1. Introduction
Cardiovascular (CVD) diseases are globally recognised as the main cause of death, and they manifest themselves in the form of myocardial infarction or heart attack. According to the WHO [
1], CVD is responsible for 17.7 million deaths. Approximately 31% of all deaths occur in poor and middle-income nations, with 75% of these deaths happening in these countries. Arrhythmias are the type of CVD that represents irregular patterns of heartbeats, such as the beating of the heart too fast or too slow. Examples of arrhythmias include: a trial Fibrillation (AF), premature ventricular contraction (PVC), ventricular fibrillation (VF), and Tachycardia. Although single cardiac arrhythmias may have little impact on one’s life, a persistent one might cause fatal problems, such as prolonged PVC that occasionally turns into Ventricular Tachycardia (VT), or Ventricular Fibrillation that can immediately lead to heart failure. Ventricular arrhythmias are one of the most prevalent types of cardiac arrhythmias that result in irregular heartbeats are responsible for nearly 80% of sudden cardiac deaths [
2,
3]. If arrhythmia conditions are detected early enough, ECG signal analysis may improve the identification of risk factors for cardiac arrest. Thus, it is fair to infer that monitoring heart rhythm regularly is crucial for avoiding CVDs. Practitioners use electrocardiographs as a diagnostic tool to detect cardiovascular diseases called arrhythmias, which detects and interprets the heart’s electrical activity during the diagnosis and is represented in ECG signals. However, ECG signals are represented in the form of waves when an ECG machine is attached to the human body; to get an exact picture of the heart, ten electrodes are needed for capturing 12 leads (signals). According to Zubair et al. [
4], 12 ECG leads are required to properly diagnose, which are divided into precocious leads (I, II, III, aVL, aVR, aVF) to precordial leads (
,
,
,
,
,
). P waves, Q waves, R waves, S waves, T waves, and U waves based on the heart’s anatomy are illustrated in
Figure 1. as positive and negative deflections from the baseline that signify a particular electrical event.
Conventionally, arrhythmia diagnosis studies focused on the filtering of noise from ECG signals [
5], waveform segmentation [
6], and manual feature extraction [
7,
8]. Various scientists have tried to classify arrhythmias using different machine learning (ML) and data mining methodologies. Here, we discuss some of the machine learning and deep learning techniques for the classification of arrhythmias.
Sahoo et al. [
9] identified QRS complex using Discrete Wavelet Transform and Empirical Mode Distribution (EMP) for noise reduction, and Support Vector Machine was used to classify 5 distinct kinds of arrhythmias with an accuracy of 98.39% and 99.87% sensitivity with an error rate of 0.42. Osowski et al. [
10] used higher order statistics (HOS) and Hermite coefficients to detect the QRS complex. The performance of the proposed approach [
10] is compared with spectral power density algorithm, genetic algorithm, and SVM for classification of 5 different types of arrhythmia, which provides an average accuracy of 98.7%, 98.85%, 93%. Although these models are quite accurate due to the manual feature extraction process, their computation cost is high. Plawiak et al. [
11] used higher-order spectra for feature extraction, PCA for dimension reduction, and SVM to identify 5 different forms of arrhythmia with the accuracy of 99.28%.
Yang et al. [
12] used HOS and Hermite functions for manual feature extraction, and they also used SVM for disease classification. Polat and Gunes et al. [
13] suggested least square SVM (LS-SVM) for the classification of arrhythmia and used PCA to reduce the dimensionality of features from 256 to 15. Melgani and Bazi et al. [
14], using SVM, carried out an experimental study to identify five types of irregular waveforms and natural beats in a database from the MIT-BIH dataset. To increase the efficiency of SVM, they adopted particle swarm optimization (PSO). The sole purpose of PSO is to fine-tune the discriminator function for selecting the best features for SVM classifier training. The authors [
14], contrasted their technique to two existing classifiers, namely K-nearest neighbours (KNN) and radial basis function neural networks, and discovered that SVM outscored them with an ≈90% accuracy. In a similar vein, to optimise the discriminator function, GA is used in [
15]. Furthermore, they used a modified SVM classifier for arrhythmia classification.
Dutta et al. [
16] suggested an LS-SVM classifier for heartbeat classification, such as normal beats, PVC beats, and other beats, by utilising the MIT-BIH arrhythmia database. Their approach yields 96.12% accuracy for classification. Desai et al. [
17], considering 48 records of MIT-BIH arrhythmias, suggested a classifier using SVM for the classification and Discrete Wavelet Transformation (DWT) for feature representation. They categorised five types of arrhythmia beats: (i) Non-ectopic (N), (ii) Supraventricular ectopic (S), (iii) Ventricular ectopic (V), (iv) Fusion (F), and (v) Unknown (U). Their approach resulted in an accuracy of 98.49%. Furthermore, the authors performed statistical analysis using ANOVA to validate the efficacy of the proposed approach.
Kumaraswamy et al. [
18], considering the MIT-BIH arrhythmia database, proposed a new classifier for the classification of heartbeats that is useful for the detection of arrhythmias. In particular, they used a random forest tree classifier and discrete cosine transform (DCT) for discovering R-R intervals (an essential pattern that helps in detecting arrhythmias) as features.
Park et al. [
19] proposed a classifier for detecting 17 different types of heartbeats that can be used to detect arrhythmias. They carried out a two-step experimental study: (a) in the first step, P waves and the QRS complex are identified using the Pan-Tompkins method, and (b) in the second step, KNN classifier is used to classify them. This model was validated using MIT-BIH arrhythmias and performed with a sensitivity of 97.1% and specificity of 96.9%. Jun et al. [
20] used a high-performance GPU-based cloud system for arrhythmia detection. Similar to [
19], they used the Pan-Tompkins algorithm and KNN for identification and classification.
Machine learning paradigms are heavily influenced by feature architecture and a focus on feature extraction and filtering. The underlying concept behind learning is to include all of the data in signals so that the machine learning algorithm can learn and choose functions. This theory also underpins the deep learning paradigm, especially CNN and its 1-D equivalents [
21]. Because of the potential and prospect of deep learning techniques, researchers [
21,
22,
23,
24,
25] have adopted these techniques for the detection/classification of various types of chronic diseases. In this direction, Acharya et al. [
22,
23,
26,
27] conducted a detailed study for arrhythmia classification using deep learning. In [
26], they proposed an automated classifier for arrhythmias. In [
23], a CNN architecture is built to predict myocardial infarction with a 95.22% accuracy. In [
27], an automated CNN model for the categorization of shockable and non-shockable ventricular arrhythmias is proposed. The suggested model outperformed with an accuracy of 93.18%, 95.32% sensitivity, and 94.04% specificity.
Kachuee et al. [
24] utilized deep residual CNN for classification, and t-SNE was used for visualization. They employed their approach to identify five distinct arrhythmias by the AAMI EC57 norm and offered better results than ALTAN et al. [
25] on the same database of MIT-BIH arrhythmias with an accuracy of 95.9%. Xia et al. [
28] utilised short-term Fourier transform (STFT) and stationary wavelet transform (SWT) for the detection of arterial fibrillation arrhythmias. Further, to evaluate ECG segments, STFT and SWT outputs are input to two separate deep CNN models. As a result, CNN outperformed with 98.29% for STFT and 98.63% for SWT. Savalia et al. [
29] proposed a model based on 5-layer CNN that can classify five types of arrhythmias. Although these models are quite accurate for minimising the loss function in backpropagation, they still suffer from the vanishing gradient problem of exploding gradients. For the classification of cardiac arrest during CPR, Jekova et al. [
30] employed an end-to-end CNN model entitled CNN3-CC-ECG, which they validated on the independent database OHCA. With automatic feature extraction, the model performed well, with a sensitivity of 89% for ventricular fibrillation (VF), a specificity of 91.7% for non-shockable organised rhythms (OR), and a specificity of 91.11% for asystole, but found moderate results with noisy data. Krasteva et al. [
31] applied convolution DNN for the classification of shockable and non-shockable rhythms, and to optimise the model they used random search optimization. Elola et al. [
32] utilized two DNN architectures to classify pulseless electrical activity (PEA) and pulse-generating rhythm (PR) using short ECG reading segments (5s) on an independent database (OHCA). Additionally, the model was optimised using Bayesian Optimization. Both architectures work well with balanced accuracy of 93.5% as compared to other state-of-the art algorithms. Although, 1D CNN outperformed with automatic feature extraction [
33] and more accurate than clinicians [
34], but recurrent neural networks (RNNs) are effective deep learning methods as they can handle time dependencies and variable-length segmented data [
35]. Although RNNs face certain difficulties, such as the vanishing gradient problem [
36], in which the gradient decreases with backpropagation and its value becomes too small, Consequently, diminished gradient values do not contribute much to learning. The reason for this is that with RNN, the layers that receive a diminished gradient to upgrade weights stop learning; as a result, these layers do not remember and may not be able to recall the longer sequences used. Thus, these layers have a short-term memory, which impacts negatively predicted problems. Nevertheless, this vanishing gradient issue can be resolved by using LSTM or GRU with ReLU, which allows capturing the impact of the earliest available data. Actually, by tuning the burden value, the vanishing gradient problem can be avoided. Altan et al. [
25] presented a classification network based on a four-layer deep belief network to classify five kinds of arrhythmias with an accuracy of 94%. Furthermore, they used DFT for feature extraction.
Ping et al. [
37] have presented an 8CSL approach for the identification of atrial fibrillation that includes shortcut connections in CNN, which aids in boosting data transmission speed, and 1 layer of LSTM, which aids in reducing long-term dependencies between data. To further test the proposed methodology, he compared it to the RNN and the multi-scale convolutional neural network (MCNN), and he discovered that 8CSL extracted features better when compared to the other two methods in terms of F1-score (84.89%, 89.55%, 85.64%) with different data segment lengths (5 s, 10 s, 20 s). When it comes to heartbeat detections, Ullah et al. [
38] used three different algorithms: CNN, CNN+LSTM and CNN+LSTM+attention model for the classification of five different types of arrhythmia in heartbeat detections over two well known databases, MIT-BIH arrhythmias and the PTB Diagnostic ECG Database. They discovered that CNN had 99.12%, CNN+LSTMhad 99.3%, and CNN+LSTM+attention model had 99.29%, all of which were statistically significant. Kang et al. [
39] tries to classify mental stress data using the CNN-LSTM model, where he has trained his model using the ST Change Database and WESAD Database, and he has converted the 1D ECG signals into the time and frequency domain in order to train the proposed method. On testing, he achieved an accuracy of 98.3%.
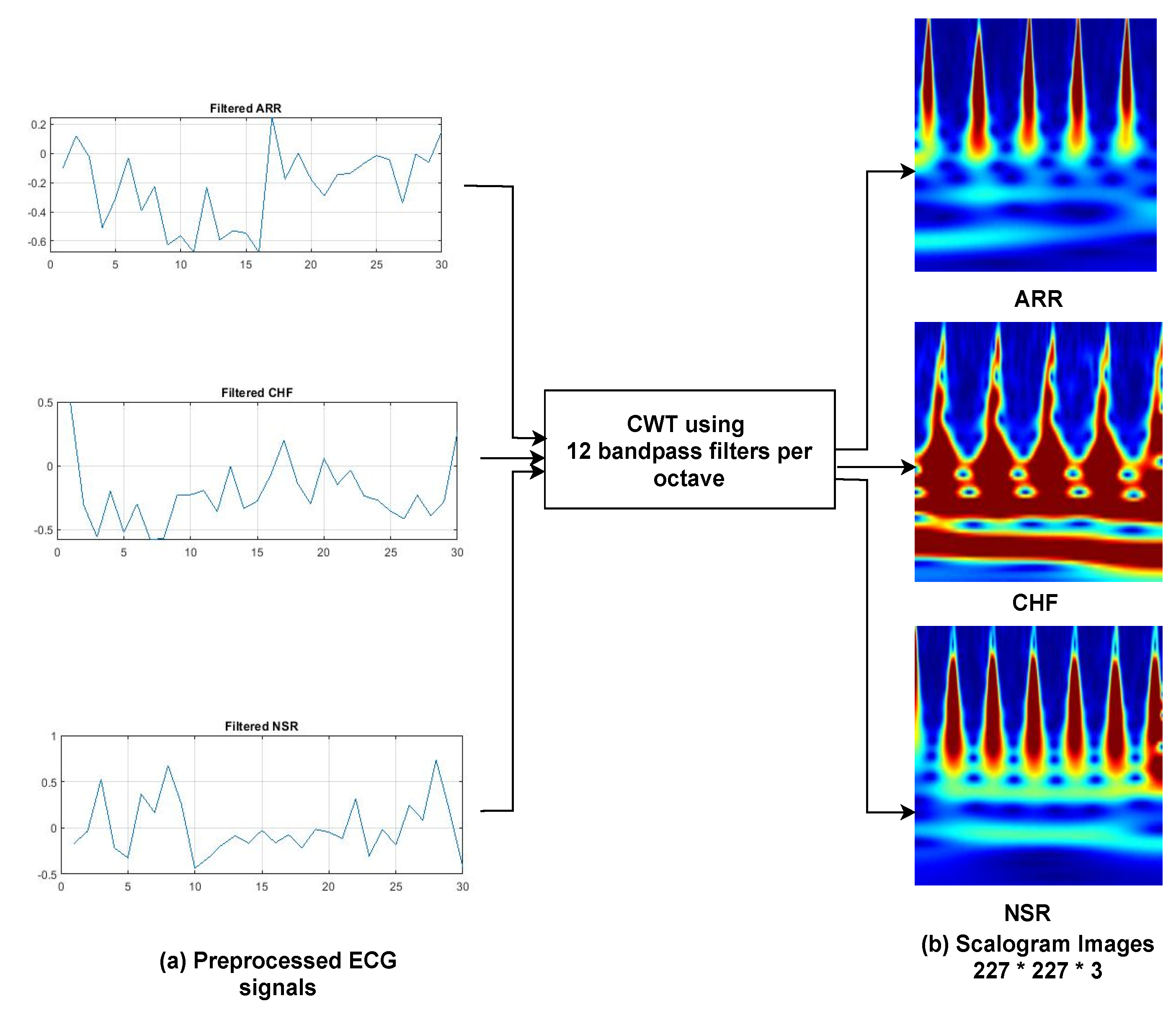
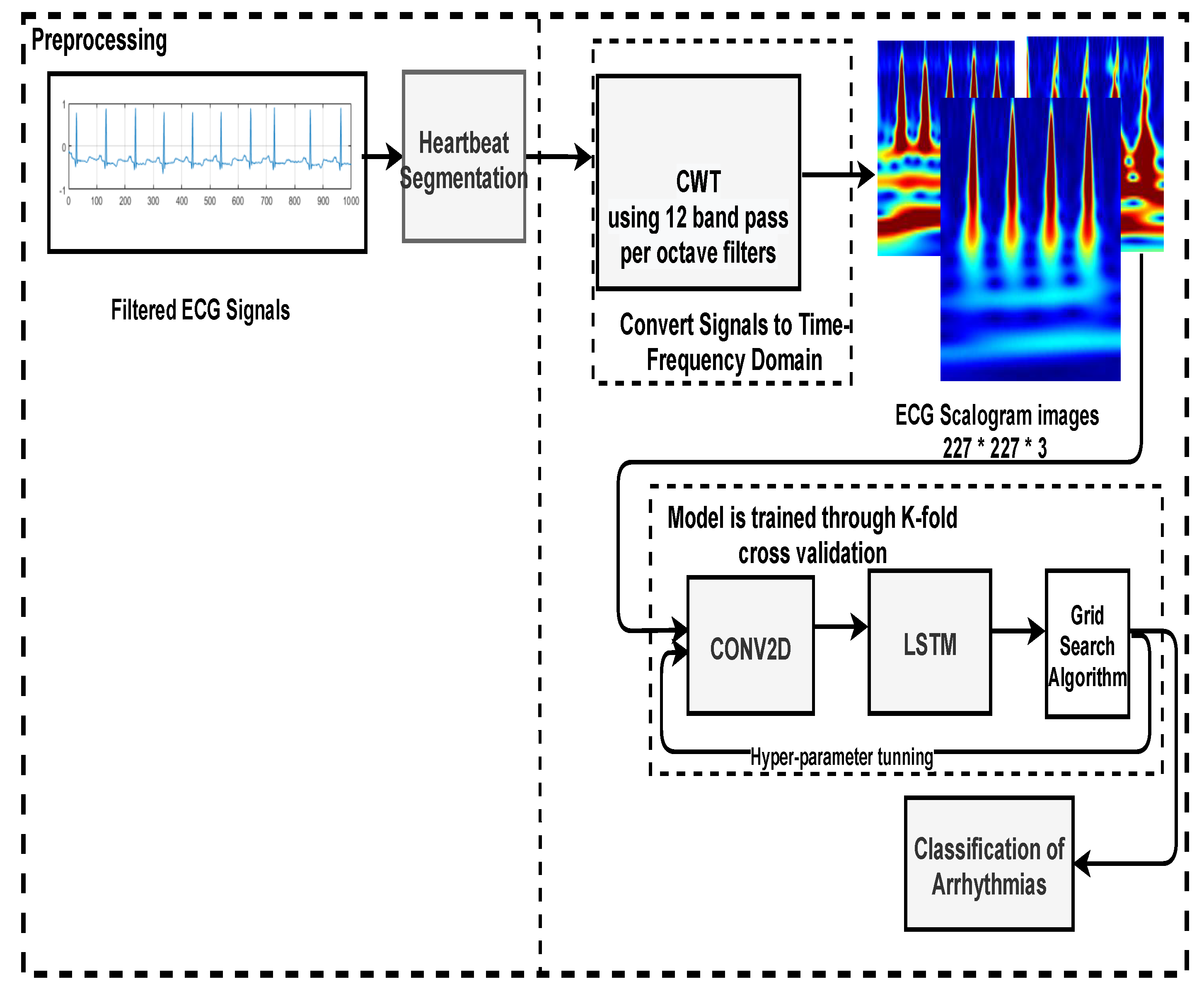
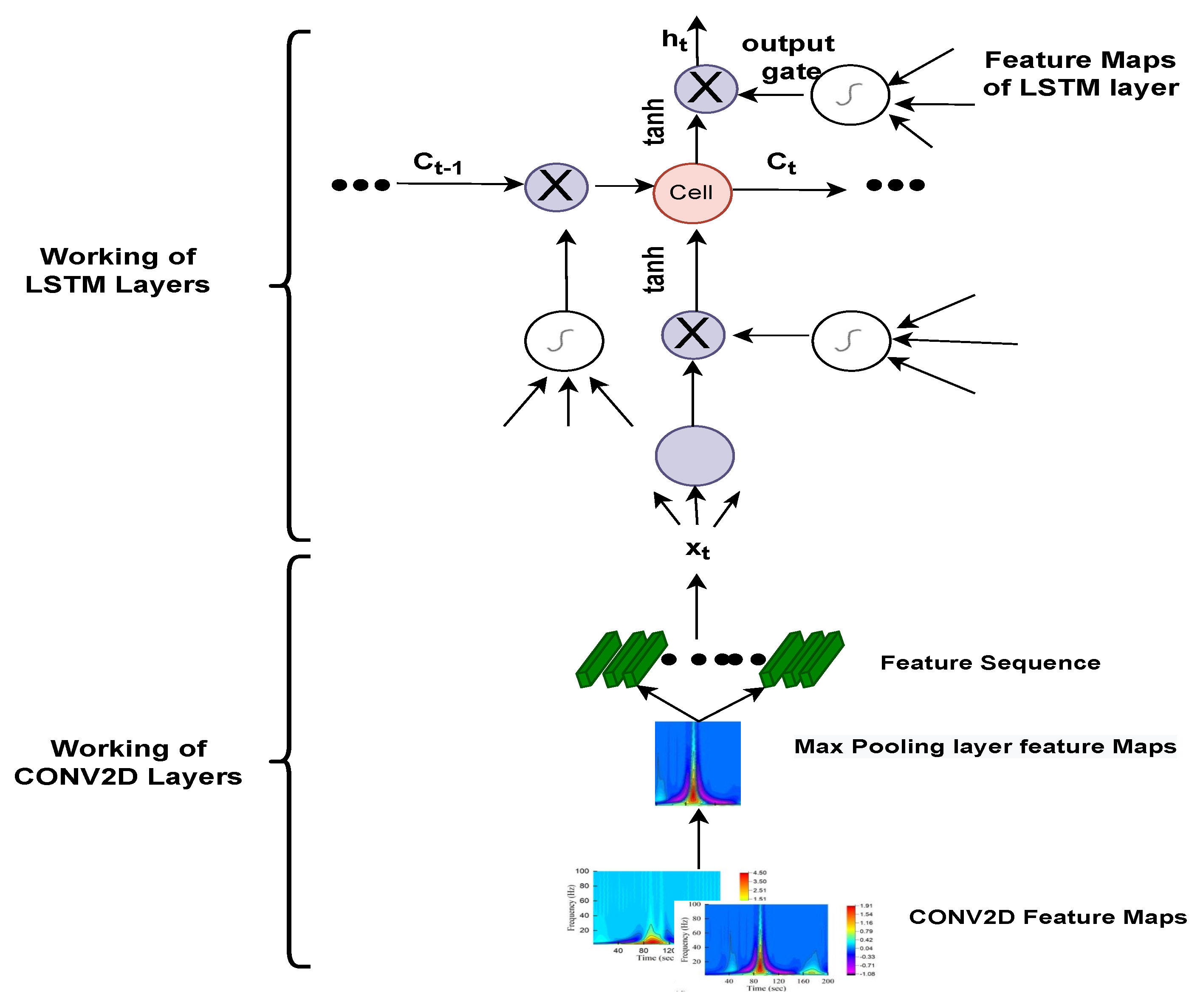
The majority of past work has trained models using a 1D ECG output, which contains a lot of noise, such as baseline wandering effects, power line interference’s, electromyographic noises [
40], and artefacts. Filtering and extracting features requires numerous preprocessing processes, which can jeopardise data integrity and model accuracy. Thus, the main aim of this study is to automate the detection and classification process. For that, 1-D ECG signals are translated into 2-D colored scalogram images using the Continuous Wavelet Transform (CWT) with a resolution of 227 × 227 × 3. Thereafter, a
CNN is adopted for automatic feature extraction and an LSTM is adopted for classification purposes. In sum, the following are the paper’s key contributions. (1) We proposed a hybrid approach combining the power of CNN and LSTM. In the initial phase, we converted all the ECG signals to images using CWT, so that the CNN model executes effectively, and then applied LSTM to classify arrhythmias. The approach was developed using the deep learning library on the MATLAB platform. The performance of the proposed approach is validated using the well-known MIT–BIH Arrhythmia database in two ways: firstly, by dividing the dataset into 75% for training and 25% for testing; and secondly, by training the model using K-fold cross validation. Additionally, the parameters of the proposed model are hyper-trained with the grid search algorithm.
Rest of the paper is organised in the following manner: The detailed description of the methodology, including MIT-BIH arrhythmias, dataset preprocessing steps to filter data and conversion of ECG signals into scalogram images, is given in
Section 2. Experimental results and performance evaluation are reported in
Section 3. Discussion is mentioned in
Section 4; Finally,
Section 5 concludes the paper.
3. Experimental Results and Performance Evaluation
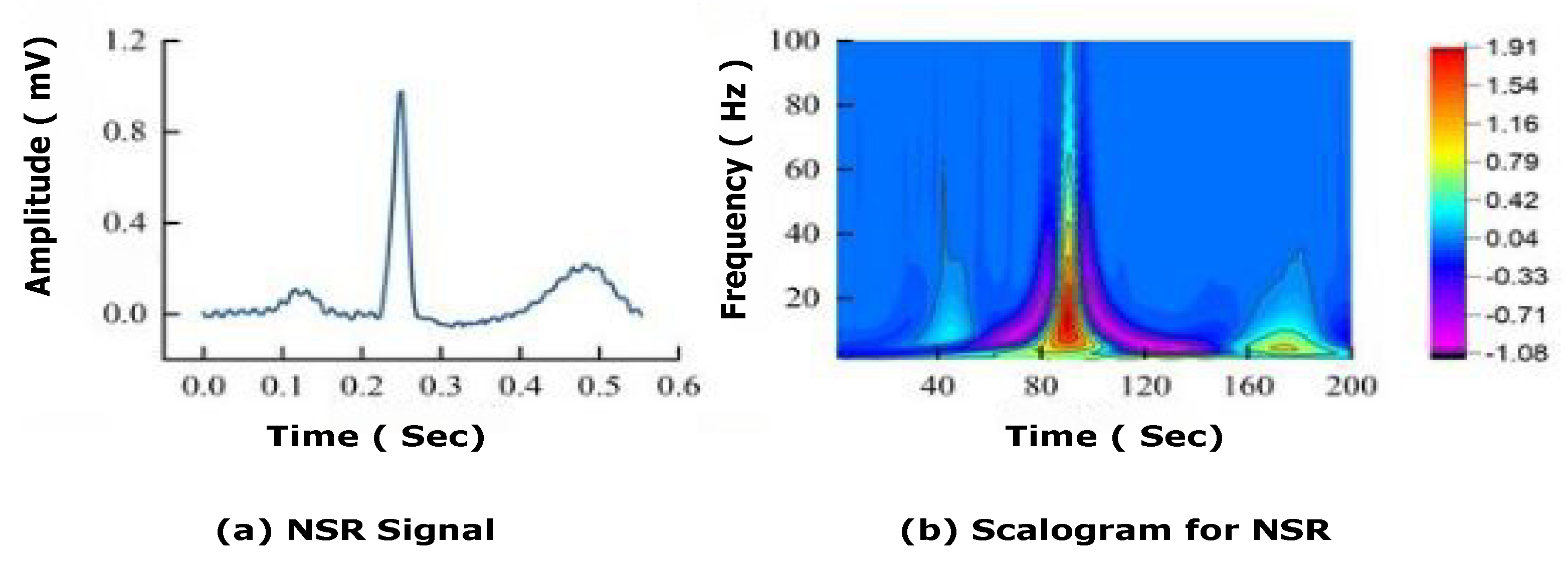
In this study, experiments were carried out on international standard ECG databases such as MIT-BIH cardiac arrhythmia (ARR), BIDMC congestive heart failure (CHF), and MIT-BIH normal sinus rhythm (NSR), which have detailed expert notation that is frequently utilised in contemporary ECG research. In this experiment, the dataset is divided using K-fold cross validation. Experimental tests were carried out on the workstation with an Intel Xeon gold 5215 10c 2.50 GHz processor and 16GB with training options: (Referenced from
Table 3) MiniBatchSize as 10, MaxEpochs as 20, InitialLearnRate as 1 × 10
−1, Learn Rate drop period as 3, Gradient Threshold as 1 and the total elapsed time for the training of the model is 6 min 30 s. These parameters are utilised using a grid search hyperparameter optimization algorithm with the highest mean test score of 92.35. Here, segmented 1D ECG signals were transformed into 2D scalogram images of size
, and trained over the 2DCNN-LSTM model. Experiments were carried out on Matlab 2021a.
To do a qualitative evaluation of the proposed 2D-CNN-LSTM, we utilise six metrics [
63] which are described as follows.
Sensitivity: determines the capacity of the model to accurately detect the actuality of the cases studied.
Specificity: used to determine the ability of the model to distinguish between individual negative samples.
Precision: defines the number of patients accurately classified by the model.
Recall: quantifies the number of positive class projections made from all positive cases in the data set
Accuracy: defines the percentage of correct classifications.
F-measure: offers a single score that combines both the accuracy and recall.
The mathematical formulas for the above-mentioned metrics are given in
Table 4, where True Positive (
TP) represents the number of positive patients who have been assessed as positive for ARR. The True Negative (
TN) represents the number of patients who are anticipated to be negative for ARR. Both of these matrices (
TP and
TN) indicate that the classification is accurate. A False Positive (
FP) represents the percentage of patients who are categorised as positive but are negative for ARR. A False Negative (
FN) is used to calculate the percentage of positive patients observed to be negative for ARR. However, both of the matrices (
FP,
FN) indicate a classification error. All three matrices (accuracy, sensitivity, and specificity) indicate the overall classification of the network model. The larger the value, the better the classification results will be.
Here, in this study, in order to evaluate the proposed model, we conducted two experiments. Firstly, the dataset was divided into two portions, 70% for the training dataset and 30% for the testing dataset, and by utilising hyperparameters (Referenced in
Table 3) such as kernel size as [3 3]; number of filters as 64; batch-size as 10; regularization dropout as 0.05; optimizer as
, maximum pool Size as [2 2]; loss-method as binary cross entropy; InitialLearnRate as 1 × 10
−1; decay as 0.0; epochs as 20 with 32 hidden units and found CNN-LSTM classifies three types of arrhythmias with an accuracy of ARR of 98%, precision of 0.97, recall of 0.96, F1-score of 0.98, sensitivity of 0.96, and specificity of 1, and for CHF class, accuracy of 77%, precision of 0.5, recall of 1, F1-score of 0.66, sensitivity of 1, and specificity of 0.69, and for the NSR class accuracy of 99%, precision of 0.98, recall of 1, F1-score of 0.66, sensitivity of 1, and specificity of 0.98, as shown in
Figure 10 and referenced in
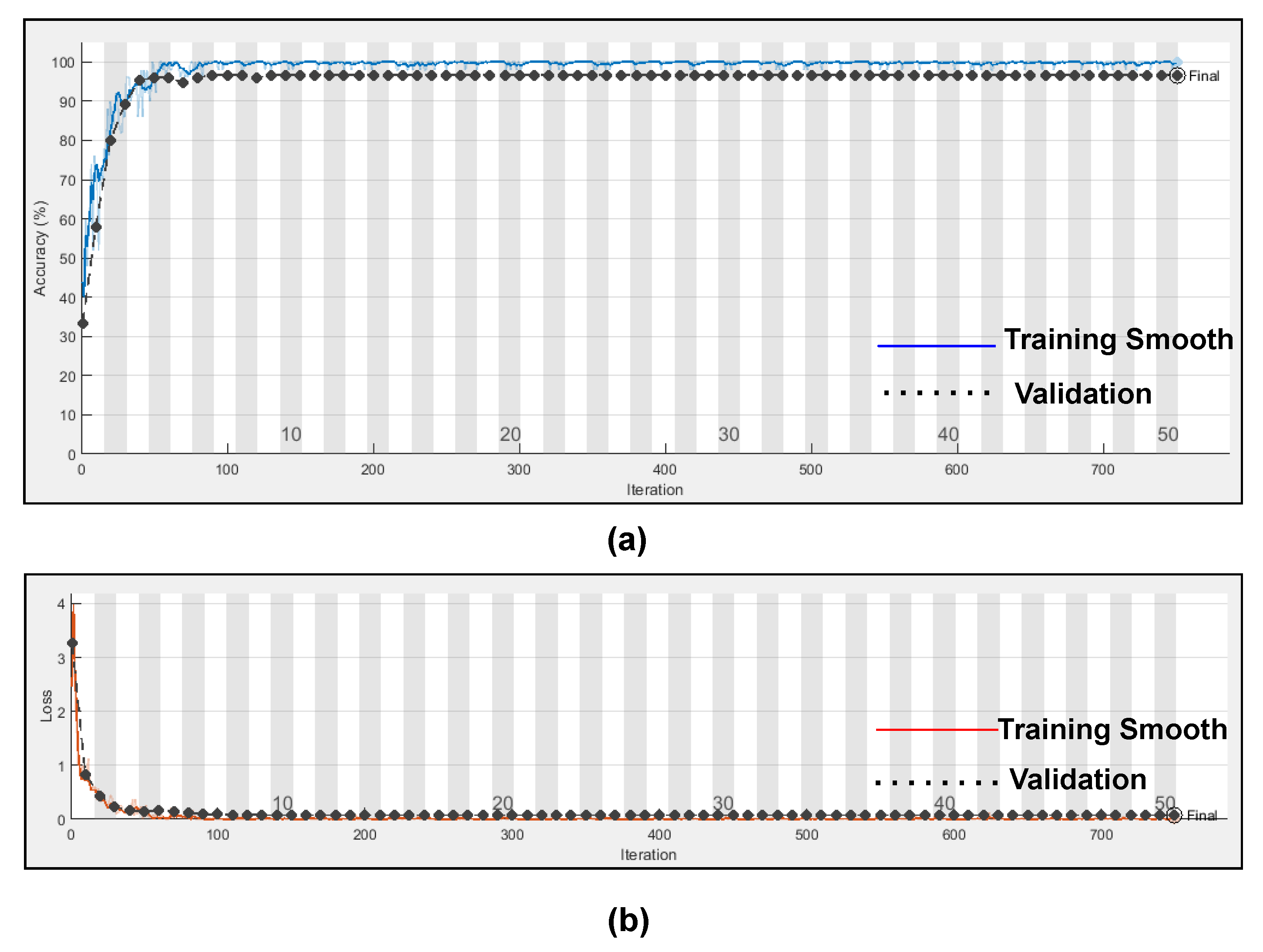
Table 5. Training accuracy and training loss of the proposed model without K-fold validation are shown in
Figure 11. Although the suggested models’ accuracy is acceptable, However, the model has under-fitting and over-fitting issues, which appear when the model has learnt less than or more than 20 epochs. The over-fitting issue model has a tendency to remember data and is unable to generalise new data, while the under-fitting model has a difficult time testing but is capable of generalising new data. We trained our models at 20 epochs and over-optimized parameters using 10-fold cross-validation to eliminate over-fitting and under-fitting, and the accuracy of proposed model was raised, (Referenced from
Table 6) where the accuracy of ARR is 98.7%, precision is 1, recall is 0.98, F1-score is 0.98, sensitivity is 0.98, and specificity is 0.98, and for CHF class accuracy is 99%, precision is 0.97, recall is 0.96, F1-score is 0.96, sensitivity is 0.96, and specificity is 0.99 and for the NSR class accuracy is 99%, precision is 0.97, recall is 1, F1-score is 0.98, sensitivity is 0.97, and specificity is 0.99 as shown in
Figure 12.
4. Discussion
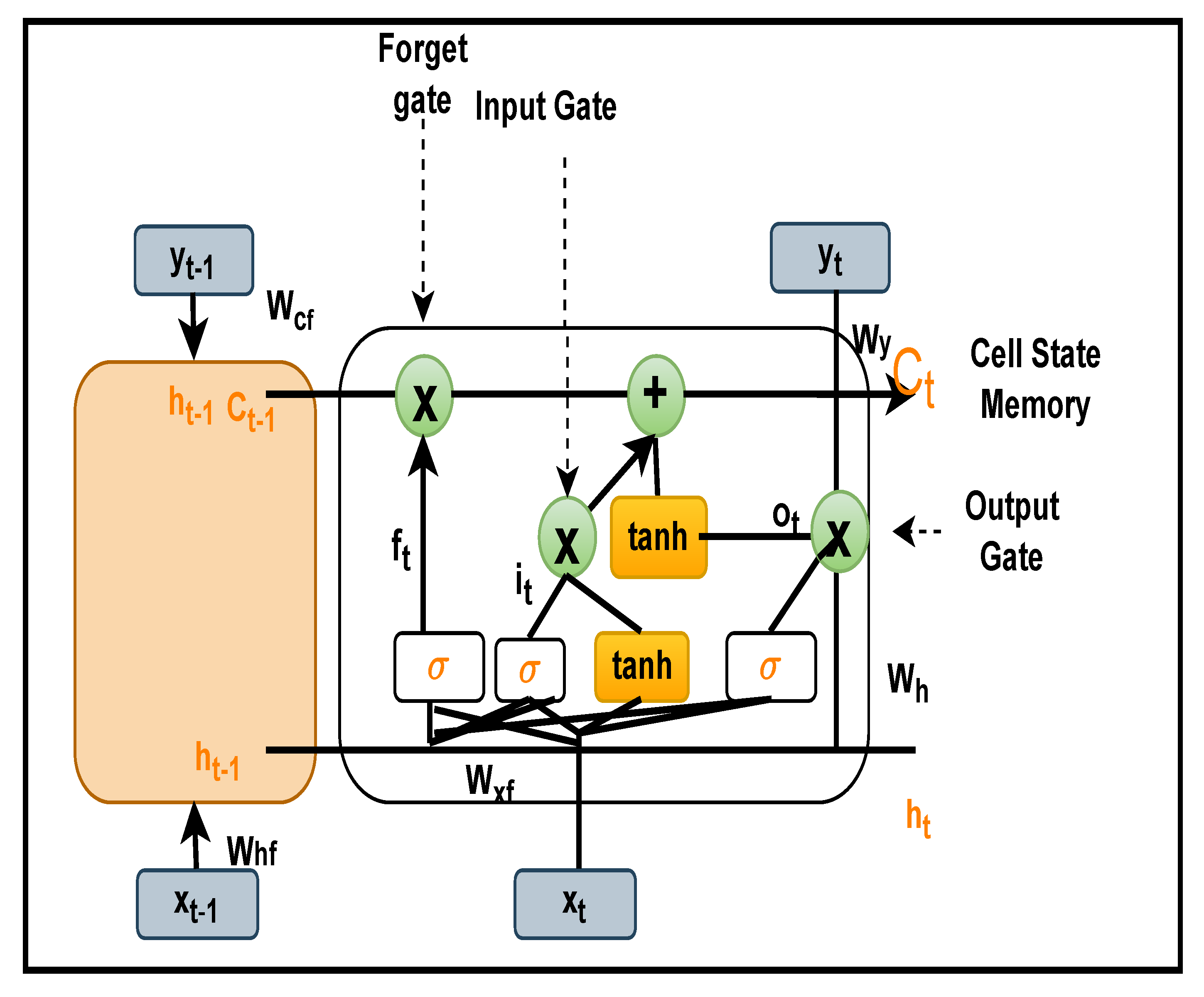
In this study, an optimised ensemble model was developed using a combination of 2DCNN, which is used for automatic feature extractions, and LSTM, which has additional cell state memory and uses its previous information to predict new data. The goal was to improve the performance of arrhythmia classification while also avoiding overfitting. An optimization approach known as “grid search” was employed to tweak the model’s hyperparameters in order to optimise it. When compared to the random search method, it provides the best hyperparameters, despite the fact that it is computationally expensive. Additional K-fold cross validations were performed on international standard ECG databases such as the MIT-BIH cardiac arrhythmia (ARR), BIDMC congestive heart failure (CHF), and MIT-BIH normal sinus rhythm (NSR) in order to train the model. These databases contain detailed expert notation that is frequently used in contemporary ECG research. As part of our research, we employed the continuous wavelet transform (CWT), which is responsible for turning 1D ECG data into 2D scalogram images of size 227 × 227 × 3, which reflect signals in the time and frequency domain, and which we used for both training and validating the model. A confusion matrix, and other performance metrics were used to assess the classifier’s performance after it was enhanced using layers such as batch normalisation, a flattening layer, and a fully connected layer.
Further, we evaluated two distinct experiments with k-fold cross validation in the presence and absence of dropout regularisation. In Scheme A, dropout regularisation was not applied during the training process. Here, all weights were utilised in the learning process. However, in Scheme B, we applied dropout regularisation with 0.5 dropouts; as a result, 50% of the information was discarded, and only 50% of data is retained for learning. Results of both experimental schemes are visualized through
Figure 13.
In the absence of dropout regularization, the results obtained from scheme A indicate high classification due to the overfitting of weights during training. The average accuracy is 99.8%, the average sensitivity is 99.77%, and the average specificity is 99.78%. However, in our suggested model where we have applied 0.5 of dropout regularization, only 50% of information is retained for learning. Hence, our suggested model has an average validation accuracy of 99%, average sensitivity as 99.33%, and average specificity as 98.35%, respectively, (Referenced from
Table 7). However, validation accuracy for the classification of ARR is 98.7%, validation accuracy for CHF is 99%, and validation accuracy for NSR is 99% while sensitivity and specificity for ARR is 0.98%, 0.98% while sensitivity and specificity for CHF is 0.96% and 0.99%, and for NSR sensitivity and specificity is 0.97% and 0.99% respectively, (Referenced form
Table 6).
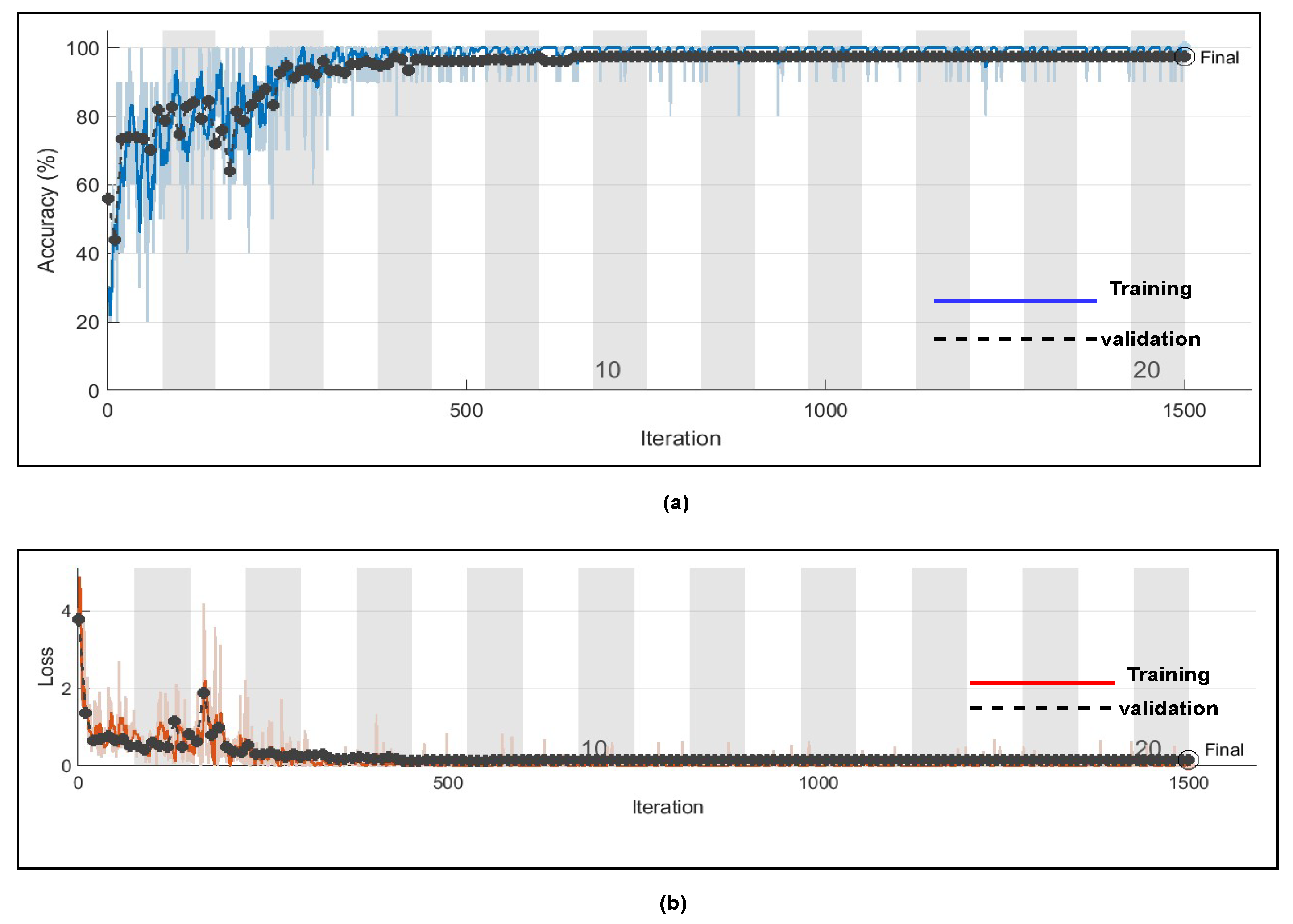
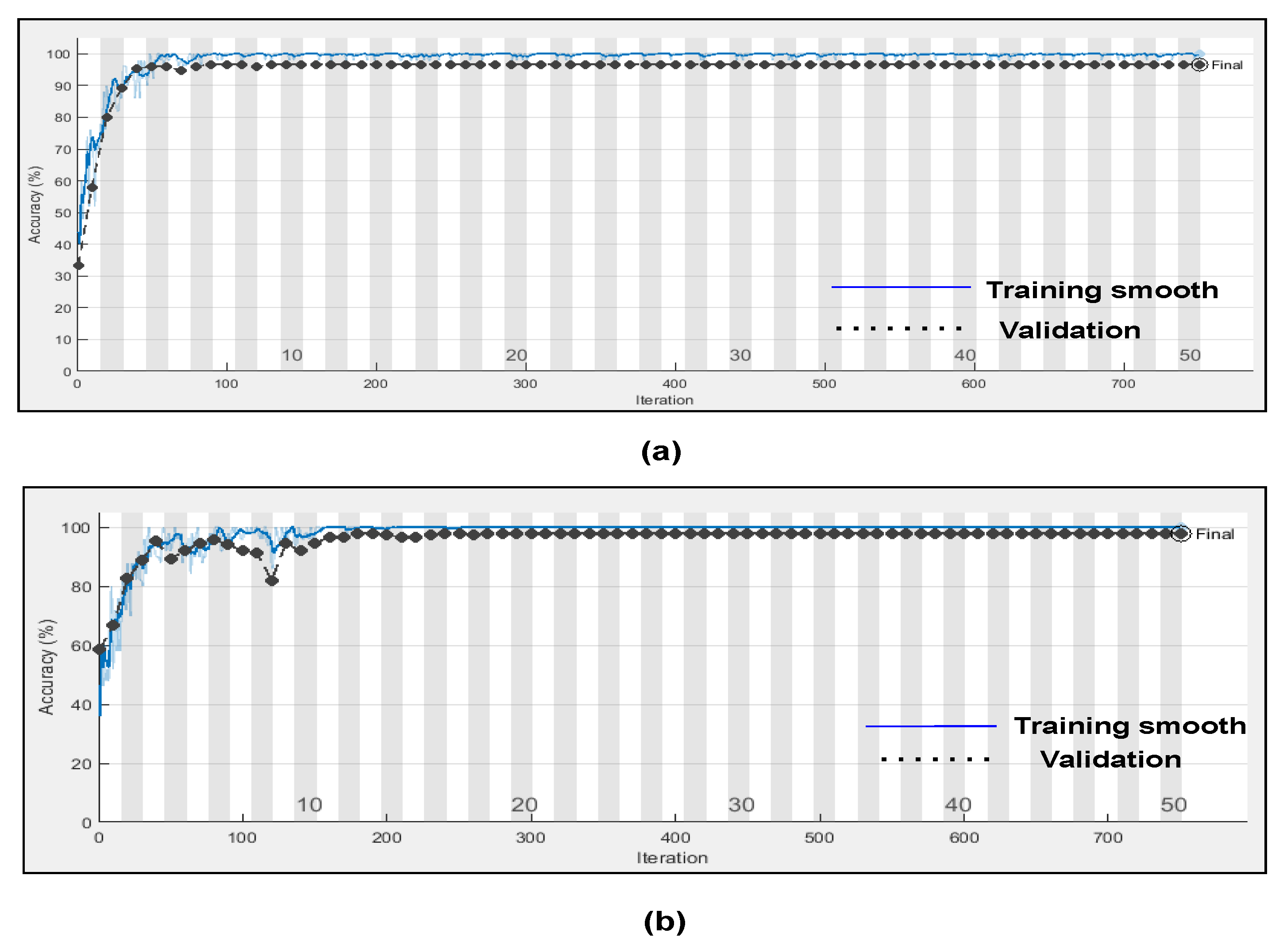
Figure 14 represents the training progress and validation accuracy, as well as the training progress and validation loss. As seen in the graphs, training and validation errors were established at a value close to zero after 100 epochs, while training and validation accuracy stabilised at 98.76%, as seen in the graphs. These outcomes are very promising and had a high degree of precision (Referenced from
Table 6).
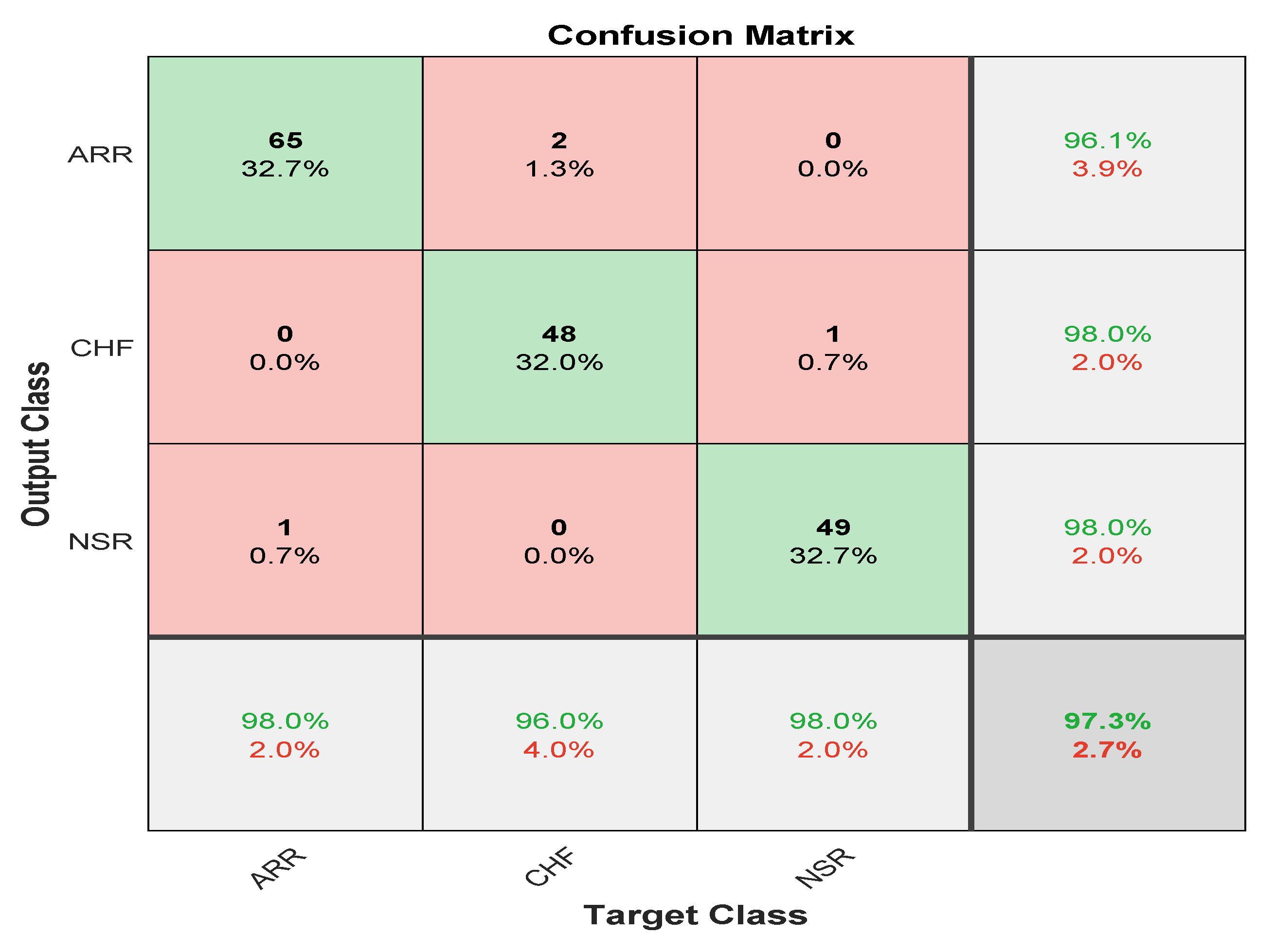
The confusion matrix is derived from the training of a proposed model for classifying three types of arrhythmias. As evidenced by the confusion matrix, the model performs better for the categorization of CHF and NSR than ARR, which is average. This may be due to the slight morphological differences in the waveforms generated during the learning process. The testing dataset’s generated confusion matrix shows 99% accuracy for the normal sinus rhythm, 98.7% for cardiac arrhythmia, and 99% for congestive heart failure, as shown in
Figure 12.
To validate our proposed methodology, we compared our results with the current and standard methods in terms of feature extraction methods, methodology, accuracy, and other statistical classifications as presented in
Table 8. It is worth mentioning that the difference between the proposed work and the state-of-the-art described is rather promising in terms of accuracy and computing cost compared to other models, as seen in
Figure 15. Knowing the potential and prospects of the proposed methodology, it would be interesting to use our proposed approach for diagnosing different critical diseases, such as gastrointestinal diseases, and distinguishing between neoplastic and non-neoplastic tissues.
6. Further Details
In this section, we present the different functions used in the training of various deep learning models.
ReLU: Activation is an incredibly critical function of a neural network. These activation functions determine if the information neuron receiving is sufficient for the information or can be overlooked. We use ReLU as an activation mechanism in this study.
Batch Normalization: is the most critical layer used to normalize the performance of the previous layer and is often used as a regularization to prevent overfitting the model.
Softmax Function: is used to quantify the probability distribution of occurrences over n distinct events in multi-class classification. This function measures the probability of each target class of the whole target class, and this likelihood range from 0–1. Later, the probabilities help decide the target class with a greater likelihood for the target class.
Sigmoid Activation Function: is a logistic function its value ranges from 0–1 it is mainly used for Binary Classification.
Tanh Activation Function: It is similar to sigmoid activation function and its values ranges from .
Dropout: is used for regularization, which is used for controlling overfitting problems.
Learning Rate: its value ranges from 0–1 it defines how fast the model adapts the problem and how many weights are modified in the loss gradient model.
Hidden units: are the no of perceptrons used in neural network its values entirely lies on activation function.
Bias: modifies the activation function by applying a constant (i.e., a given bias) to the input. It is analogous to the position of constant in a linear function.