1. Introduction
The analysis of online handwritten tasks has emerged these last years as a pertinent behavioral modality to aid clinicians in the diagnosis of neurodegenerative pathologies. Indeed, digital tablets allow capturing handwritten traces as spatiotemporal signals (“online” signals) that carry fine dynamic information about the writing process. Recent surveys in the field demonstrate the importance of this noninvasive modality to assess the health status of a person, by providing additional information to cognitive tests [
1,
2,
3].
Initiated with memory loss, Alzheimer’s disease (AD) impairs motor functions progressively. Fine motor skills acquired in early life decline as the disease advances. In the literature, additionally to handwritten tasks with semantic content, different elementary graphic tasks have been analyzed to extract AD markers: straight lines in different orientations [
4,
5] that require coordination of fingers and wrist, connected cursive loops [
6], and concentric superimposed circles [
7]. On the other hand, another graphic task, Archimedes spiral, was proposed by Pullman [
8] to assess tremors on a digitizer. Interestingly, the spiral task has been intensively studied for Parkinson’s disease (PD) [
9,
10,
11,
12,
13] and Essential Tremor detection [
14,
15], but hardly for AD. In [
9,
13], the spiral task was analyzed in a standalone mode for PD detection; but more recently, it has also been fused to other tasks (such as handwritten text, isolated words, and bigrams) in works exploiting deep learning [
16,
17,
18,
19,
20,
21]. In [
16,
18], we note a significantly higher accuracy with the spiral task compared to other tasks on the same population. This result emphasizes that the spiral task is particularly well suited for decision aid to clinicians in detecting motor dysfunction related to PD. Additionally, those works analyzing the spiral task in a standalone manner also point out its capability of characterizing stages of the disease that are difficult to detect with some standard clinical tests [
10].
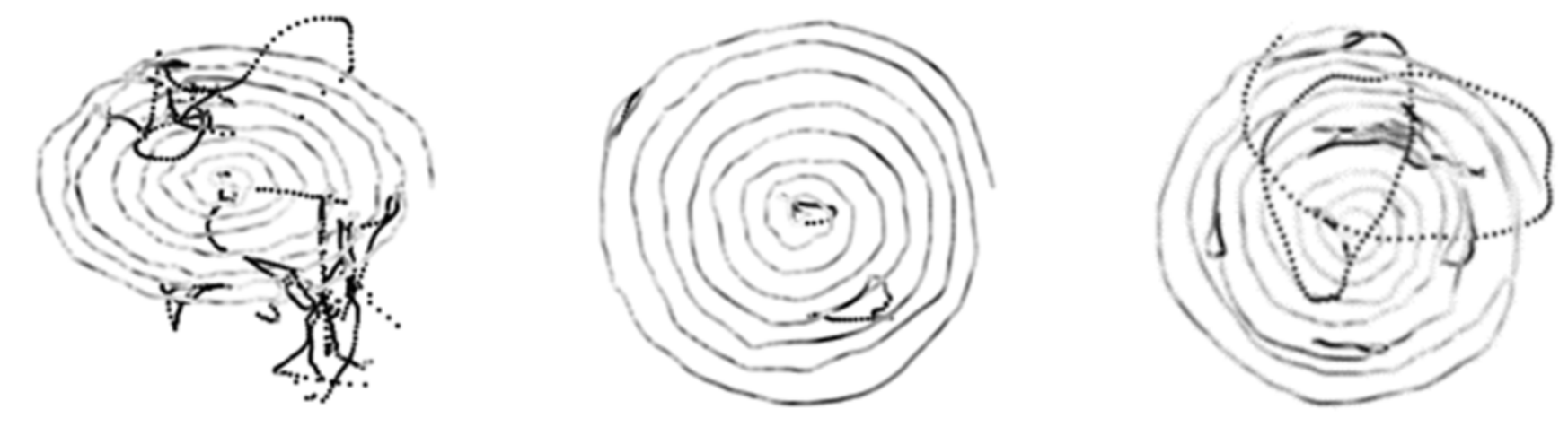
For these reasons, in this paper, we focus on the analysis of Archimedes’ spiral for early-stage AD detection. We exploit deep learning (DL) techniques to extract automatically features on spiral images generated from online sequences.
1.1. Positioning with Regard to the State-of-the-Art in Deep Learning
Two main trends emerge in the literature addressing neurodegenerative diseases’ detection with DL techniques on handwritten graphical tasks. First, works exploiting transfer learning for high-level feature extraction; such works use images generated from online handwriting [
16,
21,
22] while keeping the shape of the pen trajectory and assessing the health status of the subject with another classifier. Second, we find works that train DL classifiers on time series-based images [
17,
18,
20], which do not convey information about the shape of the pen trajectory. More precisely, in [
17,
18], raw six-dimensional time series are captured with a smart pen that contains six sensors. Such time series or temporal functions are converted into a grey-level image, in which rows are time in milliseconds and columns represent the six time series. In [
20], spectrogram images are constructed in grey level for each time series and given as input to the DL model. We note that the approaches in [
16,
17,
18,
20] do not allow taking advantage simultaneously of visual content and dynamic information of the drawing. In [
21], the authors propose to include, in pen trajectory images, in-air trajectories and to some extent local velocity, in two ways: the first is binary, since the in-air trajectory, displayed in grey, encodes the presence of a pen-up in the drawing (pen-downs remain in black). Second, to encode pen velocity, the sampled points of the spiral trajectory are not connected: as the sampling rate of the digitizer is fixed, when the pen moves faster on the writing surface, the spacing between consecutive captured points increases, and accordingly, decreases when the pen moves slower. Thus, velocity is encoded through points’ spacing on the image of the handwriting, instead of an explicit value belonging to a given discrete range. This work shows two main drawbacks: first, it proposes a simultaneous encoding of velocity and pen-ups in the same handwriting image, which makes it difficult to analyze separately the impact of each dynamic information (pen-ups’ occurrence and changes in velocity) for pathology detection. Second, the dynamic range of any other parameter, such as pen pressure, pen inclination, or higher order derivatives of pen position, is difficult to exploit through this pure spatial encoding on the pen trajectory image.
In this context, we propose a novel framework to embed dynamic information in spiral images generated from online sequences, for enhancing discrimination between HC and AD populations. This framework allows taking advantage simultaneously of static and dynamic characteristics of the spiral drawing. Our approach proposes instead to embed each dynamic parameter in a devoted spiral image, explicitly, as a value in the [0, 255] interval, in greyscale. Contrary to the above-mentioned works [
17,
18,
21], our approach allows assessing the impact of each dynamic parameter separately, as a particular “view” of the drawn spiral; we are thus able to study which dynamic information is the most pertinent for discriminating between AD and HC subjects.
By exploiting transfer learning, we compare off-the-shelf automatically extracted features, at different levels of representation, in terms of their capability to discriminate between AD and HC populations. In [
16,
21,
22], deep representations are extracted at the highest layer before the fully connected ones, and later fed to classifiers. To our knowledge, within the field of neurodegenerative diseases’ detection through handwritten graphical tasks analysis, our work is the first to carry out a comprehensive study using representations extracted at lower layers of the deep network, especially in terms of sensitivity and specificity metrics. This trend in transfer learning is of high potential, as shown in other image processing tasks [
23]: according to the field, very deep networks may indeed provide high-quality off-the-shelf features in early layers. In the area of neurodegenerative diseases’ assessment, transfer learning is of particular interest: most of the time, very little data is available; the heterogeneity between subjects is important, especially at an early stage of the disease; finally, it avoids the strong difficulty of tailoring by hand features to a population. AlexNet [
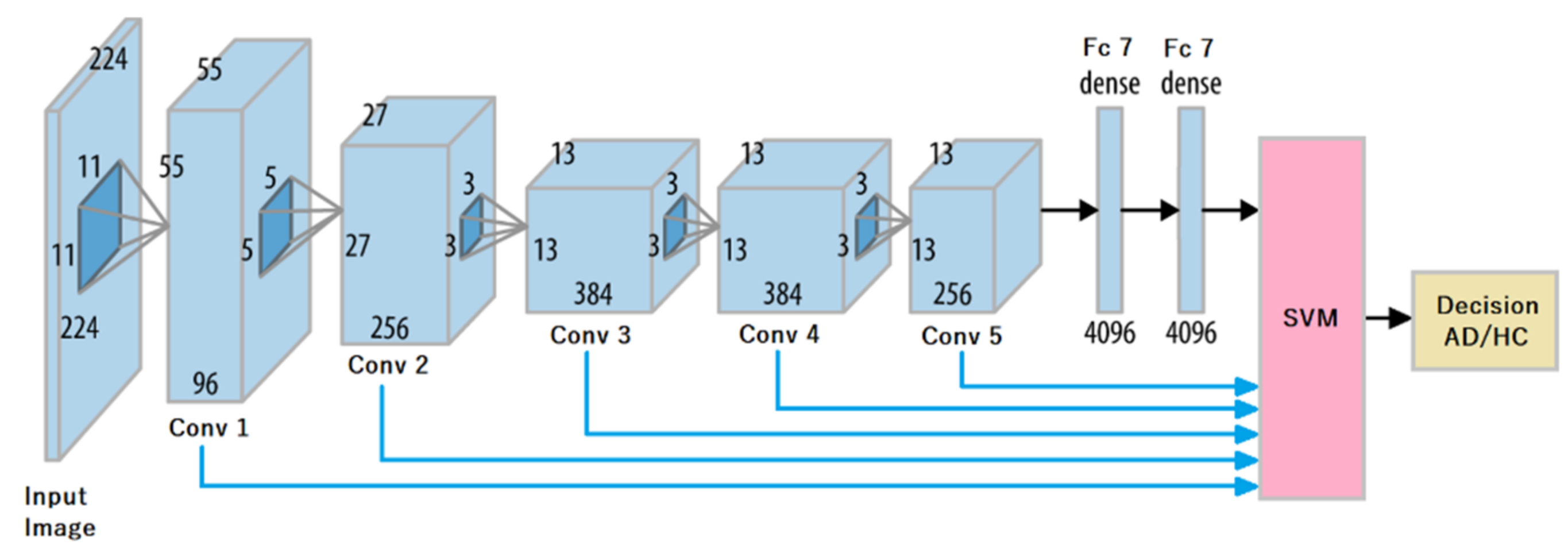
24] has proven its capacity to extract pertinent features on static images of handwriting at a high level (fc7 layer) [
16]. One main objective of the present work is to study how our novel approach to embed dynamic parameters impacts the quality of representations extracted from AlexNet at lower or intermediate layers.
To enhance pathology detection, usually, experts (classifiers) are trained on different handwritten tasks and cooperate through decision fusion [
16,
17,
18,
20,
21]. Thanks to our approach providing for each dynamic parameter a specific “view” of the spiral drawing, our work investigates, through different fusion frameworks, which are the most pertinent dynamic information discriminating AD and HC populations, based on only one task.
An overview of the main contributions of this work is presented in the next subsection.
1.2. Overview of Our Contributions
Our study clarifies the potential of the spiral task as a dynamic gesture for early AD detection and defines the most pertinent methodology for exploiting transfer learning in this context. More precisely, in this work, we propose:
- (i)
A novel embedding of dynamic parameters, each encoded separately in different images of a same spiral, providing different “views” of the spiral image (different types of “hybrid“ images);
- (ii)
A comprehensive study on which dynamic parameters are more discriminant at the classification step, for AD detection. Indeed, we study in this framework of transfer learning different dynamic parameters never studied before in the literature;
- (iii)
A comparative study on features automatically extracted at different levels of representation by transfer learning; this study is carried out on a reference architecture in the area of neurodegenerative diseases’ assessment, namely AlexNet [
24];
- (iv)
A complete study on the scope of fusion. Our approach allows new possibilities for fusing dynamic information: at input images (low-level fusion), at the feature level (on representations extracted with AlexNet), and at the experts’ decision level.
1.3. Organization of the Paper
This paper is organized as follows: in
Section 2, we detail the database, its associated acquisition protocol, and present our methodology for analyzing dynamically enriched spiral trajectory images through transfer learning.
Section 3 describes our experimental setup and the obtained results.
Section 4 states our conclusions and perspectives for future work.
4. Discussion and Conclusions
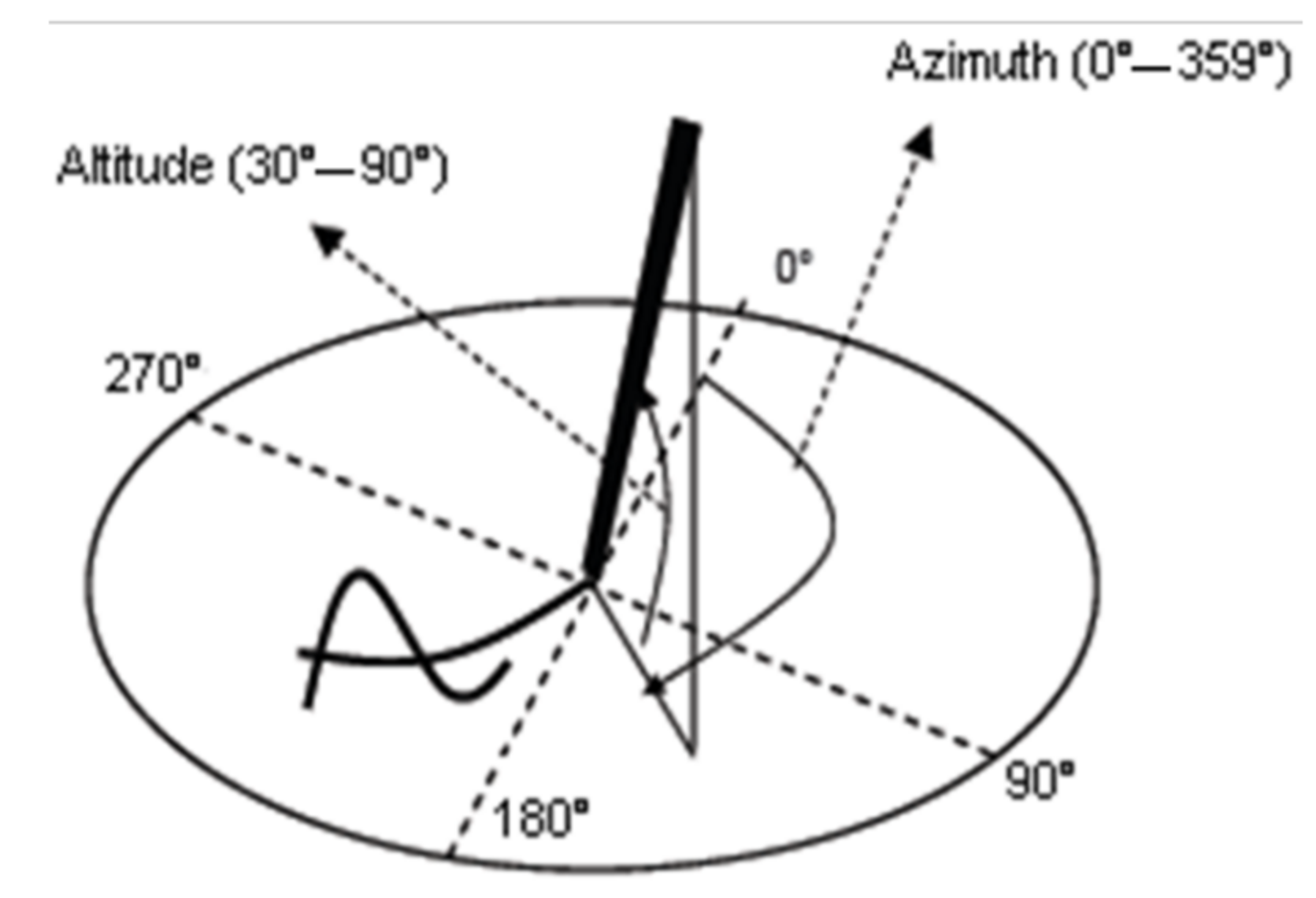
The present study proposes a novel scheme for the automatic assessment of early-stage AD, based on Archimedes’ spiral. It exploits transfer learning for feature extraction on “hybrid” spiral images, which convey dynamic information of the handwritten gesture. Such images, generated from online sequences captured by a digitizer, embed in the spiral trajectory image, in a pointwise manner, dynamic information encoded in greyscale. We consider four types of hybrid images, embedding separately pressure, altitude, velocity, and acceleration values in the spiral trajectory, both when the pen is on paper and in-air.
Our experiments demonstrate that this hybrid encoding of the spiral gesture leads to more discriminant features, extracted by transfer learning, compared to those obtained on raw spiral images that only convey spatial information of the trajectory. We actually assess the discriminant power of such features with the accuracy, sensitivity, and specificity metrics obtained after training an SVM classifier on feature vectors extracted from different layers of AlexNet.
When embedding dynamic parameters in the spiral trajectory, we first remark a strong improvement of the average sensitivity, from 62% on the raw image (
Table 1), to 77% with pressure (
Table 2), to 81% with altitude (
Table 3), to 76% with velocity (
Table 4), and to 71% with acceleration (
Table 5). Our methodology coupling hybrid images with transfer learning thus enhances the detection of AD at an early stage. Moreover, our methodology leads to more balanced average sensitivity and specificity values: on raw images, we note a difference of 25% between specificity (87%) and sensitivity (62%), while on hybrid images such difference is significantly reduced, as it appears in
Table 2,
Table 3,
Table 4 and
Table 5. This means that our approach enhances significantly sensitivity while maintaining high specificity values.
Our work also shows that such improvements required studying representations extracted by the network at different layers. Indeed, on raw images, fc7 gives the best accuracy, with high specificity at the price of a very low sensitivity. For hybrid images, in general, we obtain instead Conv3 discriminant descriptors that strike a better balance between sensitivity and specificity: it is the case for pressure, altitude, and velocity images. For acceleration, we obtain the best performance at Conv5.
Concerning dynamic parameters, pressure, altitude, and velocity lead to more discriminant features between AD and HC, comparatively to acceleration. Actually, acceleration leads to the lowest average accuracy (74.5%), the same obtained on raw images, but with a better balance between sensitivity and specificity. The highest average accuracies are obtained with pressure images (77% in
Conv3 and 79.5% in
Conv5) and altitude (77.5% in
Conv3), followed by velocity images (73.5% in
Conv3). This confirms our previous result in [
28], obtained on the same population, but on signature samples: pressure and altitude convey a high discriminant power for early AD detection. More precisely, the altitude angle leads to the highest sensitivity (81%), highlighting that the way the pen is held by the writer during the spiral gesture, is of significant importance for AD detection. This fact has also an impact on pen pressure, explaining why pressure images also show a high sensitivity (78%).
We confirm these results when fusing by majority voting the decisions of experts trained on descriptors extracted from AlexNet. Given our previous analysis of pressure and altitude descriptors, we compare two configurations both including such descriptors: one fusing decisions of experts trained on pressure, altitude and velocity descriptors, and another fusing decisions of experts trained on pressure, altitude, and acceleration descriptors. The first outperforms all previous results: the accuracy reaches 81.5% on average and sensitivity increases to 84%, also reducing the standard deviation over the 10 samplings (
Table 6). Compared to the classification performance obtained on descriptors from raw images, there is an absolute improvement in sensitivity of 22% (from 62% in
Table 1 to 84% in
Table 6), and in accuracy of 7%, confirming the power of our approach. This result, which is the best, is obtained with intermediate-level descriptors from the third convolutional layer (
Conv3), confirming the trend already observed in most types of hybrid images.
Moreover, we note that accuracy and sensitivity both increase when extracting descriptors in the first layers, since Conv2; instead, on descriptors of higher layers, accuracy and sensitivity decrease. For this reason, we studied majority voting with more experts, each trained respectively on descriptors from fc7 and lower layers, such as Conv2 and Conv3, or on higher ones, such as Conv3 and Conv5. None of such fusion schemes improves classification results, leading to the conclusion that Conv3 already extracts the most discriminant descriptors for our specific classification task on spiral images.
Finally, decision fusion outperforms low-level fusion, namely when giving as input to AlexNet each type of hybrid image through one channel and extracting descriptors at different layers. Low-level fusion of the best combination of hybrid images (pressure, velocity, and altitude) led indeed to lower accuracy (77.5% in
Table 10 versus 81.5% in
Table 6) and lower sensitivity (76% in
Table 10 versus 84% in
Table 6), with a larger standard deviation over the 10 samplings. Automatic feature extraction is indeed efficient when considering as input to AlexNet only one type of hybrid image.
With regard to the literature on degenerative diseases, most works (on Parkinson’s disease) exploit decision fusion successfully, but on several tasks [
16,
17,
18,
20,
21]. Our work focuses on one task in order to show its intrinsic potential for AD detection. This study demonstrates that the digitized spiral task can be used successfully in a standalone mode for early AD detection, with a precise transfer learning methodology.
Furthermore, our methodology allows confirming observed trends in our previous works about the most pertinent dynamic parameters for AD detection: altitude, pen pressure, and velocity convey precious information on the wrist-hand-finger system for pathology detection. The best representations for dynamic information can be extracted by AlexNet automatically at Conv3, and decision fusion is the most reliable paradigm for discriminating between the two populations. The high performance obtained on the spiral with this fusion scheme makes it possible to consider other short tasks as signatures or other graphomotor gestures for improving detection.
In the future, we aim at studying transfer learning based on other DL models following the same methodology. The objective is to analyze the effectiveness of descriptors extracted from more complex architectures compared to those obtained with AlexNet. Moreover, we will go further in studying other strategies for encoding dynamic information in visual contents, in order to enhance the benefit of exploiting transfer learning.