Simplified Convolutional Neural Network Application for Cervix Type Classification via Colposcopic Images
Abstract
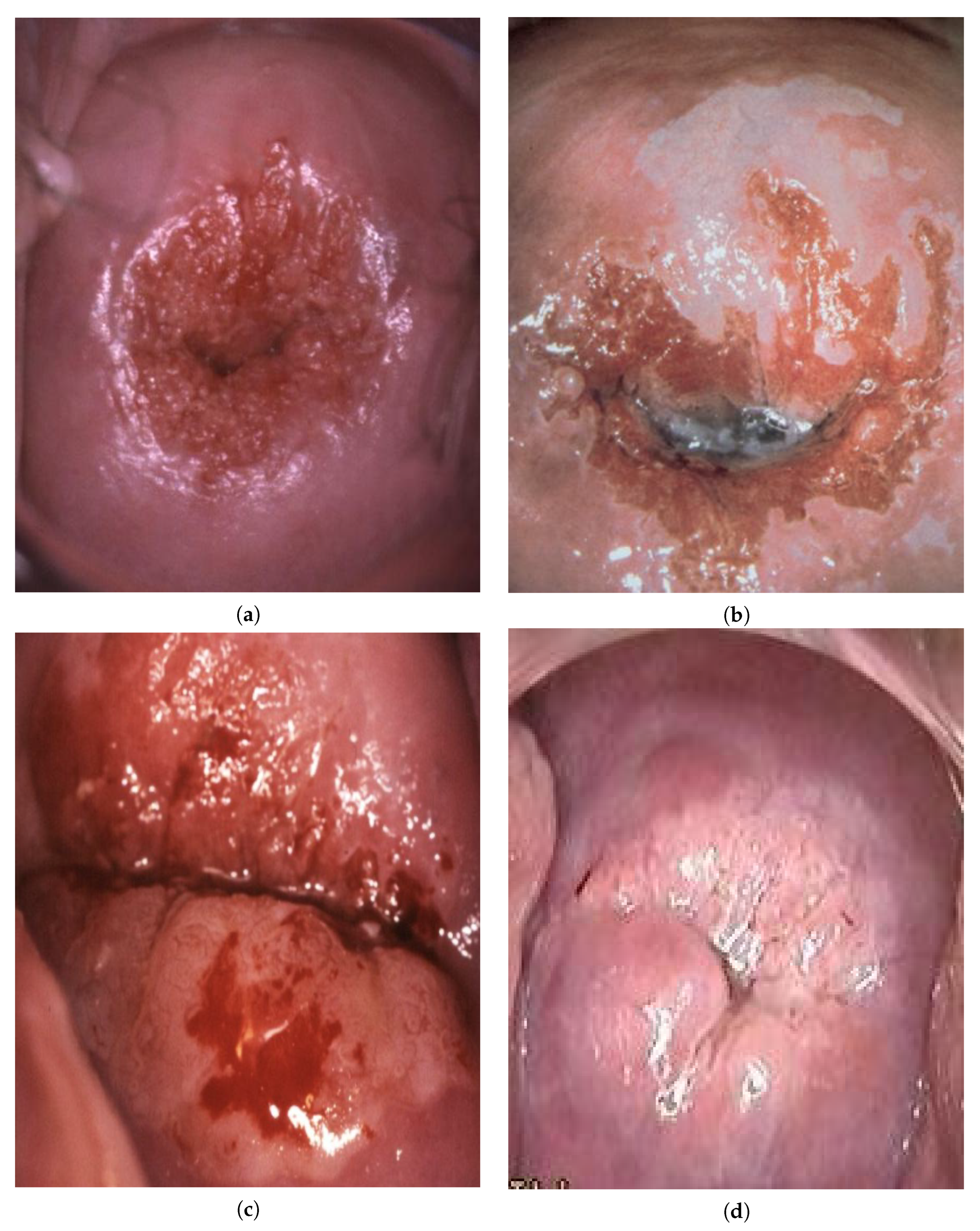
:1. Introduction
2. Related Work
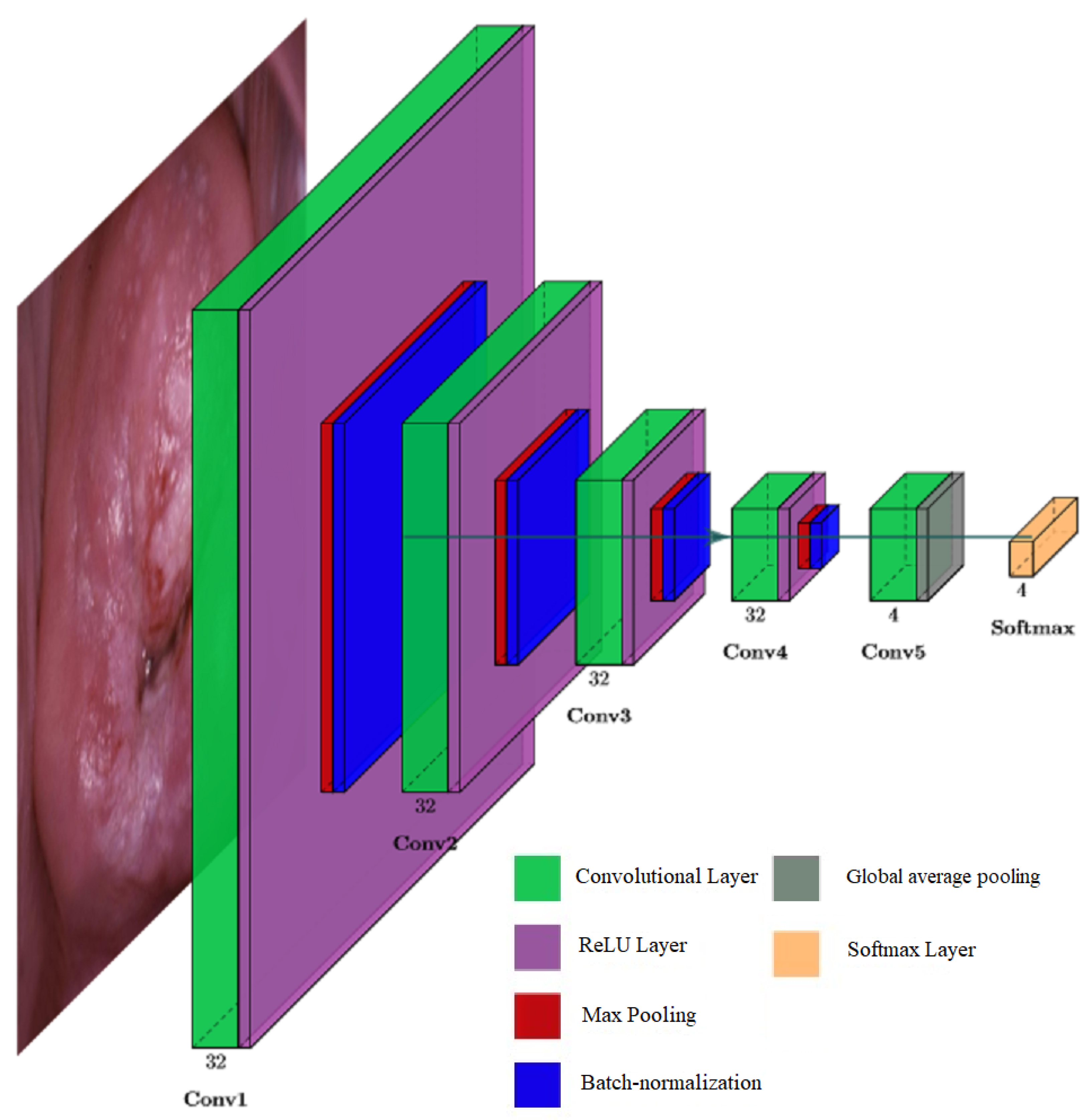
3. Methodology
- An image is converted into grayscale;
- A grayscale image binarization is performed to select glares;
- Convolutional layer is a layer that uses convolution operation for extracting features from the input.
- ReLU is a rectified linear unit activation function. It is less prone to the vanishing gradient problem because its derivative is always 1 for the positive values of the argument.
- Max-pooling is a subsampling layer using the maximum value. It is used to increase the receptive field.
- Batch normalization is a method that improves performance and stabilizes the operation of neural networks. The idea of the method is that the layers of the neural network are fed with data that has been preprocessed and have zero mathematical expectation and unit variance.
- Global average pooling is a layer that can replace a fully connected layer. As shown in [40], this layer has the same functionality as traditional fully connected layers. One of the advantages of global pooling over fully connected layers is that its structure is similar to convolutional layers, ensuring correspondence between feature maps and object categories. Additionally, unlike a fully connected layer, which requires many training and tuning parameters, reducing the spatial parameters will make the model more robust and resist overfitting.
- Softmax is a layer for predicting the probabilities associated with a categorical distribution.
4. Results
5. Discussion
- –
- The training set is two times smaller than the test set, which has a positive effect in conditions of a shortage of colposcopic images in the field of cervical cancer;
- –
- The neural network showed good learnability with the maximum achieved classification accuracy of 94.68% on the tested data set;
- –
- The computational complexity is 5 times less than that of existing solutions [14]. This is especially important since the placement of neural networks in medical tools, such as a colposcope, imposes performance requirements with limited resources of the computer system embedded in the tool.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ANN | Artificial Neural Network |
| AUC | Area Under the Curve |
| APC | Affinity Propagation Clustering |
| CC | Cervical Cancer |
| CIN | Cervical Intraepithelial Neoplasia |
| CNN | Convolutional Neural Network |
| DSS | Decision Support System |
| HPV | Human Papillomavirus |
| HSIL | High-Grade Squamous Intraepithelial Lesion |
| IOMT | Internet of Medical Things |
| LAST | Lower Anogenital Squamous Cell Terminology |
| LSIL | Low-Grade Squamous Intraepithelial Lesion |
| WCE | Wireless Capsule Endoscopy |
| WHO | World Health Organization |
References
- Sung, H.; Ferlay, J.; Siegel, R.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Franco, E.; Schlecht, N.; Saslow, D. The epidemiology of cervical cancer. Cancer J. 2003, 9, 348–359. [Google Scholar] [CrossRef]
- Lei, J.; Ploner, A.; Elfström, K.; Wang, J.; Roth, A.; Fang, F.; Sundström, K.; Dillner, J.; Sparén, P. HPV vaccination and the risk of invasive cervical cancer. N. Engl. J. Med. 2020, 383, 1340–1348. [Google Scholar] [CrossRef]
- Simms, K.; Steinberg, J.; Caruana, M.; Smith, M.; Lew, J.; Soerjomataram, I.; Castle, P.; Bray, F.; Canfell, K. Impact of scaled up human papillomavirus vaccination and cervical screening and the potential for global elimination of cervical cancer in 181 countries, 2020–99: A modelling study. Lancet Oncol. 2019, 20, 394–407. [Google Scholar] [CrossRef]
- Barchuk, A.; Bespalov, A.; Huhtala, H.; Chimed, T.; Laricheva, I.; Belyaev, A.; Bray, F.; Anttila, A.; Auvinen, A. Breast and cervical cancer incidence and mortality trends in Russia 1980–2013. Cancer Epidemiol. 2018, 55, 73–80. [Google Scholar] [CrossRef]
- Goldie, S.; Grima, D.; Kohli, M.; Wright, T.; Weinstein, M.; Franco, E. A comprehensive natural history model of HPV infection and cervical cancer to estimate the clinical impact of a prophylactic HPV-16/18 vaccine. Int. J. Cancer 2003, 106, 896–904. [Google Scholar] [CrossRef]
- World Health Organization. Human papillomavirus vaccines: WHO position paper = Vaccins anti-papillomavirus humain: Note d’information de l’OMS. Wkly. Epidemiol. Rec. 2009, 84, 118–131. [Google Scholar]
- Finnish Cancer Registry. Cancer Statistics Site. Available online: https://syoparekisteri.fi/tilastot/tautitilastot/ (accessed on 21 August 2018).
- Demarco, M.; Egemen, D.; Raine-Bennett, T.; Cheung, L.; Befano, B.; Poitras, N.; Lorey, T.; Chen, X.; Gage, J.; Castle, P.; et al. A Study of Partial Human Papillomavirus Genotyping in Support of the 2019 ASCCP Risk-Based Management Consensus Guidelines. J. Low. Genit. Tract Dis. 2020, 24, 144–147. [Google Scholar] [CrossRef]
- Bosch, X.; Harper, D. Prevention strategies of cervical cancer in the HPV vaccine era. Gynecol. Oncol. 2006, 103, 21–24. [Google Scholar] [CrossRef]
- Nobbenhuis, M.; Helmerhorst, T.; van den Brule, A.; Rozendaal, L.; Voorhorst, F.; Bezemer, P.; Verheijen, R.; Meijer, C. Cytological regression and clearance of high-risk human papillomavirus in women with an abnormal cervical smear. Lancet 2001, 358, 1782–1783. [Google Scholar] [CrossRef]
- Ho, G.; Bierman, R.; Beardsley, L.; Chang, C.; Burk, R. Natural history of cervicovaginal papillomavirus infection in young women. N. Engl. J. Med. 1998, 338, 423–428. [Google Scholar] [CrossRef]
- Moore, E.; Danielewski, J.; Garland, S.; Tan, J.; Quinn, M.; Stevens, M.; Tabrizi, S. Clearance of Human Papillomavirus in Women Treated for Cervical Dysplasia. Obstet. Gynecol. 2011, 338, 101–108. [Google Scholar] [CrossRef]
- Rodríguez, A.; Schiffman, M.; Herrero, R.; Wacholder, S.; Hildesheim, A.; Castle, P.; Solomon, D.; Burk, R.; Proyecto Epidemiológico Guanacaste Group. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J. Natl. Cancer Inst. 2008, 100, 512–517. [Google Scholar] [CrossRef] [Green Version]
- Ferris, D.; Litaker, M. Interobserver Agreement for Colposcopy Quality Control Using Digitized Colposcopic Images During the ALTS Trial. J. Low. Genit. Tract Dis. 2008, 9, 29–35. [Google Scholar] [CrossRef]
- Bornstein, J.; Bentley, J.; Bösze, P.; Girardi, F.; Haefner, H.; Menton, M.; Perrotta, M.; Prendiville, W.; Russell, P.; Sideri, M.; et al. Colposcopic terminology of the International Federation for Cervical Pathology and Colposcopy. Obstet. Gynecol. 2012, 120, 166–172. [Google Scholar] [CrossRef] [Green Version]
- Gage, J.; Hanson, V.; Abbey, K.; Dippery, S.; Gardner, S.; Kubota, J.; Schiffman, M.; Solomon, D.; Jeronimo, J. ASCUS LSIL Triage Study (ALTS) Group. Number of cervical biopsies and sensitivity of colposcopy. Obstet. Gynecol. 2006, 108, 264–272. [Google Scholar] [CrossRef]
- Pretorius, R.G.; Zhang, W.H.; Belinson, J.L.; Huang, M.N.; Wu, L.Y.; Zhang, X.; Qiao, Y.L. Colposcopically directed biopsy, random cervical biopsy, and endocervical curettage in the diagnosis of cervical intraepithelial neoplasia II or worse. Obstet. Gynecol. 2004, 191, 430–434. [Google Scholar] [CrossRef]
- Topol, E. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Jha, S.; Topol, E.J. Adapting to Artificial Intelligence: Radiologists and Pathologists as Information Specialists. JAMA 2016, 316, 2353–2354. [Google Scholar] [CrossRef]
- Sato, M.; Horie, K.; Hara, A.; Miyamoto, Y.; Kurihara, K.; Tomio, K.; Yokota, H. Application of deep learning to the classification of images from colposcopy. Oncol. Lett. 2018, 15, 3518–3523. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, K.; Cardoso, J.; Fernandes, J. Automated Methods for the Decision Support of Cervical Cancer Screening Using Digital Colposcopies. IEEE Access 2018, 6, 33910–33927. [Google Scholar] [CrossRef]
- AI-Powered Radiology Platform. Available online: https://botkin.ai/ (accessed on 30 January 2022).
- Ngan, T.; Tuan, T.; Son, L.; Minh, N.; Dey, N. Decision making based on fuzzy aggregation operators for medical diagnosis from dental X-ray images. J. Med. Syst. 2016, 40, 280. [Google Scholar] [CrossRef]
- Mofidi, R.; Duff, M.; Madhavan, K.; Garden, O.; Parks, R. Identification of severe acute pancreatitis using an artificial neural network. Surgery 2007, 141, 59–66. [Google Scholar] [CrossRef]
- Andersson, B.; Andersson, R.; Ohlsson, M.; Nilsson, J. Prediction of severe acute pancreatitis at admission to hospital using artificial neural networks. Pancreatology 2011, 11, 328–335. [Google Scholar] [CrossRef]
- Chen, W.; Cockrell, C.; Ward, K.; Najarian, K. Intracranial Pressure Level Prediction in Traumatic Brain Injury by Extracting Features from Multiple Sources and Using Machine Learning Methods. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine (BIBM ’10), Hong Kong, China, 18–21 December 2010; pp. 510–515. [Google Scholar]
- Davuluri, P.; Wu, J.; Ward, K.; Cockrell, C.; Najarian, K.; Hobson, R. An Automated Method for Hemorrhage Detection in Traumatic Pelvic Injuries. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 5108–5111. [Google Scholar]
- Shin, Y.; Qadir, H.; Aabakken, L.; Bergsland, J.; Balasingham, I. Automatic Colon Polyp Detection Using Region Based Deep CNN and Post Learning Approaches. IEEE Access 2018, 6, 40950–40962. [Google Scholar] [CrossRef]
- Owais, M.; Arsalan, M.; Choi, J.; Mahmood, T.; Park, K. Artificial intelligence-based classification of multiple gastrointestinal diseases using endoscopy videos for clinical diagnosis. Clin. Med. 2019, 8, 986. [Google Scholar] [CrossRef] [Green Version]
- Alaskar, H.; Hussain, A.; Al-Aseem, N.; Liatsis, P.; Al-Jumeily, D. Application of Convolutional Neural Networks for Automated Ulcer Detection in Wireless Capsule Endoscopy Images. Sensors 2019, 19, 1265. [Google Scholar] [CrossRef] [Green Version]
- Park, H.; Kim, Y.; Lee, S. Adenocarcinoma Recognition in Endoscopy Images Using Optimized Convolutional Neural Networks. Appl. Sci. 2020, 10, 1650. [Google Scholar] [CrossRef] [Green Version]
- Cho, B.J.; Choi, Y.J.; Lee, M.J.; Kim, J.H.; Son, G.H.; Park, S.H.; Kim, H.B.; Joo, Y.J.; Cho, H.Y.; Kyung, M.S.; et al. Classification of cervical neoplasms on colposcopic photography using deep learning. Sci. Rep. 2020, 10, 13652. [Google Scholar] [CrossRef]
- Bing, B.; Yongzhao, D.; Peizhong, L.; Pengming, S.; Ping, L.; Yuchun, L. Detection of cervical lesion region from colposcopic images based on feature reselection. Biomed. Signal Process. Control 2020, 57, 101785. [Google Scholar]
- Xue, P.; Tang, C.; Li, Q.; Li, Y.; Shen, Y.; Zhao, Y.; Chen, J.; Wu, J.; Li, L.; Wang, W.; et al. Development and validation of an artificial intelligence system for grading colposcopic impressions and guiding biopsies. BMC Med. 2020, 18, 406. [Google Scholar] [CrossRef]
- Kaggle. Intel & MobileODT Cervical Cancer Screening. Available online: https://www.kaggle.com/c/intel-mobileodt-cervical-cancer-screening (accessed on 30 January 2022).
- Kim, J.; Lee, J.; Lee, K. Accurate Image Super-Resolution Using Very Deep Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 1646–1654. [Google Scholar]
- Telea, A. An Image Inpainting Technique Based on the Fast Marching Method. J. Graph. Tools 2004, 9, 23–34. [Google Scholar] [CrossRef]
- OpenCV Site. Available online: https://opencv.org/ (accessed on 30 January 2022).
- Lin, M.; Chen, Q.; Yan, S. Network In Network. arXiv 2013, arXiv:1312.4400v3. [Google Scholar]
- Velichko, E.; Nepomnyashchaya, E.; Baranov, M.; Galeeva, M.; Pavlov, V.; Zavjalov, S.; Savchenko, E.; Pervunina, T.; Govorov, I.; Komlichenko, E. Concept of Smart Medical Autonomous Distributed System for Diagnostics Based on Machine Learning Technology. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2019; pp. 515–524. [Google Scholar]
- Tataria, H.; Shafi, M.; Molisch, A.; Dohler, M.; Sjöland, H.; Tufvesson, F. 6G Wireless Systems: Vision, Requirements, Challenges, Insights, and Opportunities. Proc. IEEE 2021, 109, 1166–1199. [Google Scholar] [CrossRef]
- Akyildiz, I.; Kak, A.; Nie, S. 6G and Beyond: The Future of Wireless Communications Systems. IEEE Access 2020, 8, 133995–134030. [Google Scholar] [CrossRef]
- MobileODT Site. Available online: https://www.mobileodt.com/ (accessed on 30 January 2022).
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Noh, H.; Hong, S.; Han, B. Learning deconvolution network for semantic segmentation. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1520–1528. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6230–6239. [Google Scholar]
- Chen, L.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Zhang, H.; Dana, K.; Shi, J.; Zhang, Z.; Wang, X.; Tyagi, A.; Agrawal, A. Context Encoding for Semantic Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7151–7160. [Google Scholar]
- Bolya, D.; Zhou, C.; Xiao, F.; Lee, Y. YOLACT: Real-Time Instance Segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Korea, 27 October–2 November 2019; pp. 9156–9165. [Google Scholar]
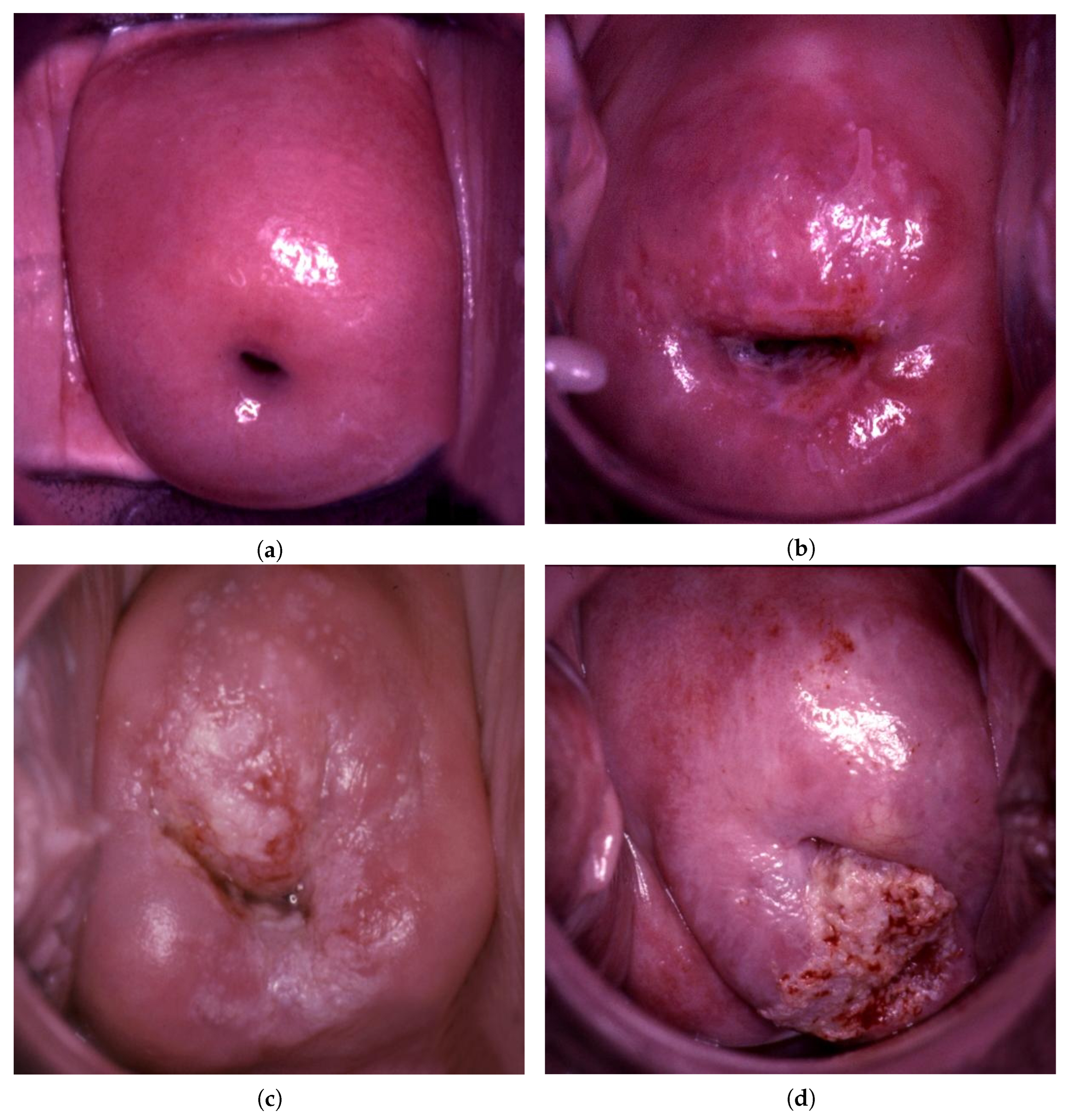
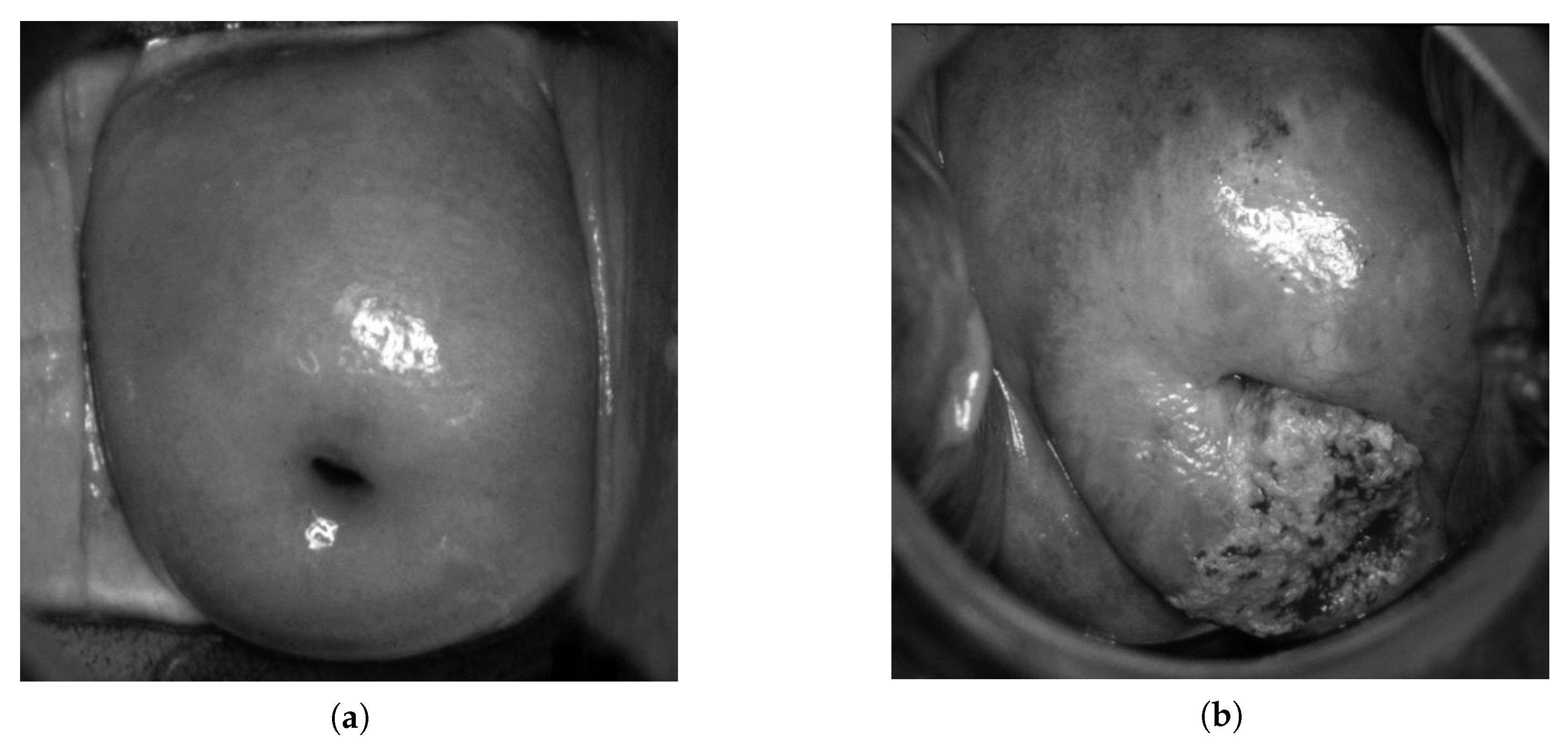
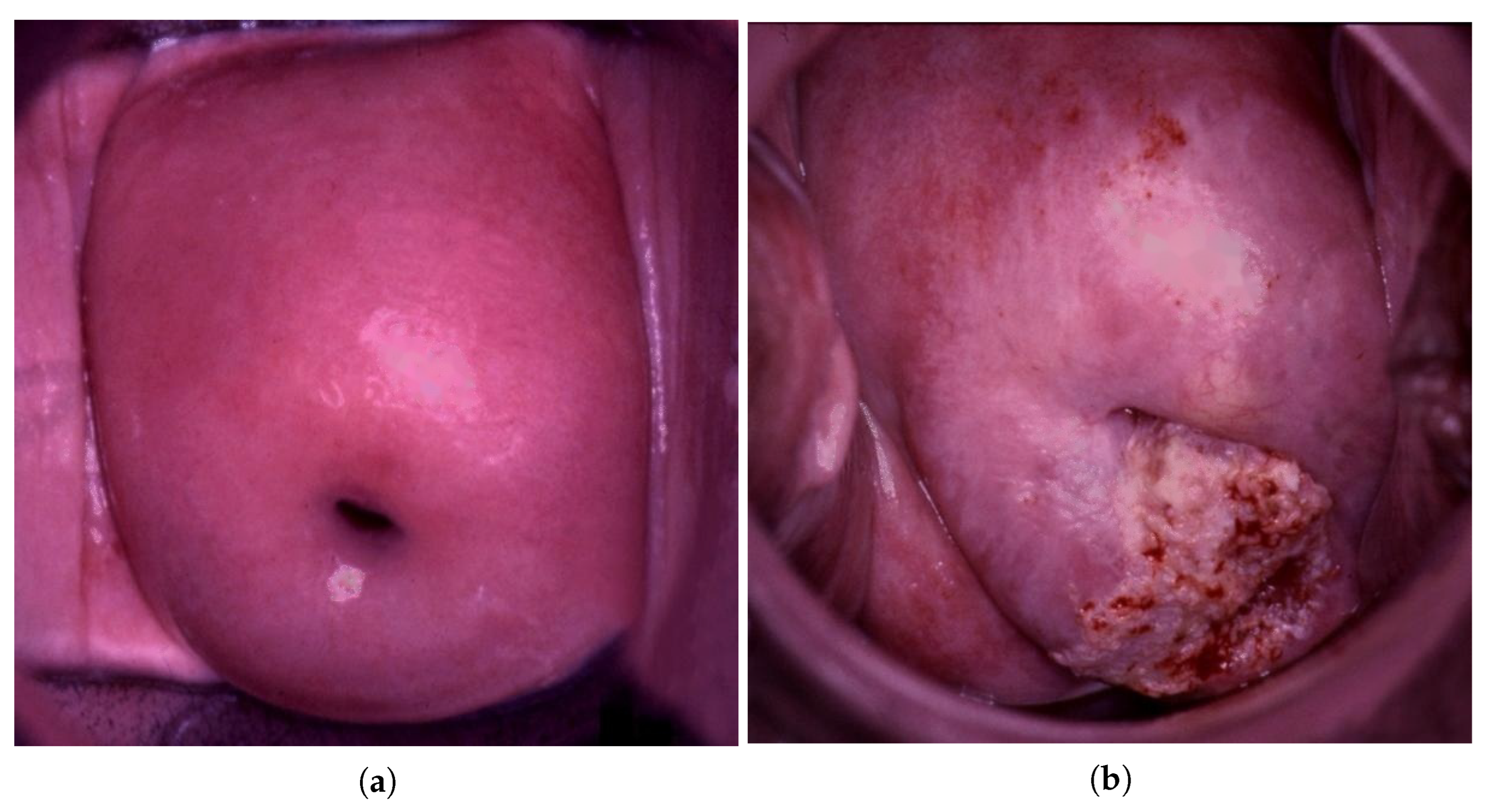


| Data Type | Normal | LSIL | HSIL | Suspicious for Invasion |
|---|---|---|---|---|
| Train | 657 | 63 | 133 | 38 |
| Test | 1323 | 94 | 1046 | 379 |
| Normal | LSIL | HSIL | Suspicious for Invasion | All |
|---|---|---|---|---|
| 95.46% | 79.78% | 94.16% | 97.09% | 94.68% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlov, V.; Fyodorov, S.; Zavjalov, S.; Pervunina, T.; Govorov, I.; Komlichenko, E.; Deynega, V.; Artemenko, V. Simplified Convolutional Neural Network Application for Cervix Type Classification via Colposcopic Images. Bioengineering 2022, 9, 240. https://doi.org/10.3390/bioengineering9060240
Pavlov V, Fyodorov S, Zavjalov S, Pervunina T, Govorov I, Komlichenko E, Deynega V, Artemenko V. Simplified Convolutional Neural Network Application for Cervix Type Classification via Colposcopic Images. Bioengineering. 2022; 9(6):240. https://doi.org/10.3390/bioengineering9060240
Chicago/Turabian StylePavlov, Vitalii, Stanislav Fyodorov, Sergey Zavjalov, Tatiana Pervunina, Igor Govorov, Eduard Komlichenko, Viktor Deynega, and Veronika Artemenko. 2022. "Simplified Convolutional Neural Network Application for Cervix Type Classification via Colposcopic Images" Bioengineering 9, no. 6: 240. https://doi.org/10.3390/bioengineering9060240
APA StylePavlov, V., Fyodorov, S., Zavjalov, S., Pervunina, T., Govorov, I., Komlichenko, E., Deynega, V., & Artemenko, V. (2022). Simplified Convolutional Neural Network Application for Cervix Type Classification via Colposcopic Images. Bioengineering, 9(6), 240. https://doi.org/10.3390/bioengineering9060240







