Laser Bioprinting of Cells Using UV and Visible Wavelengths: A Comparative DNA Damage Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Culture and Sample Preparation
2.2. Cell Viability Assay
2.3. Immunofluorescence (IF)
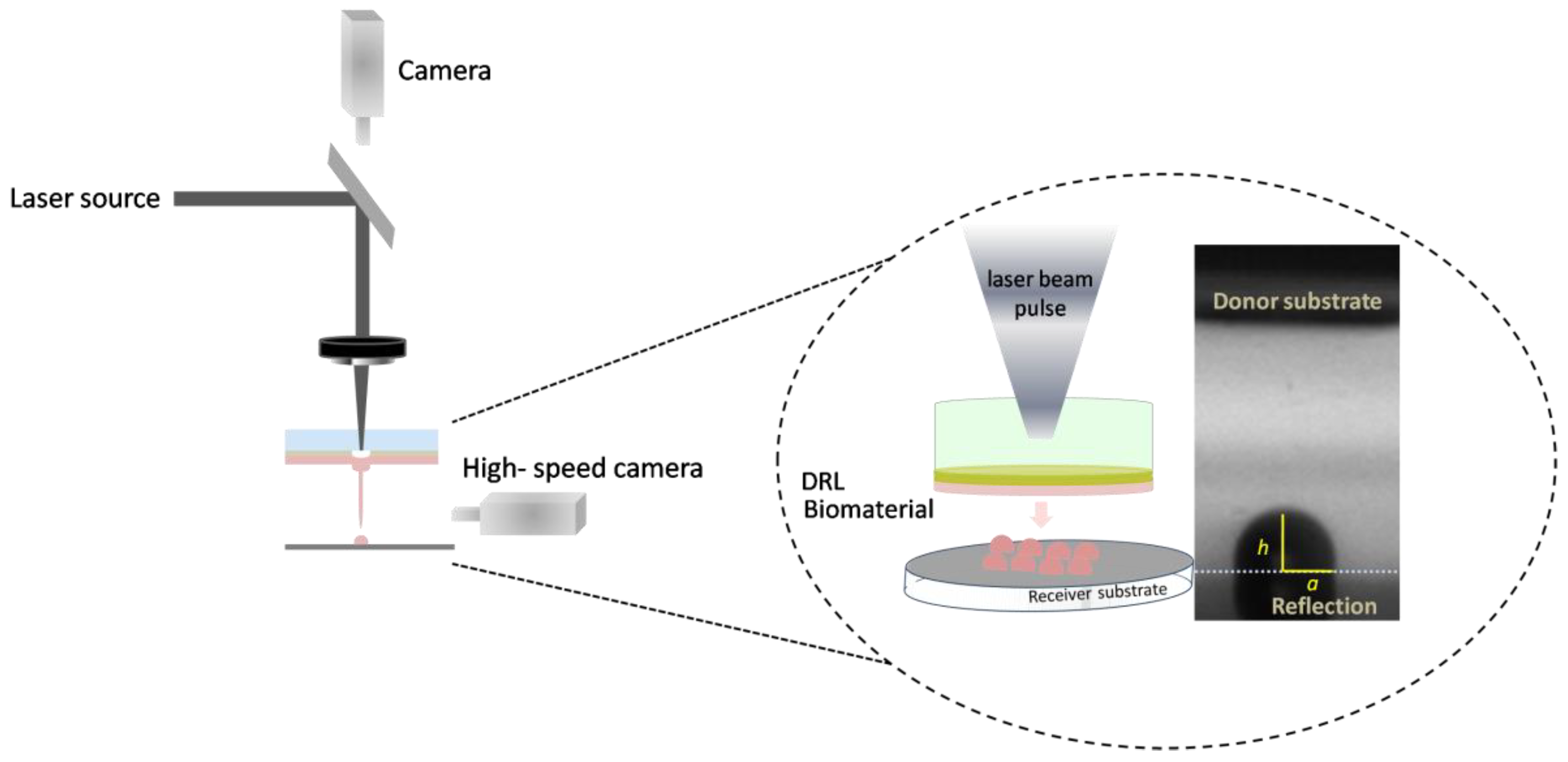
2.4. LIFT Setup and Laser Source
2.4.1. LIFT Setup, at 532 nm (System 1)
2.4.2. LIFT Setup, at 355 nm (System 2)
2.5. Measuring Droplet Diameter, Volume and Jet Velocity
2.6. High-Speed Camera Setup
2.7. Imaging and Image Analysis
2.8. Statistical Analysis
3. Results and Discussion
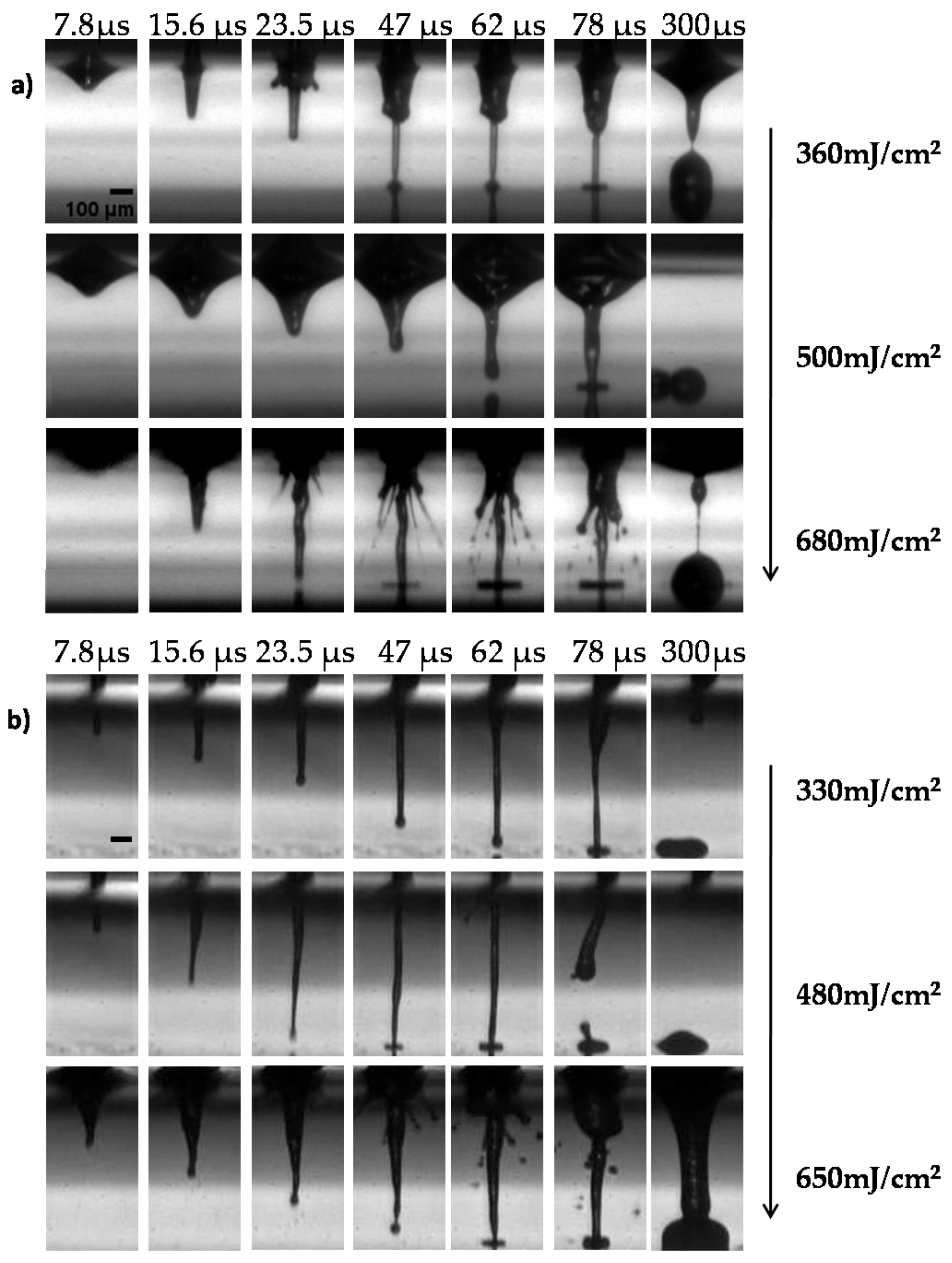
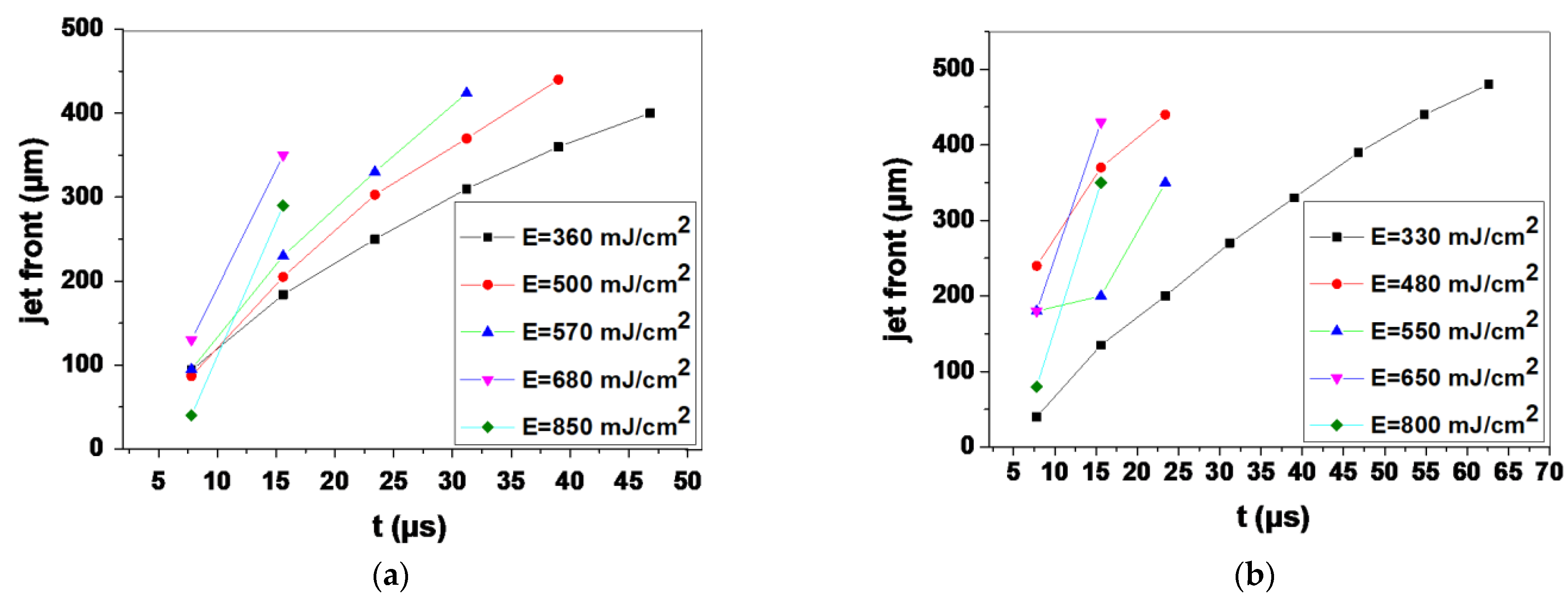
3.1. Optimizing the LIFT Process
3.2. Cell Growth Kinetics after LIFT
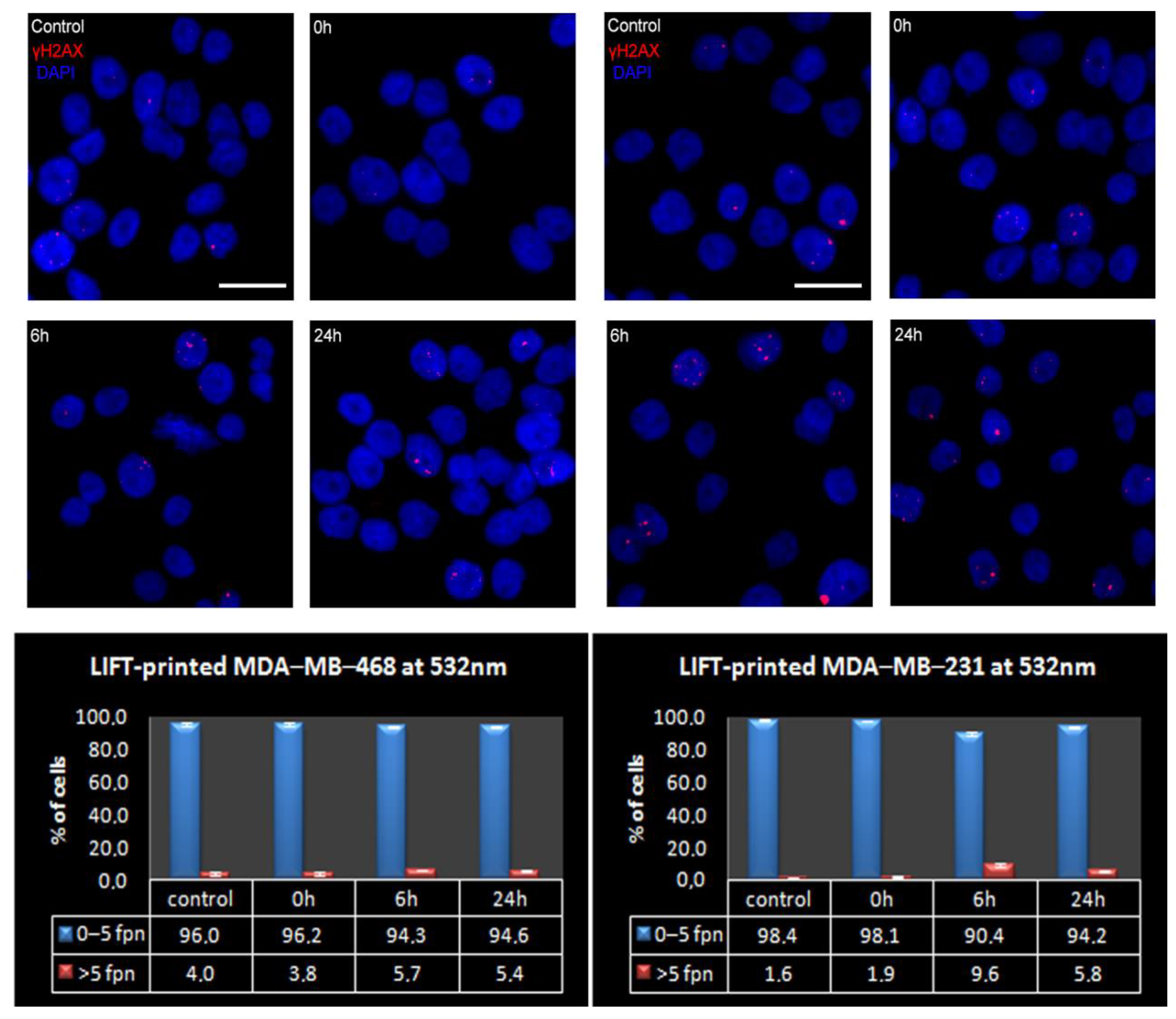
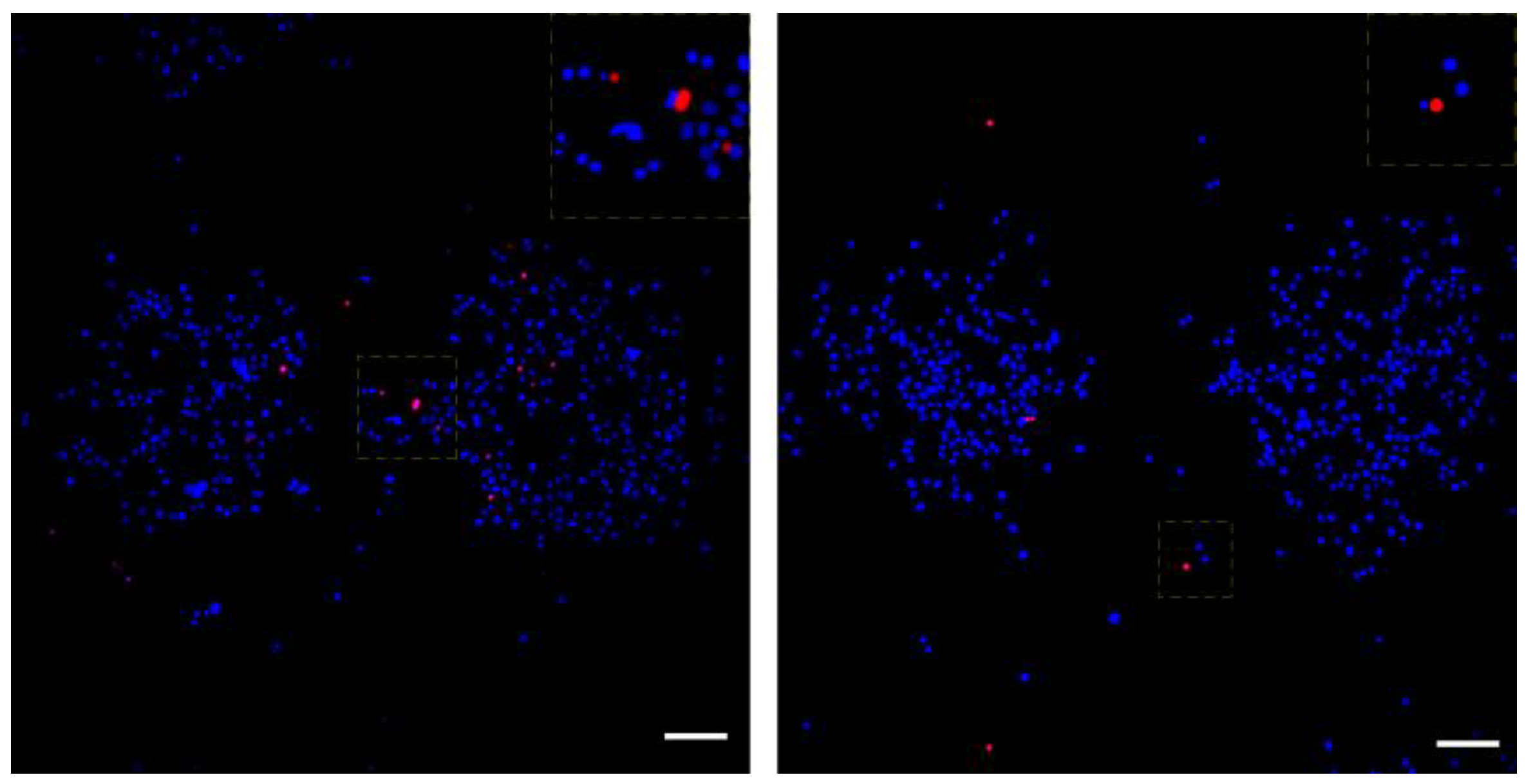
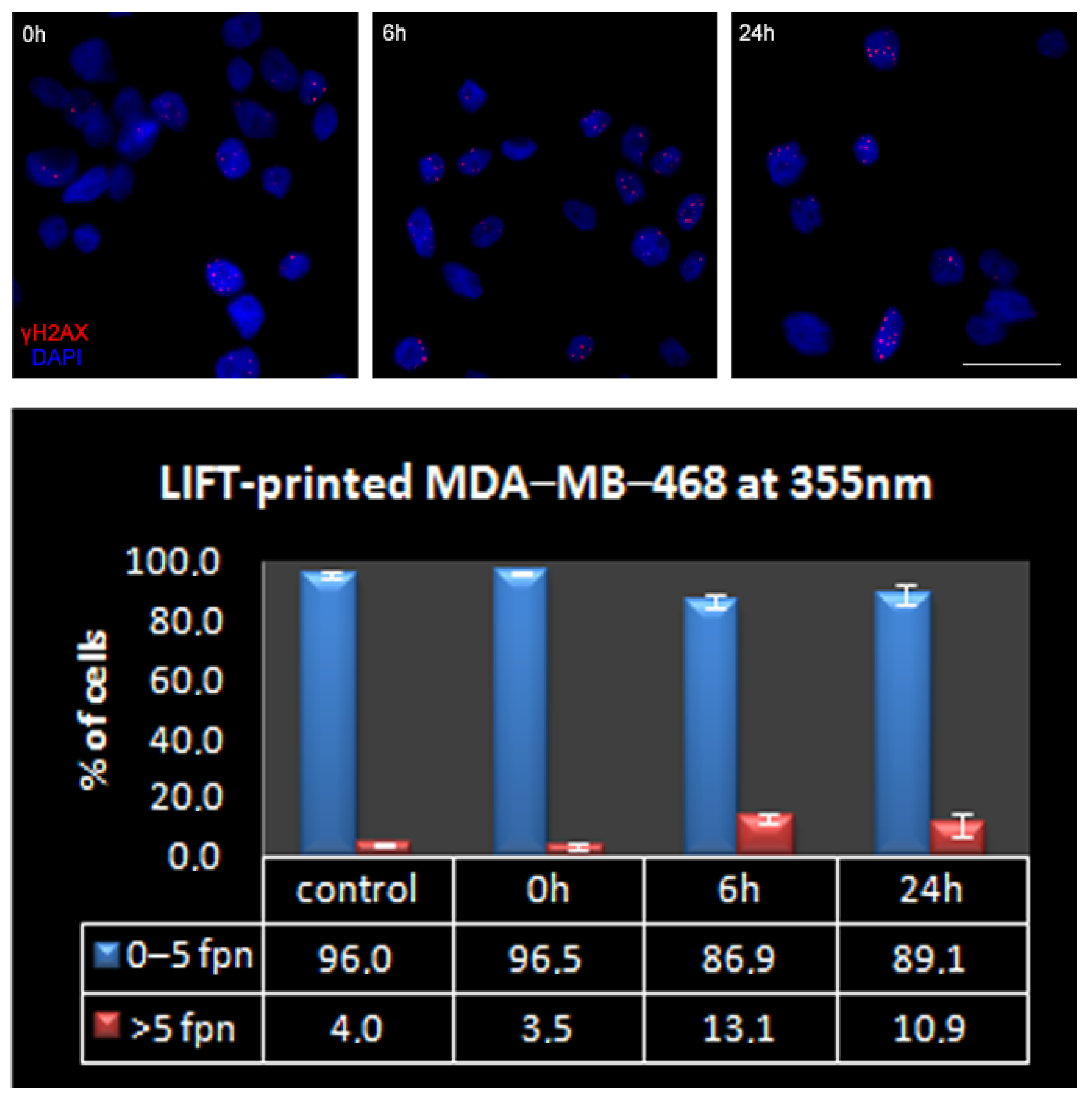
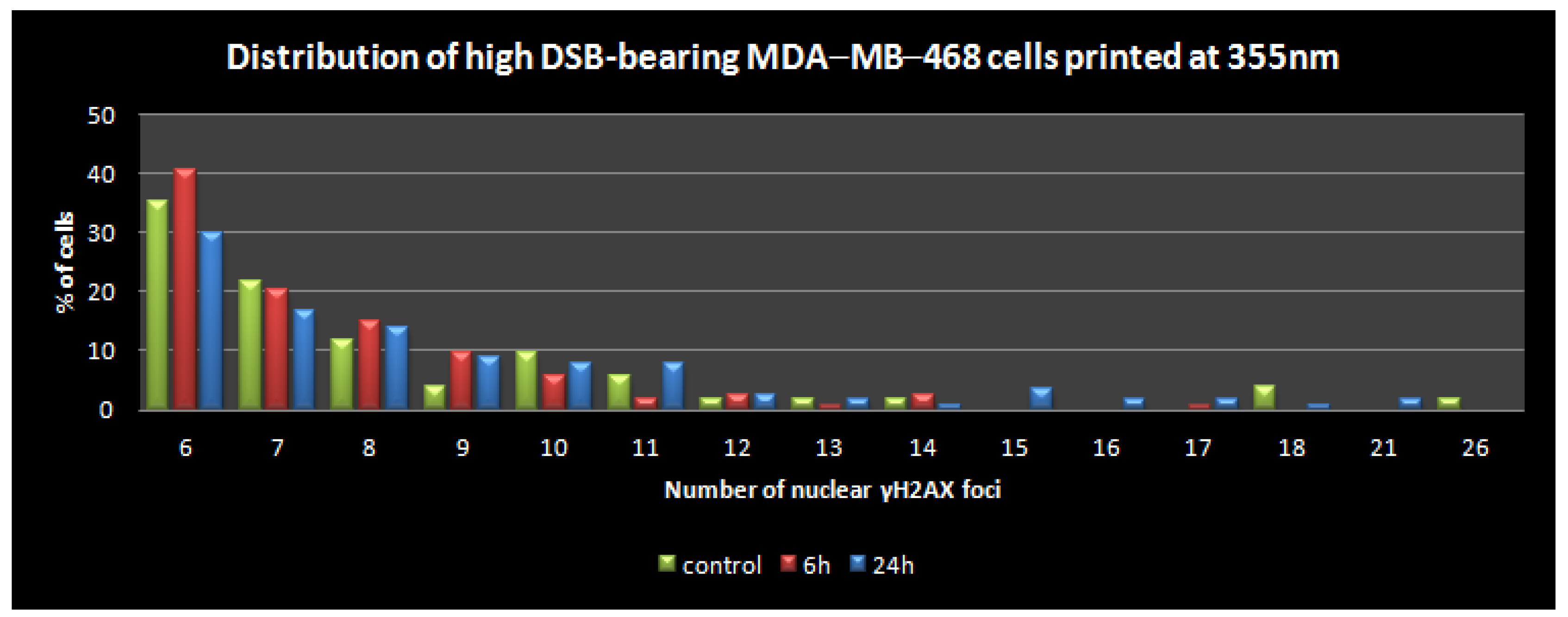
3.3. Quantitative Assessment of LIFT-Induced DNA Damage
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ringeisen, B.R.; Pirlo, R.K.; Wu, P.K.; Boland, T.; Huang, Y.; Sun, W.; Hamid, Q.; Chrisey, D.B. Cell and organ printing turn s 15: Diverse research to commercial transitions. MRS Bull. 2013, 38, 834. [Google Scholar]
- Nakamura, M.; Iwanaga, S.; Henmi, C.; Arai, K.; Nishiyama, Y. Biomatrices and biomaterials for future developments of bioprinting and biofabrication. Biofabrication 2010, 2, 014110. [Google Scholar] [CrossRef]
- Lee, H.; Young, W.K.; Miji, Y.; Kim, S.H.; Kim, G.H. Recent cell printing systems for tissue engineering. Int. J. Bioprint. 2017, 3, 004. [Google Scholar]
- Christensen, K.; Xu, C.; Chai, W.; Zhang, Z.; Fu, J.; Huang, Y. Freeform inkjet printing of cellular structures with bifurcations. Biotechnol. Bioeng. 2014, 112, 1047–1055. [Google Scholar] [CrossRef]
- Kryou, C.; Leva, V.; Chatzipetrou, M.; Zergioti, I. Bioprinting for Liver Transplantation. Bioengineering 2019, 6, 95. [Google Scholar] [CrossRef]
- Karaiskou, A.; Zergioti, I.; Fotakis, C.; Kapsetaki, M.; Kafetzopoulos, D. Microfabrication of biomaterials by the sub-ps laser-induced forward transfer process. Appl. Surf. Sci. 2003, 208, 245–249. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y. Tissue Engineering Applications of Three-Dimensional Bioprinting. Cell Biophys. 2015, 72, 777–782. [Google Scholar] [CrossRef]
- Dinca, V.; Farsari, M.; Kafetzopoulos, D.; Popescu, A.; Dinescu, M.; Fotakis, C. Patterning parameters for biomolecules microarrays constructed with nanosecond and femtosecond UV lasers. Thin Solid Film. 2008, 516, 6504–6511. [Google Scholar] [CrossRef]
- Zergioti, I.; Karaiskou, A.; Papazoglou, D.; Fotakis, C.; Kapsetaki, M.; Kafetzopoulos, D. Femtosecond laser microprinting of biomaterials. Appl. Phys. Lett. 2005, 86, 163902. [Google Scholar] [CrossRef]
- Barron, J.A.; Krizman, D.B.; Ringeisen, B.R. Laser Printing of Single Cells: Statistical Analysis, Cell Viability, and Stress. Ann. Biomed. Eng. 2005, 33, 121–130. [Google Scholar] [CrossRef]
- Leva, V.; Chatzipetrou, M.; Alexopoulos, L.; Tzeranis, D.S.; Zergioti, I. Direct laser printing of liver cells on porous collagen scaffold. JLMN J. Laser Micro Nanoeng. 2018, 13, 234–237. [Google Scholar]
- Zhang, Z.; Xu, C.; Xiong, R.; Chrisey, D.B.; Huang, Y. Effects of living cells on the bioink printability during laser printing. Biomicrofluidics 2017, 11, 034120. [Google Scholar] [CrossRef]
- Michael, S.; Sorg, H.; Peck, C.-T.; Koch, L.; Deiwick, A.; Chichkov, B.; Vogt, P.M.; Reimers, K. Tissue Engineered Skin Substitutes Created by Laser-Assisted Bioprinting Form Skin-Like Structures in the Dorsal Skin Fold Chamber in Mice. PLoS ONE 2013, 8, e57741. [Google Scholar] [CrossRef]
- Wu, P.K.; Ringeisen, B.R. Development of human umbilical vein endothelial cell (HUVEC) and human umbilical vein smooth muscle cell (HUVSMC) branch/stem structures on hydrogel layers via biological laser printing (BioLP). Biofabrication 2010, 2, 014111. [Google Scholar] [CrossRef]
- Ringeisen, B.R.; Othon, C.; Barron, J.A.; Young, D.; Spargo, B.J. Jet-based methods to print living cells. Biotechnol. J. 2006, 1, 930–948. [Google Scholar] [CrossRef]
- Gruene, M.; Unger, C.; Koch, L.; Deiwick, A.; Chichkov, B. Dispensing pico to nanolitre of a natural hydrogel by laser-assisted bioprinting. Biomed. Eng. Online 2011, 10, 19. [Google Scholar]
- Phamduy, T.B.; Raof, N.A.; Schiele, N.R.; Yan, Z.; Corr, D.T.; Huang, Y.; Xie, Y.; Chrisey, D.B. Laser direct-write of single microbeads into spatially-ordered patterns. Biofabrication 2012, 4, 025006. [Google Scholar] [CrossRef][Green Version]
- Ouyang, L.; Yao, R.; Chen, X.; Na, J.; Sun, W. 3D printing of HEK 293FT cell-laden hydrogel into macroporous constructs with high cell viability and normal biological functions. Biofabrication 2015, 7, 015010. [Google Scholar] [CrossRef]
- Murphy, S.V.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef]
- Koch, L.; Kuhn, S.; Sorg, H.; Gruene, M.; Schlie, S.; Gaebel, R.; Polchow, B.; Reimers, K.; Stoelting, S.; Ma, N.; et al. Laser Printing of Skin Cells and Human Stem Cells. Tissue Eng. Part C Methods 2010, 16, 847–854. [Google Scholar] [CrossRef]
- Guillemot, F.; Souquet, A.; Catros, S.; Guillotin, B. Laser-assisted cell printing: Principle, physical parameters versus cell fate and perspectives in tissue engineering. Nanomedicine 2010, 5, 507–515. [Google Scholar] [CrossRef]
- Ringeisen, B.R.; Kim, H.; Barron, J.A.; Krizman, D.B.; Chrisey, D.B.; Jackman, S.; Auyeung, R.Y.C.; Spargo, B.J. Laser Printing of Pluripotent Embryonal Carcinoma Cells. Tissue Eng. 2004, 10, 483–491. [Google Scholar]
- Pati, F.; Jang, J.; Ha, D.H.; Kim SWon Rhie, J.W.; Shim, J.H.; Kim, D.H.; Cho, D.W. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat. Commun. 2014, 5, 3935. [Google Scholar]
- Zhang, Z.; Chai, W.; Xiong, R.; Zhou, L.; Huang, Y. Printing-induced cell injury evaluation during laser printing of 3T3 mouse fibroblasts. Biofabrication 2017, 9, 025038. [Google Scholar]
- Xiong, R.; Zhang, Z.; Chai, W.; Huang, Y.; Chrisey, D.B. Freeform drop-ondemand laser printing of 3D alginate and cellular constructs. Biofabrication 2015, 7, 045011. [Google Scholar]
- Mardikar, S.H.; Niranjan, K. Observations onthe shear damage to different animal cells in a concentriccylinder viscometer. Biotechnol. Bioeng. 2000, 68, 697–704. [Google Scholar]
- Lin, Y.; Huang, Y.; Wang, G.; Tzeng, J.; Chrisey, D.B. Effect of laser fluence on yeast cell viability inlaser-assisted cell transfer. J. Appl. Phys. 2009, 106, 043106. [Google Scholar]
- Keriquel, V.; Oliveira, H.; Rémy, M.; Ziane, S.; Delmond, S.; Rousseau, B.; Rey, S.; Catros, S.; Amédée, J.; Guillemot, F.; et al. In situ printing of mesenchymal stromal cells, by laser-assisted bioprinting, for in vivo bone regeneration applications. Sci. Rep. 2017, 7, 1778. [Google Scholar]
- Serra, P.; Duocastella, M.; Fernández-Pradas, J.; Morenza, J. Liquids microprinting through laser-induced forward transfer. Appl. Surf. Sci. 2009, 255, 5342–5345. [Google Scholar] [CrossRef]
- Gruene, M.; Pflaum, M.; Hess, C.; Diamantouros, S.; Schlie, S.; Deiwick, A.; Koch, L.; Wilhelmi, M.; Jockenhoevel, S.; Haverich, A.; et al. Laser Printing of Three-Dimensional Multicellular Arrays for Studies of Cell–Cell and Cell–Environment Interactions. Tissue Eng. Part C Methods 2011, 17, 79–87. [Google Scholar]
- Fernández-Pradas, J.M.; Duocastella, M.; Colina, M.; Serra, P.; Morenza, J.L. Production of miniaturized biosensors through laser-induced forward transfer. Bioeng. Bioinspired Syst. III 2007, 6592, 281–288. [Google Scholar]
- Othon, C.M.; Wu, X.; Anders, J.J.; Ringeisen, B.R. Single-cell printing to form three-dimensional lines of olfactory ensheathing cells. Biomed. Mater. 2008, 3, 034101. [Google Scholar] [CrossRef]
- Chen, C.Y.; Barron, J.A.; Ringeisen, B.R. Cell patterning without chemical surface modification: Cell-cell interacftions between bovine aortic endothelial cells (BAEC) on a homogeneous cell-adherent hydrogel. Appl. Surf. Sci. 2006, 252, 8641–8645. [Google Scholar]
- Xiong, R.; Zhang, Z.; Chai, W.; Chrisey, D.B.; Huang, Y. Study of gelatin as an effective energy absorbing layer for laser bioprinting. Biofabrication 2017, 9, 024103. [Google Scholar] [CrossRef]
- Desrus, H.; Chassagne, B.; Catros, S.; Artiges, C.; Devillard, R.; Petit, S.; Deloison, F.; Fricain, J.C.; Guillemot, F.; Kling, R.; et al. Laser assisted bioprinting using a femtosecond laser with and without a gold transductive layer: A parametric study. Opt. Interact. Tissue Cells XXVII 2016, 9706, 91–108. [Google Scholar] [CrossRef]
- Petit, S.; Kérourédan, O.; Devillard, R.; Cormier, E. Femtosecond versus picosecond laser pulses for film-free laser bioprinting. Appl. Opt. 2017, 56, 8648–8655. [Google Scholar] [CrossRef]
- Catros, S.; Fricain, J.-C.; Guillotin, B.; Pippenger, B.; Bareille, R.; Remy, M.; Lebraud, E.; Desbat, B.; Amédée, J.; Guillemot, F. Laser-assisted bioprinting for creating on-demand patterns of human osteoprogenitor cells and nano-hydroxyapatite. Biofabrication 2011, 3, 025001. [Google Scholar] [CrossRef]
- Guillemot, F.; Souquet, A.; Catros, S.; Guillotin, B.; Lopez, J.; Faucon, M.; Pippenger, B.; Bareille, R.; Rémy, M.; Bellance, S.; et al. High-throughput laser printing of cells and biomaterials for tissue engineering. Acta Biomater. 2010, 6, 2494–2500. [Google Scholar] [CrossRef]
- Doraiswamy, A.; Narayan, R.; Lippert, T.; Urech, L.; Wokaun, A.; Nagel, M.; Hopp, B.; Dinescu, M.; Modi, R.; Auyeung, R.; et al. Excimer laser forward transfer of mammalian cells using a novel triazene absorbing layer. Appl. Surf. Sci. 2006, 252, 4743–4747. [Google Scholar] [CrossRef]
- Hopp, B.; Smausz, T.; Kresz, N.; Barna, N.; Bor, Z.; Kolozsvári, L.; Chrisey, D.B.; Szabó, A.; Nógrádi, A. Survival and proliferative ability of various living cell types after laser-induced forward transfer. Tissue Eng. 2005, 11, 1817–1823. [Google Scholar]
- Pagès, E.; Remy, M.; Kériquel, V.; Correa, M.M.; Guillotin, B.; Guillemot, F. Creation of Highly Defined Mesenchymal Stem Cell Patterns in Three Dimensions by Laser-Assisted Bioprinting. J. Nanotechnol. Eng. Med. 2015, 6, 021006. [Google Scholar] [CrossRef]
- Yusupov, V.; Churbanov, S.; Churbanova, E.; Bardakova, K.; Antoshin, A.; Evlashin, S.; Timashev, P.; Minaev, N. Laser-induced Forward Transfer Hydrogel Printing: A Defined Route for Highly Controlled Process. Int. J. Bioprint. 2020, 6, 271. [Google Scholar] [CrossRef]
- Nebbioso, A.; Benedetti, R.; Conte, M.; Carafa, V.; De Bellis, F.; Shaik, J.; Matarese, F.; DELLA Ventura, B.; Gesuele, F.; Velotta, R.; et al. Time-resolved analysis of DNA-protein interactions in living cells by UV laser pulses. Sci. Rep. 2017, 7, 11725. [Google Scholar] [CrossRef]
- Benedetti, R.; Bajardi, F.; Capozziello, S.; Carafa, V.; Conte, M.; Del Sorbo, M.R.; Nebbioso, A.; Singh, M.; Stunnenberg, H.G.; Valadan, M.; et al. Different Approaches to Unveil Biomolecule Configurations and Their Mutual Interactions. Anal. Lett. 2021, 54, 40–56. [Google Scholar]
- Barron, J.; Spargo, B.; Ringeisen, B. Biological laser printing of three dimensional cellular structures. App. Phys. A 2004, 79, 1027–1030. [Google Scholar] [CrossRef]
- Barron, J.A.; Ringeisen, B.R.; Kim, H.; Spargo, B.J.; Chrisey, D.B. Application of laser printing to mammalian cells. Thin Solid Film. 2004, 453–454, 383–387. [Google Scholar] [CrossRef]
- Barron, J.A.; Wu, P.; Ringeisen, B.R. Biological laser printing: A novel technique for creating heterogeneous 3-dimensional cell patterns. Biomed. Microdev. 2004, 6, 139–147. [Google Scholar]
- Hopp, B.; Smausz, T.; Anta, Z.; Kresz, N.; Bor, Z.; Chrisey, D. Absorbing film assisted laser induced forward transfer of fungi (Trichoderma conidia). J. Appl. Phys. 2004, 96, 3478–3481. [Google Scholar]
- Catros, S.; Guillotin, B.; Bačáková, M.; Fricain, J.-C.; Guillemot, F. Effect of laser energy, substrate film thickness and bioink viscosity on viability of endothelial cells printed by Laser-Assisted Bioprinting. Appl. Surf. Sci. 2011, 257, 5142–5147. [Google Scholar]
- Riester, D.; Budde, J.; Gach, C.; Gillner, A.; Wehner, M. High Speed Photography of Laser Induced Forward Transfer (LIFT) of Single and Double-layered Transfer Layers for Single Cell Transfer. J. Laser Micro/Nanoeng. 2016, 11, 199–203. [Google Scholar]
- Schiele, N.R.; Chrisey, D.B.; Corr, D.T. Gelatin-Based Laser Direct-Write Technique for the Precise Spatial Patterning of Cells. Tissue Eng. Part C Methods 2011, 17, 289–298. [Google Scholar] [CrossRef]
- Meerbreya, K.L.; Hu, G.; Kessler, J.D.; Roarty, K.; Li, M.Z.; Fang, J.E.; Herschkowitz, J.I.; Burrows, A.E.; Ciccia, A.; Sun, T.; et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 3665–3670. [Google Scholar]
- Kryou, C.; Theodorakos, I.; Karakaidos, P.; Klinakis, A.; Hatziapostolou, A.; Zergioti, I. Parametric Study of Jet/Droplet Formation Process during LIFT Printing of Living Cell-Laden Bioink. Micromachines 2021, 12, 1408. [Google Scholar] [CrossRef]
- Rampias, T.; Karagiannis, D.; Avgeris, M.; Polyzos, A.; Kokkalis, A.; Kanaki, Z.; Kousidou, E.; Tzetis, M.; Kanavakis, E.; Stravodimos, K.; et al. The lysine-specific methyltransferase KMT 2C/ MLL 3 regulates DNA repair components in cancer. EMBO Rep. 2019, 20, e46821. [Google Scholar] [CrossRef]
- Barron, J.; Rosen, R.; Jones-Meehan, J.; Spargo, B.; Belkin, S.; Ringeisen, B. Biological laser printing of genetically modified Escherichia coli for biosensor applications. Biosens. Bioelectron. 2004, 20, 246–252. [Google Scholar] [CrossRef]
- Koch, L.; Brandt, O.; Deiwick, A.; Chichkov, B. Laser assisted bioprinting at different wavelengths and pulse durations with a metal dynamic release layer: A parametric study. Int. J. Bioprint. 2017, 3, 42–53. [Google Scholar] [CrossRef]
- Duocastella, M.; Fernández-Pradas, J.M.; Serra, P.; Morenza, J.L. Sessile droplet formation in the laser-induced forward transfer of liquids: A time-resolved imaging study. Thin Solid Film. 2010, 518, 5321–5325. [Google Scholar]
- Hopp, B.; Smausz, T.; Barna, N.; Vass Cs Antal Zs Kredics, L.; Chrisey, D.B. Time-resolved study of absorbing film assisted laser induced forward transfer of trichoderma longibrachiatum conidia. J. Phys. D Appl. Phys. 2005, 38, 833–837. [Google Scholar]
- Young, D.; Auyeung, R.C.Y.; Piqué, A.; Chrisey, D.B.; Dlott, D.D. Time-resolved optical microscopy of a laser-based forward transfer process. Appl. Phys. Lett. 2001, 78, 3169–3171. [Google Scholar] [CrossRef]
- Wang, W.; Huang, Y.; Grujicic, M.; Chrisey, U.B. Study of Impact-Induced Mechanical Effects in Cell Direct Writing Using Smooth Particle Hydrodynamic Method. ASME J. Manuf. Sci. Eng. 2008, 130, 021012. [Google Scholar] [CrossRef]
- Leverett, L.B.; Hellums, J.D.; Alfrey, C.P.; Lynch, E.C. Red Blood Cell Damage by Shear Stress. Biophys. J. 1972, 12, 257–273. [Google Scholar] [CrossRef]
- Thoumine, O.; Ott, A.; Louvard, D. Critical centrifugal forces induce adhesion rupture or structural reorganization in cultured cells. Cell Motil. Cytoskelet. 1996, 33, 276–287. [Google Scholar]
- Abercrombie, M. Contact inhibition and malignancy. Nature 1979, 281, 259–262. [Google Scholar] [CrossRef]
- Klinakis, A.; Karagiannis, D.; Rampias, T. Targeting DNA repair in cancer: Current state and novel approaches. Experientia 2019, 77, 677–703. [Google Scholar] [CrossRef]
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM Phosphorylates Histone H2AX in Response to DNA Double-strand Breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef]
- Tian, X.; Liu, C.; Wang, X.; Wang, F.; Wang, L.; Xu, L.; Ma, J.; Gao, Y.; Bao, Y.; Wang, F.; et al. Sei-1 promotes double minute chromosomes formation through activation of the PI3K/Akt/BRCA1-Abraxas pathway and induces double-strand breaks in NIH-3T3 fibroblasts. Cell Death Dis. 2018, 9, 341. [Google Scholar] [CrossRef]
- Cai, Z.; Chen, Z.; Bailey, K.E.; Scollard, D.A.; Reilly, R.M.; Vallis, K.A. Relationship Between Induction of Phosphorylated H2AX and Survival in Breast Cancer Cells Exposed to 111In-DTPA-Hegf. J. Nucl. Med. 2008, 49, 8. [Google Scholar]
- Adhikari, J.; Roy, A.; Das, A.; Ghosh, M.; Thomas, S.; Sinha, A.; Kim, J.; Saha, P. Effects of Processing Parameters of 3D Bioprinting on the Cellular Activity of Bioinks. Macromol. Biosci. 2020, 21, e2000179. [Google Scholar] [CrossRef]
- Antoshin, A.A.; Churbanov, S.N.; Minaev, N.V.; Zhang, D.; Zhang, Y.; Shpichka, A.I.; Timashev, P.S. LIFT-bioprinting, is it worth it? Bioprinting 2020, 20, e00114. [Google Scholar]




| Laser | System 1 | System 2 |
|---|---|---|
| Wavelength (nm) | 532 | 355 |
| Pulse duration (ns) | 0.6 | 10 |
| Pulse energy (μJ) | 10–18 | 9–15 |
| Foci Per Nucleus: | 0 | 1 | 2 | 3 | 4 | 5 | >5 | |
|---|---|---|---|---|---|---|---|---|
| MDA-MB-468 | Mean (%) | 30.1 | 34.9 | 16.4 | 7.5 | 4.9 | 2.1 | 4.0 |
| SEM | 3.6 | 4.8 | 0.5 | 1.5 | 0.8 | 0.7 | 1.0 | |
| MDA-MB-231 | Mean (%) | 24.9 | 57.0 | 10.1 | 3.7 | 1.9 | 0.9 | 1.6 |
| SEM | 8.9 | 2.8 | 3.2 | 1.9 | 0.3 | 0.6 | 0.5 | |
| 0 h | 6 h | 24 h | |
|---|---|---|---|
| Measured cells | 7758 | 3739 | 12,449 |
| Apoptotic cells (mean%) | 1.22 | 1.85 | 0.81 |
| SEM | 0.08 | 0.15 | 0.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karakaidos, P.; Kryou, C.; Simigdala, N.; Klinakis, A.; Zergioti, I. Laser Bioprinting of Cells Using UV and Visible Wavelengths: A Comparative DNA Damage Study. Bioengineering 2022, 9, 378. https://doi.org/10.3390/bioengineering9080378
Karakaidos P, Kryou C, Simigdala N, Klinakis A, Zergioti I. Laser Bioprinting of Cells Using UV and Visible Wavelengths: A Comparative DNA Damage Study. Bioengineering. 2022; 9(8):378. https://doi.org/10.3390/bioengineering9080378
Chicago/Turabian StyleKarakaidos, Panagiotis, Christina Kryou, Nikiana Simigdala, Apostolos Klinakis, and Ioanna Zergioti. 2022. "Laser Bioprinting of Cells Using UV and Visible Wavelengths: A Comparative DNA Damage Study" Bioengineering 9, no. 8: 378. https://doi.org/10.3390/bioengineering9080378
APA StyleKarakaidos, P., Kryou, C., Simigdala, N., Klinakis, A., & Zergioti, I. (2022). Laser Bioprinting of Cells Using UV and Visible Wavelengths: A Comparative DNA Damage Study. Bioengineering, 9(8), 378. https://doi.org/10.3390/bioengineering9080378








