Modification of Fatty Acid Composition of Escherichia coli by Co-Expression of Fatty Acid Desaturase and Thioesterase from Arabidopsis thaliana
Abstract
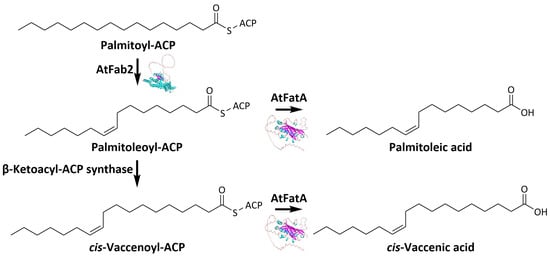
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Culture Media and Conditions
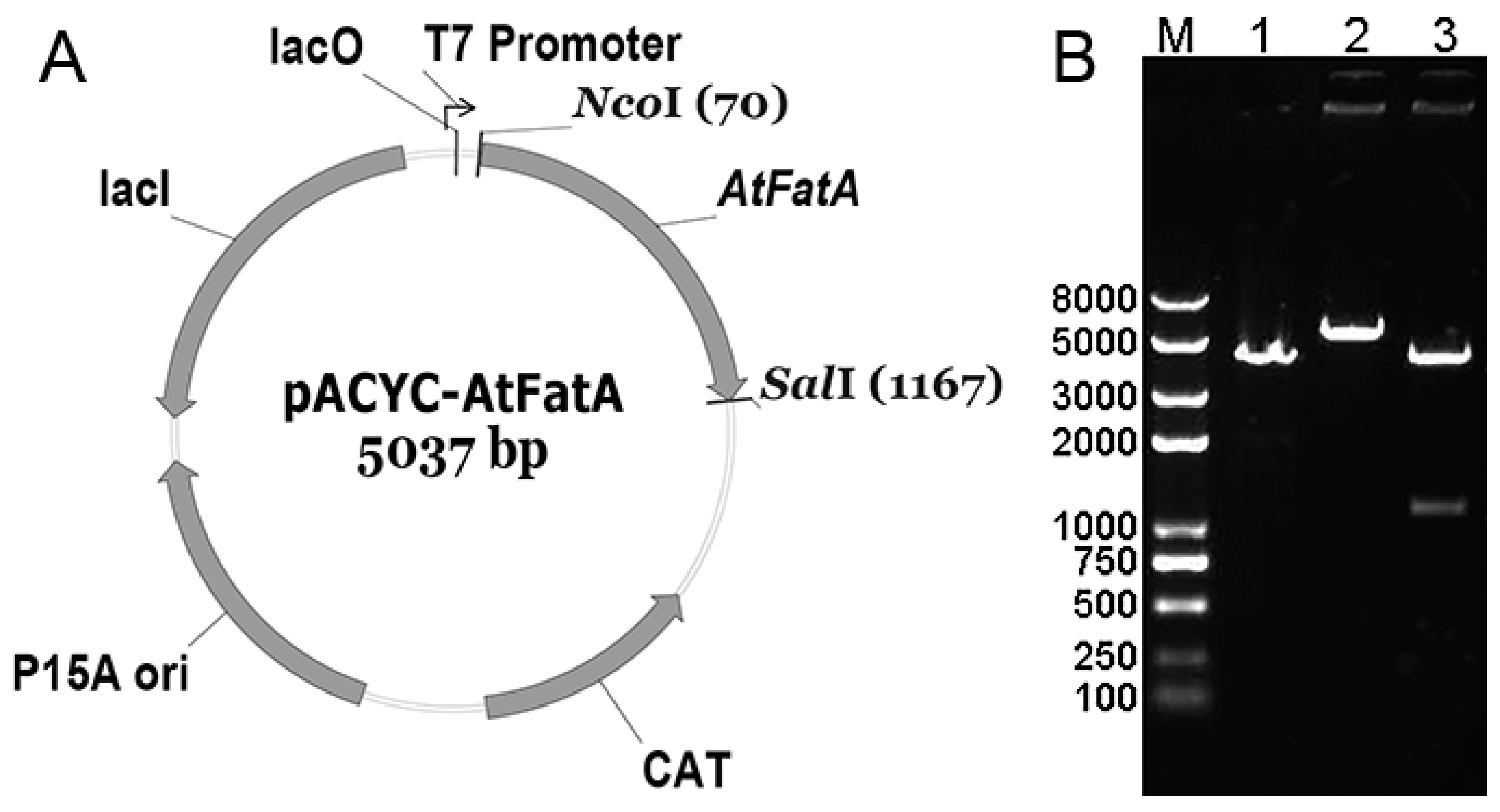
2.2. Plasmids Construction
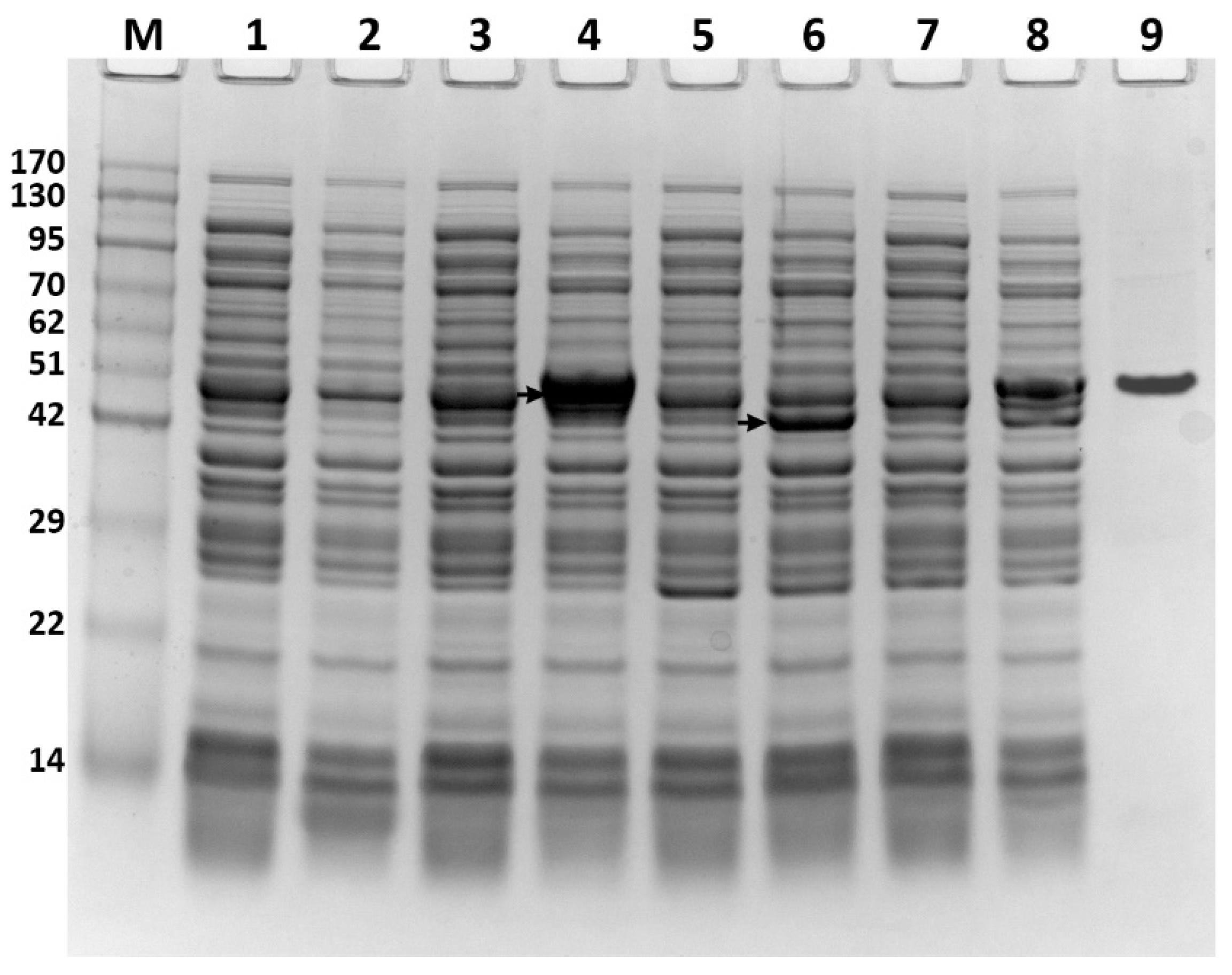
2.3. Protein Expression and Gel Electrophoresis Analysis
2.4. Protein Purification
2.5. Molecular Modeling of the Enzymes
2.6. Lipid Extraction and Thin Layer Chromatography Analysis
2.7. FAMEs Preparation and Gas Chromatography Analysis
3. Results
3.1. Expression of ATFab2 and AtFatA Proteins in E. coli
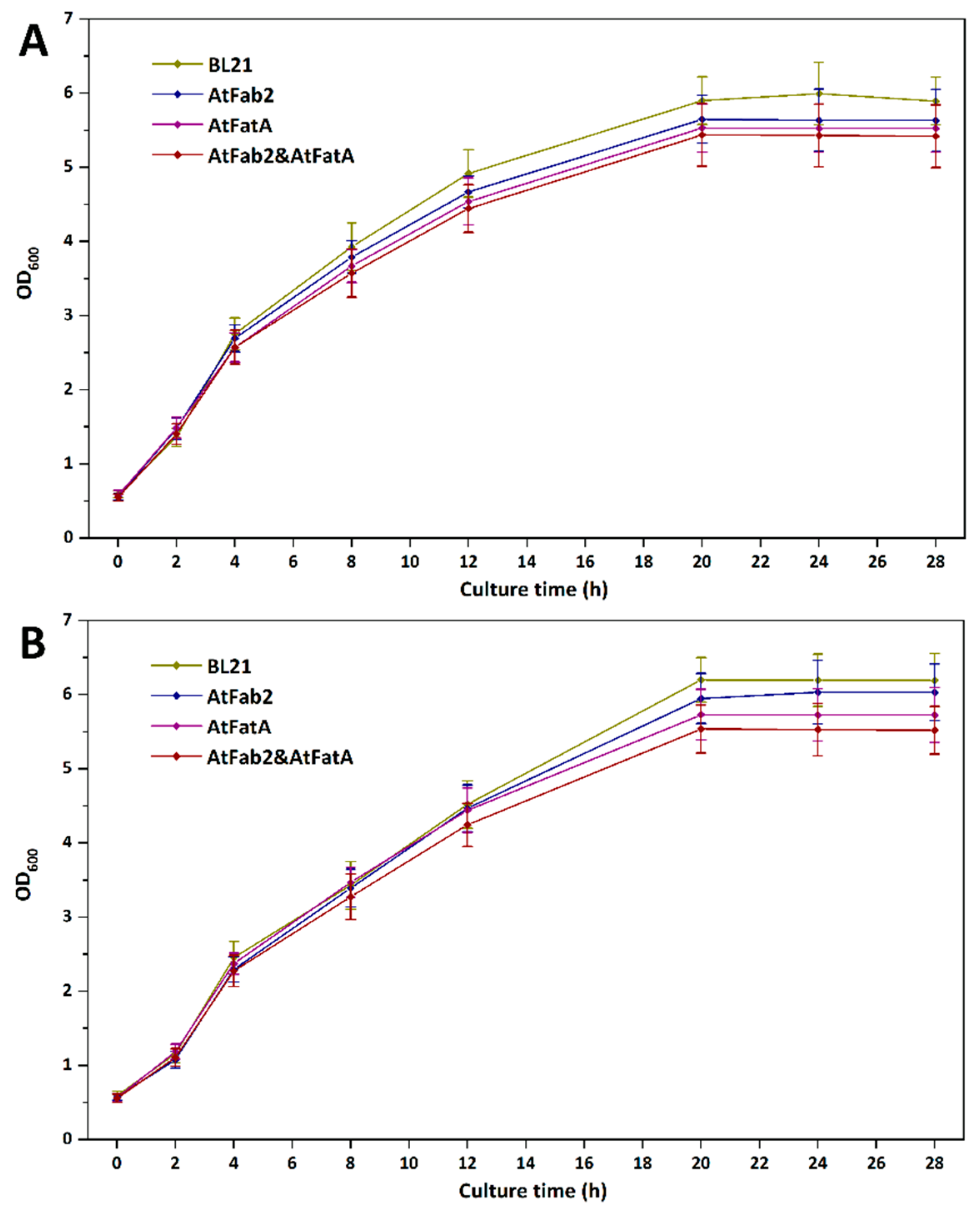
3.2. Effects of ATFab2 and AtFatA on Cell Growth
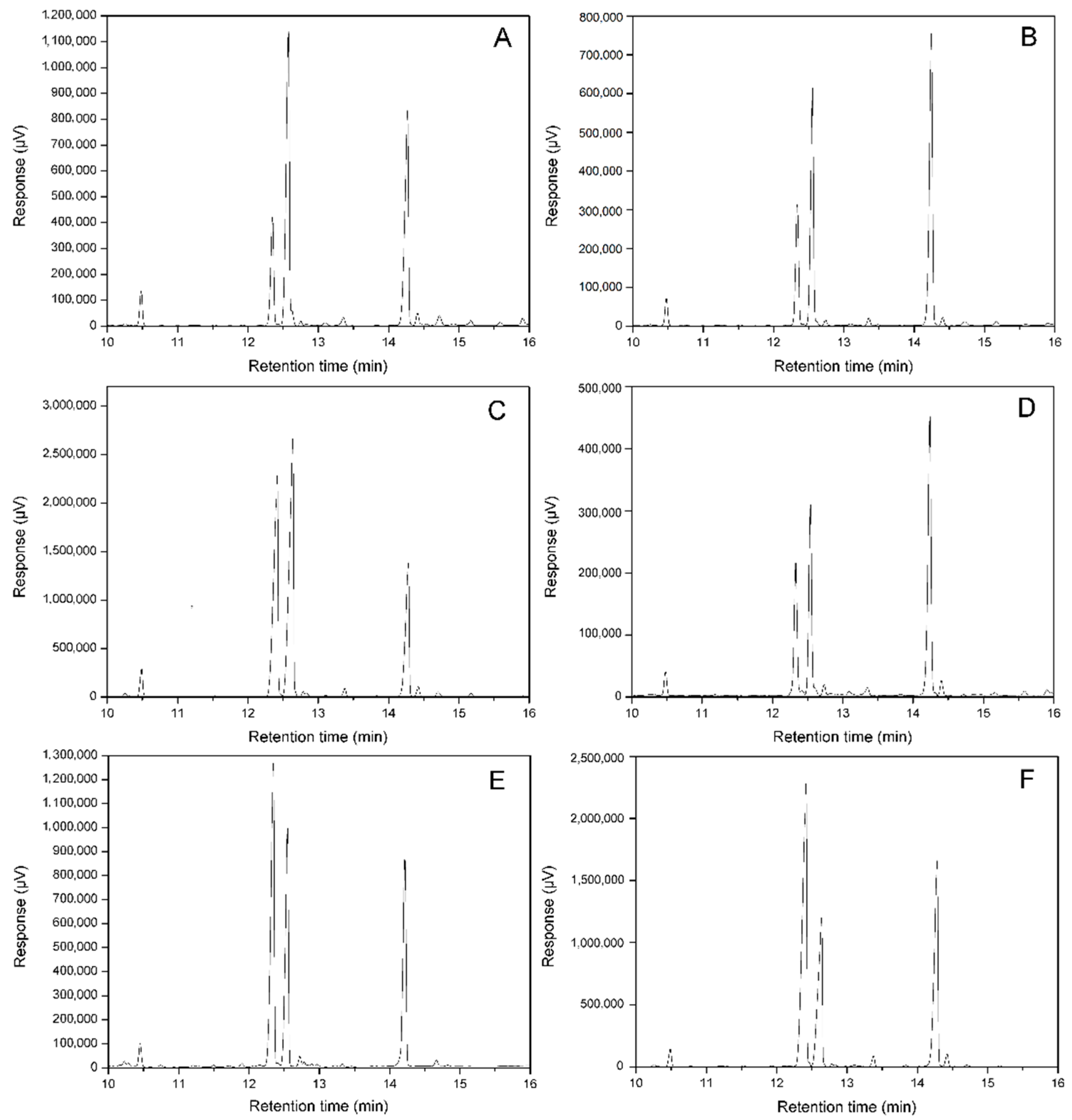
3.3. Changes of Fatty Acids Composition by Expressing ATFab2 and AtFatA
3.4. Changes of Free Fatty Acids Composition by Expressing ATFab2 and AtFatA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, J.I.; Kim, S.S.; Kang, D.H. Susceptibility of Escherichia coli O157:H7 grown at low temperatures to the krypton-chlorine excilamp. Sci. Rep. 2019, 9, 563. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, K.; Jackowski, S.; Rock, C.O.; Cronan, J.E. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 1993, 57, 522–542. [Google Scholar] [CrossRef] [PubMed]
- Marrakchi, H.; Zhang, Y.M.; Rock, C.O. Mechanistic diversity and regulation of Type II fatty acid synthesis. Biochem. Soc. Trans. 2002, 30, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Halim, N.F.A.A.; Ali, M.S.M.; Leow, A.T.C.; Rahman, R.N.Z.R.A. Membrane fatty acid desaturase: Biosynthesis, mechanism, and architecture. Appl. Microbiol. Biotechnol. 2022, 106, 5957–5972. [Google Scholar] [CrossRef]
- Los, D.A.; Murata, N. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta-Lipids Lipid Metab. 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Whittle, E.; Cahoon, E.B.; Subrahmanyam, S.; Shanklin, J. A multifunctional acyl-acyl carrier protein desaturase from Hedera helix L. (English ivy) can synthesize 16-and 18-carbon monoene and diene products. J. Biol. Chem. 2005, 280, 28169–28176. [Google Scholar] [CrossRef]
- Gummeson, P.O.; Lenman, M.; Lee, M.; Singh, S.; Stymne, S. Characterisation of acyl-ACP desaturases from Macadamia integrifolia Maiden & Betche and Nerium oleander L. Plant Sci. 2000, 154, 53–60. [Google Scholar] [CrossRef]
- Cahoon, E.B.; Coughlan, S.J.; Shanklin, J. Characterization of a structurally and functionally diverged acyl-acyl carrier protein desaturase from milkweed seed. Plant Mol. Biol. 1997, 33, 1105–1110. [Google Scholar] [CrossRef]
- Cao, Y.; Xian, M.; Yang, J.; Xu, X.; Liu, W.; Li, L. Heterologous expression of stearoyl-acyl carrier protein desaturase (S-ACP-DES) from Arabidopsis thaliana in Escherichia coli. Protein Expr. Purif. 2010, 69, 209–214. [Google Scholar] [CrossRef]
- Jones, A.; Davies, H.M.; Voelker, T.A. Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell 1995, 7, 359–371. [Google Scholar] [CrossRef]
- Cho, H.S.; Cronan, J.E. Escherichia coli thioesterase, I, molecular cloning and sequencing of the structural gene and identification as a periplasmic enzyme. J. Biol. Chem. 1993, 268, 9238–9245. [Google Scholar] [CrossRef] [PubMed]
- Spencer, A.K.; Greenspan, A.D.; Cronan, J.E. Thioesterases I and II of Escherichia coli. J. Biol. Chem. 1978, 253, 5922–5926. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Wang, Y.; Liu, J.; Liu, Y.; Cao, X.; Xue, S. Tuning of acyl-ACP thioesterase activity directed for tailored fatty acid synthesis. Appl. Microbiol. Biotechnol. 2018, 102, 3173–3182. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Pérez, A.J.; Sánchez-García, A.; Salas, J.J.; Garcés, R.; Martínez-Force, E. Acyl-ACP thioesterases from macadamia (Macadamia tetraphylla) nuts: Cloning, characterization and their impact on oil composition. Plant Physiol. Biochem. 2011, 49, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.T.; Pirtle, R.M.; Chapman, K.D. Expression of a Gossypium hirsutum cDNA encoding a FatB palmitoyl-acyl carrier protein thioesterase in Escherichia coli. Plant Physiol. Biochem. 2002, 40, 1–9. [Google Scholar] [CrossRef]
- Jha, J.K.; Maiti, M.K.; Bhattacharjee, A.; Basu, A.; Sen, P.C.; Sen, S.K. Cloning and functional expression of an acyl-ACP thioesterase FatB type from Diploknema (Madhuca) butyracea seeds in Escherichia coli. Plant Physiol. Biochem. 2006, 44, 645–655. [Google Scholar] [CrossRef]
- Voelker, T.A.; Davies, H.M. Alteration of the specificity and regulation of fatty-acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J. Bacteriol. 1994, 176, 7320–7327. [Google Scholar] [CrossRef]
- Bonaventure, G.; Bao, X.; Ohlrogge, J.; Pollard, M. Metabolic responses to the reduction in palmitate caused by disruption of the FATB gene in Arabidopsis. Plant Physiol. 2004, 135, 1269–1279. [Google Scholar] [CrossRef][Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Valeur, A.; Tunlid, A.; Odham, G. Differences in lipid composition between free-living and initially adhered cells of a Gram-negative bacterium. Arch. Microbiol. 1988, 149, 521–526. [Google Scholar] [CrossRef]
- Prabhune, A.; Fox, S.R.; Ratledge, C. Transformation of arachidonic acid to 19-hydroxy- and 20-hydroxy-eicosatetraenoic acids using Candida bombicola. Biotechnol. Lett. 2002, 24, 1041–1044. [Google Scholar] [CrossRef]
- Lounds, C.; Eagles, J.; Carter, A.T.; MacKenzie, D.A.; Archer, D.B. Spore germination in Mortierella alpina is associated with a transient depletion of arachidonic acid and induction of fatty acid desaturase gene expression. Arch. Microbiol. 2007, 188, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, Y.; Huang, W.; Schneider, G.; Shanklin, J. Crystal structure of delta9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J. 1996, 15, 4081–4092. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, Y.; Liu, J.; Liu, Y.; Cao, X.; Xue, S. Structural insight into acyl-ACP thioesterase toward substrate specificity design. ACS Chem. Biol. 2017, 12, 2830–2836. [Google Scholar] [CrossRef] [PubMed]
- Nagao, K.; Murakami, A.; Umeda, M. Structure and function of Δ9-fatty acid desaturase. Chem. Pharm. Bull. 2019, 67, 327–332. [Google Scholar] [CrossRef]
- Srikanta Dani, K.G.; Hatti, K.S.; Ravikumar, P.; Kush, A. Structural and functional analyses of a saturated acyl ACP thioesterase, type B from immature seed tissue of Jatropha curcas. Plant Biol. 2011, 13, 453–461. [Google Scholar] [CrossRef]
- Weber, M.H.W.; Klein, W.; Müller, L.; Niess Ulf, M.; Marahiel, M.A. Role of the Bacillus subtilis fatty acid desaturase in membrane adaptation during cold shock. Mol. Microbiol. 2001, 39, 1321–1329. [Google Scholar] [CrossRef]
- Cao, Y.; Yang, J.; Xian, M.; Xu, X.; Liu, W. Increasing unsaturated fatty acid contents in Escherichia coli by coexpression of three different genes. Appl. Microbiol. Biotechnol. 2010, 87, 271–280. [Google Scholar] [CrossRef]
- Aguilar, P.S.; de Mendoza, D. Control of fatty acid desaturation: A mechanism conserved from bacteria to humans. Mol. Microbiol. 2006, 62, 1507–1514. [Google Scholar] [CrossRef]
- Annous, B.A.; Kozempel, M.F.; Kurantz, M.J. Changes in membrane fatty acid composition of Pediococcus sp. strain NRRL B-2354 in response to growth conditions and its effect on thermal resistance. Appl. Environ. Microbiol. 1999, 65, 2857–2862. [Google Scholar] [CrossRef] [PubMed]
- Hazel, J.R.; Eugene Williams, E. The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog. Lipid Res. 1990, 29, 167–227. [Google Scholar] [CrossRef] [PubMed]
- Leekumjorn, S.; Cho, H.J.; Wu, Y.; Wright, N.T.; Sum, A.K.; Chan, C. The role of fatty acid unsaturation in minimizing biophysical changes on the structure and local effects of bilayer membranes. Biochim. Biophys. Acta-Biomembr. 2009, 1788, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Ohlrogge, J.; Browse, J. Lipid biosynthesis. Plant Cell 1995, 7, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, P.; Shanklin, J.; Shah, J.; Whittle, E.J.; Klessig, D.F. A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 2001, 98, 9448–9453. [Google Scholar] [CrossRef]
- Salas, J.J.; Ohlrogge, J.B. Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch. Biochem. Biophys. 2002, 403, 25–34. [Google Scholar] [CrossRef]
- De Carvalho, C.C.C.R.; Caramujo, M.J. The various roles of fatty acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef]

| Strains or Plasmids | Genotype/Description | Sources |
|---|---|---|
| Strains | ||
| E. coli DH5α | huA2 lac(del)U169 phoA glnV44 Φ80’ lacZ(del)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | TransGen |
| E. coli BL21(DE3) | F− ompT hsdSB (rB− mB−) gal dcm rne131 (DE3) | TransGen |
| Plasmids | ||
| pACYCDuet-1 | Cmr oriP15A lacIq T7p | Novagen |
| pET30a(+) | Kanr oripBR322 lacIq T7p | Novagen |
| pUCm-T | Ampr oripUC lacZ | Sangon |
| pET-AtFab2 | pET30a(+) harboring A. thaliana fatty acid desaturase | [7] |
| pUCm-T-AtFatA | pUCm-T harboring A. thaliana thioesterase | This study |
| pACYC-AtFatA | pACYCDuet-1 harboring A. thaliana thioesterase | This study |
| Strains | Myristic Acid | Palmitoleic Acid | Palmitic Acid | cis-Vaccenic Acid |
|---|---|---|---|---|
| Fatty Acids (%) | ||||
| BL21 | 4.1 ± 0.3 | 20.2 ± 0.9 | 44.7 ± 1.0 | 31.0 ± 1.0 |
| BL21/AtFab2 | 3.1 ± 0.4 | 19.6 ± 0.9 | 34.4 ± 0.8 | 42.9 ± 0.8 |
| BL21/AtFatA | 2.9 ± 0.3 | 35.6 ± 0.6 | 38.7 ± 0.7 | 22.8 ± 1.4 |
| BL21/AtFab2&AtFatA | 3.0 ± 0.2 | 21.0 ± 1.0 | 29.6 ± 1.0 | 46.4 ± 1.7 |
| Free fatty acids (%) | ||||
| BL21/AtFatA | 1.4 ± 0.2 | 46.3 ± 0.9 | 31.4 ± 0.6 | 20.9 ± 0.7 |
| BL21/AtFab2&AtFatA | 0.9 ± 0.1 | 47.1 ± 0.7 | 22.9 ± 0.5 | 29.1 ± 0.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pu, Y.; Cao, Y.; Xian, M. Modification of Fatty Acid Composition of Escherichia coli by Co-Expression of Fatty Acid Desaturase and Thioesterase from Arabidopsis thaliana. Bioengineering 2022, 9, 771. https://doi.org/10.3390/bioengineering9120771
Pu Y, Cao Y, Xian M. Modification of Fatty Acid Composition of Escherichia coli by Co-Expression of Fatty Acid Desaturase and Thioesterase from Arabidopsis thaliana. Bioengineering. 2022; 9(12):771. https://doi.org/10.3390/bioengineering9120771
Chicago/Turabian StylePu, Yihan, Yujin Cao, and Mo Xian. 2022. "Modification of Fatty Acid Composition of Escherichia coli by Co-Expression of Fatty Acid Desaturase and Thioesterase from Arabidopsis thaliana" Bioengineering 9, no. 12: 771. https://doi.org/10.3390/bioengineering9120771
APA StylePu, Y., Cao, Y., & Xian, M. (2022). Modification of Fatty Acid Composition of Escherichia coli by Co-Expression of Fatty Acid Desaturase and Thioesterase from Arabidopsis thaliana. Bioengineering, 9(12), 771. https://doi.org/10.3390/bioengineering9120771