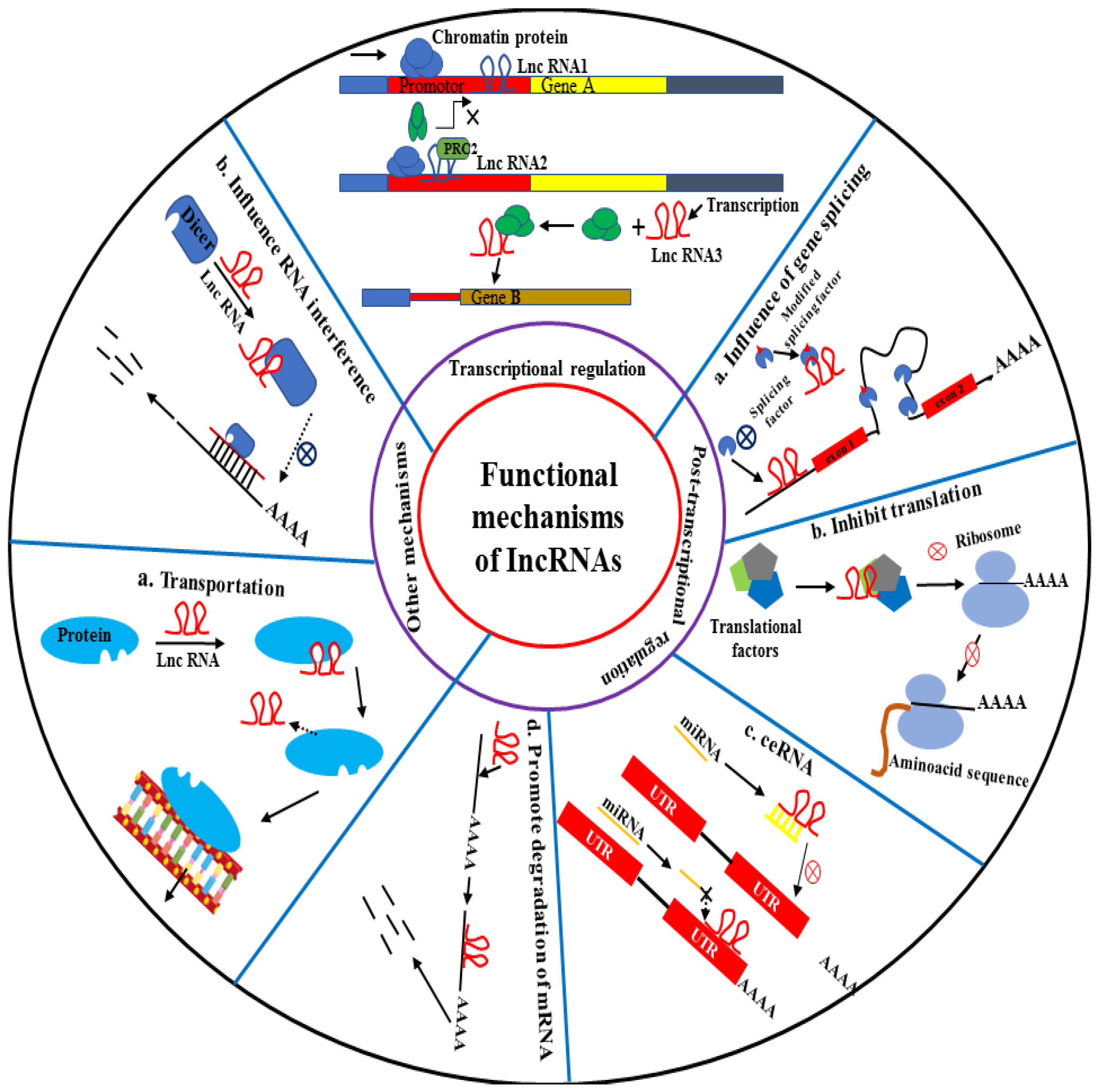

| LncRNAs | Target Pathways/ Mechanisms | Biological Functions in Cancers | Type of Cancer | Reference |
|---|---|---|---|---|
| LncRNA 00665 | miR-224-5p/VMA21 | Promoting proliferation, invasion, and migration of cancer cells | Melanoma | [146] |
| lncRNA RACGAP1P | miR-345-5p/RACGAP1 | Breast cancer | [145] | |
| SNHG20 | miR-148a/ROCK1 | Ovarian cancer | [153] | |
| UCA1 | miR-206 | Cervical cancer | [154] | |
| VCAN-AS1 | p53 | Gastric cancer | [155] | |
| LINC01559 | YAP | Pancreatic cancer | [156] | |
| SNHG4 | ZIC5 | Prostate cancer | [157] | |
| TTN-AS1 | KLF15 | Colorectal cancer | [158] | |
| LINC00673 | miR-515-5p/MARK4/Hippo | Breast cancer | [159] | |
| RAIN | RUNX2 | Breast/thyroid | [160] | |
| PVT1 | Smad3/miR-140-5p | Cervical cancer | [161] | |
| FOXD2-AS1 | miR-185-5p | Thyroid cancer | [162] | |
| LINC00052 | miR-608/EGFR | Head/neck cancer | [163] | |
| TCONS-00020456 | Smad2/PKCa | Suppression of proliferation and invasion of cancer cells | Glioblastoma cancer | [164] |
| ADAMTS9-AS2 | CDH3 | Esophageal cancer | [165] | |
| ENST00000489676 | MiR-922 | Thyroid cancer | [166] | |
| OSER1-AS1 | miR-372-3p/Rab23 | Hepatocellular carcinoma | [167] | |
| HOXA-AS3 | HOXA3 | Prognosis and efficacy | NSCL cancer | [168] |
| ADAMTS9-AS2 | FUS/MDM2 | Glioblastoma cancer | [169] | |
| UCA1, H19 | 5-fluorouracil | Rectal cancer | [170] | |
| SNHG12 | ------ | Potential biomarkers | Pan-cancer | [171] |
| HOTAIR | ------ | Breast cancer | [172] | |
| SNHG11 | ------ | Colorectal cancer | [173] |
| LncRNAs | Target Pathways/Mechanisms | Biological Functions in HCC | Reference |
|---|---|---|---|
| LncRNA CYTOR | miR-125b/SEMA4C | Promoting proliferation, invasion, and migration of cancer cells Angiogenesis and metastasis Tumorigenesis and EMT Growth and metastasis Progression and angiogenesis | [181] |
| DNAJC3-AS1 | miR-27b | [182] | |
| LncRNA SNHG8 | miR-542-3p and miR-4701-5p | [175] | |
| MCM3AP-AS1 | miR-194-5p/FOXA1 axis | [183] | |
| RNA LINC00908 | Sox-4 | [184] | |
| SNHG15 | miR-490-3p/histone deacetylase 2 axis | [185] | |
| GIHCG | miR-200b/a/429 PPAR gamma | [186] | |
| ANRIL | EZH2 protein Target gene DNA | [187] | |
| TUG1 | EZH2 protein Target gene DNA | [188] | |
| UFC1 | β-catenin mRNA HuR protein | [189] | |
| MALAT1 | miR-143-3p | [190] | |
| ICR | ICAM-1 mRNA | [191] | |
| ZFAS1 | miR-150 | [192] | |
| MVIH | PGK1 protein | [193] | |
| CASC9 | HNRNPL protein | [194,195] | |
| LncCAMTA1 | CAMTA1 | [196] | |
| Ftx | PPAR gamma | [197] | |
| ATB | Autophagy-related protein | [198] | |
| PDPK2P | PDK1/AKT/Caspase 3 | [199] | |
| HOXD-AS1 | SOX4 | [200] | |
| HIS | ERK&AKT/GSK-3b | [201] | |
| HOTAIR | OGFr, miR-122, SETD2 | [202,203,204,205,206] | |
| LINC00161 | Activate ROCK2, miR-590-3p | [207] | |
| DLGAP1-AS1 | miR-26a/b-5p/IL-6/JAK2/STAT3 | [208] | |
| 91H | IGF2 | [209] | |
| MYLK-AS1 | miR-424-5p/E2F7 & activating VEGFR-2 | [210] | |
| Linc-ROR | DEPDC1 | [211] | |
| HULC | HULC/miR-383-5p/VAMP2 | [212] | |
| LINC00238 | miR-522/SFRP2/DKK1 | Suppression of proliferation, invasion, and migration Suppression of HCC progression | [213] |
| TMEM220-AS1 | TMEM220/β-catenin | [214] | |
| NBR2 | JNK/ERK | [215] | |
| lncRNA W5 | --------- | [216] | |
| GAS8-AS1 | GAS8 | [217] | |
| MIR22HG | miR-10a-5p/NCOR2 | [218] | |
| MIR31HG | microRNA-575 | [219] | |
| GAS5 | miR182/ANGPTL1 | [220] | |
| FENDRR | miR-423-5p | [221] | |
| EPB41L4A-AS2 | miR301a-5p/FOXL1 | [222] | |
| TCONS_00006195 | ENO1 | [223] | |
| Uc.134 | LATS1 | [224] | |
| SVUGP2 | MMP2 and 9 | [225] | |
| RP11-286H15.1 | PABPC4 Ubiquitination | [226] | |
| LncRNA-Dreh | Vimentin protein | [227] | |
| XIST | miR-92b | [228] | |
| LINC00221 | lncRNA–miRNA–mRNA miR-485-5p/BSG miR-195-5p/MACC1 ---- HNRNPA2B1/NF-KB Caspase-8/LSD1/H3K9me3 ------ ------ miR-195/EYA1 axis | Prognosis and efficacy | [49] [229] [230] [231] [232] [204] [82] [233] |
| LOC554202 | |||
| LncRNA DDX11-AS1 | |||
| RP11-464I1.1 | |||
| miR503HG | |||
| MALAT1, HOTAIR, MDG | |||
| HOTAIR | |||
| MIR22HG, CTC-297N7.9, | |||
| CTD-2139B15.2, RP11-589N15.2, | |||
| RP11-343N15.5, and | |||
| RP11-479G22.8 | |||
| LINC00511 | |||
| lncRNA W42 | DBN1 miR-448/ROCK1 ----- ----- ----- ----- ----- | Potential biomarkers | [67] [120] [234] [235] [236] [237] [238] |
| PITPNA-AS1 | |||
| PVT1, uc002mbe.2 e | |||
| UCA1 | |||
| RP11-486O12.2, RP11-273G15.2, RP11 863K10.7 and LINC01093 | |||
| LRB1 | |||
| ELMO1-AS1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, A.; Zhang, X. Function of the Long Noncoding RNAs in Hepatocellular Carcinoma: Classification, Molecular Mechanisms, and Significant Therapeutic Potentials. Bioengineering 2022, 9, 406. https://doi.org/10.3390/bioengineering9080406
Khan A, Zhang X. Function of the Long Noncoding RNAs in Hepatocellular Carcinoma: Classification, Molecular Mechanisms, and Significant Therapeutic Potentials. Bioengineering. 2022; 9(8):406. https://doi.org/10.3390/bioengineering9080406
Chicago/Turabian StyleKhan, Ahmad, and Xiaobo Zhang. 2022. "Function of the Long Noncoding RNAs in Hepatocellular Carcinoma: Classification, Molecular Mechanisms, and Significant Therapeutic Potentials" Bioengineering 9, no. 8: 406. https://doi.org/10.3390/bioengineering9080406
APA StyleKhan, A., & Zhang, X. (2022). Function of the Long Noncoding RNAs in Hepatocellular Carcinoma: Classification, Molecular Mechanisms, and Significant Therapeutic Potentials. Bioengineering, 9(8), 406. https://doi.org/10.3390/bioengineering9080406






