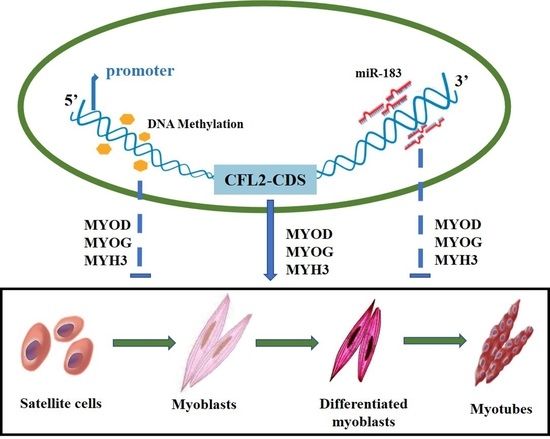
New Insight into Muscle-Type Cofilin (CFL2) as an Essential Mediator in Promoting Myogenic Differentiation in Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Collection and Cell Culturing
2.2. Overexpression or Knock-Down Vector Construction of CFL2
2.3. Cell Treatment and Cell Differentiation
2.4. Dual Luciferase Reporter Assay
2.5. Quantitative Real-Time PCR and Western Blot Assay
2.6. Bisulfite Sequencing Polymerase Chain Reaction and Combined Bisulfite Restriction Analysis
2.7. Statistical Data Analysis
3. Results
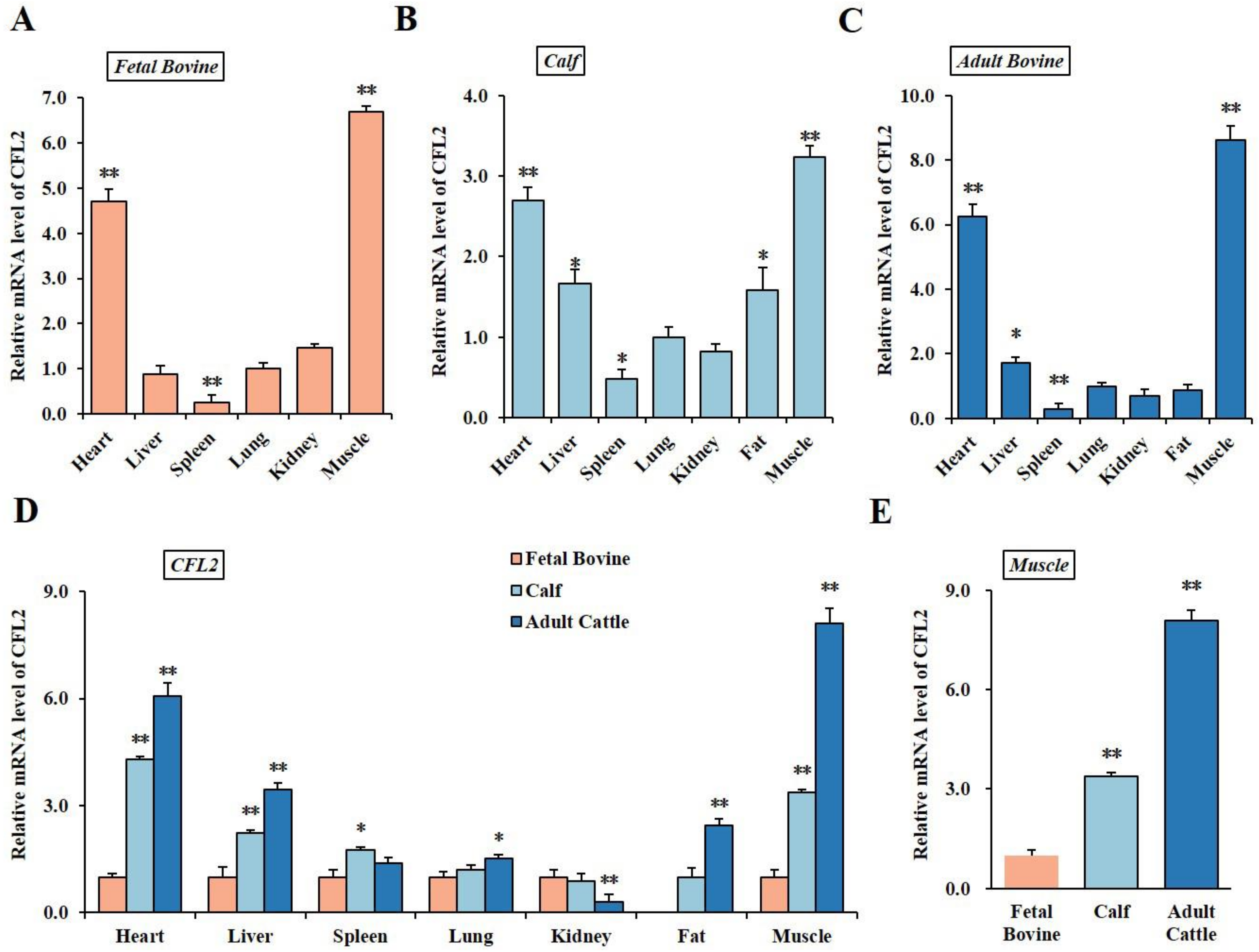
3.1. Spatiotemporal Expression Profiles of CFL2 Gene in Different Tissues
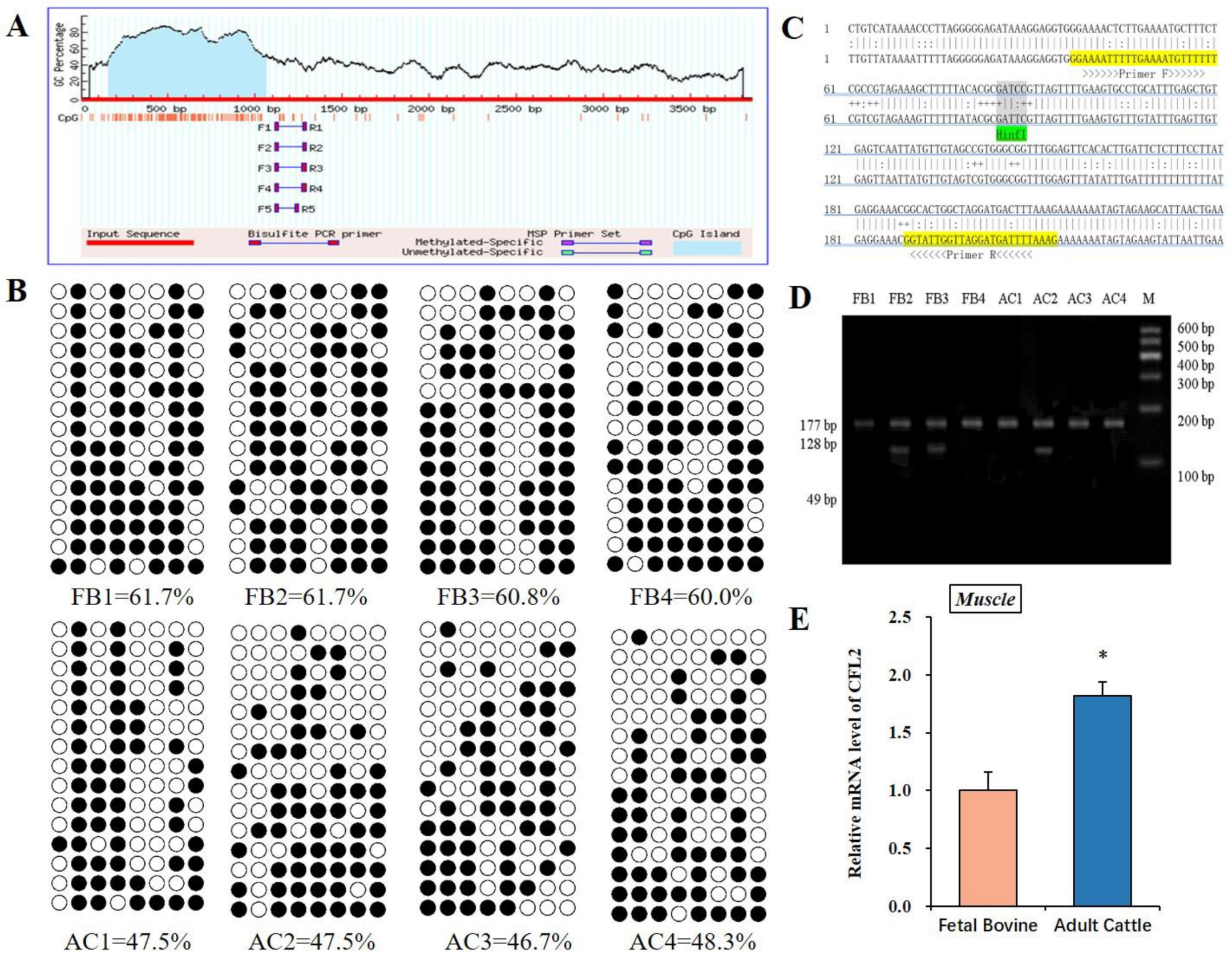
3.2. DNA Methylation Analysis of CFL2 Promoter Region in Muscle Tissues
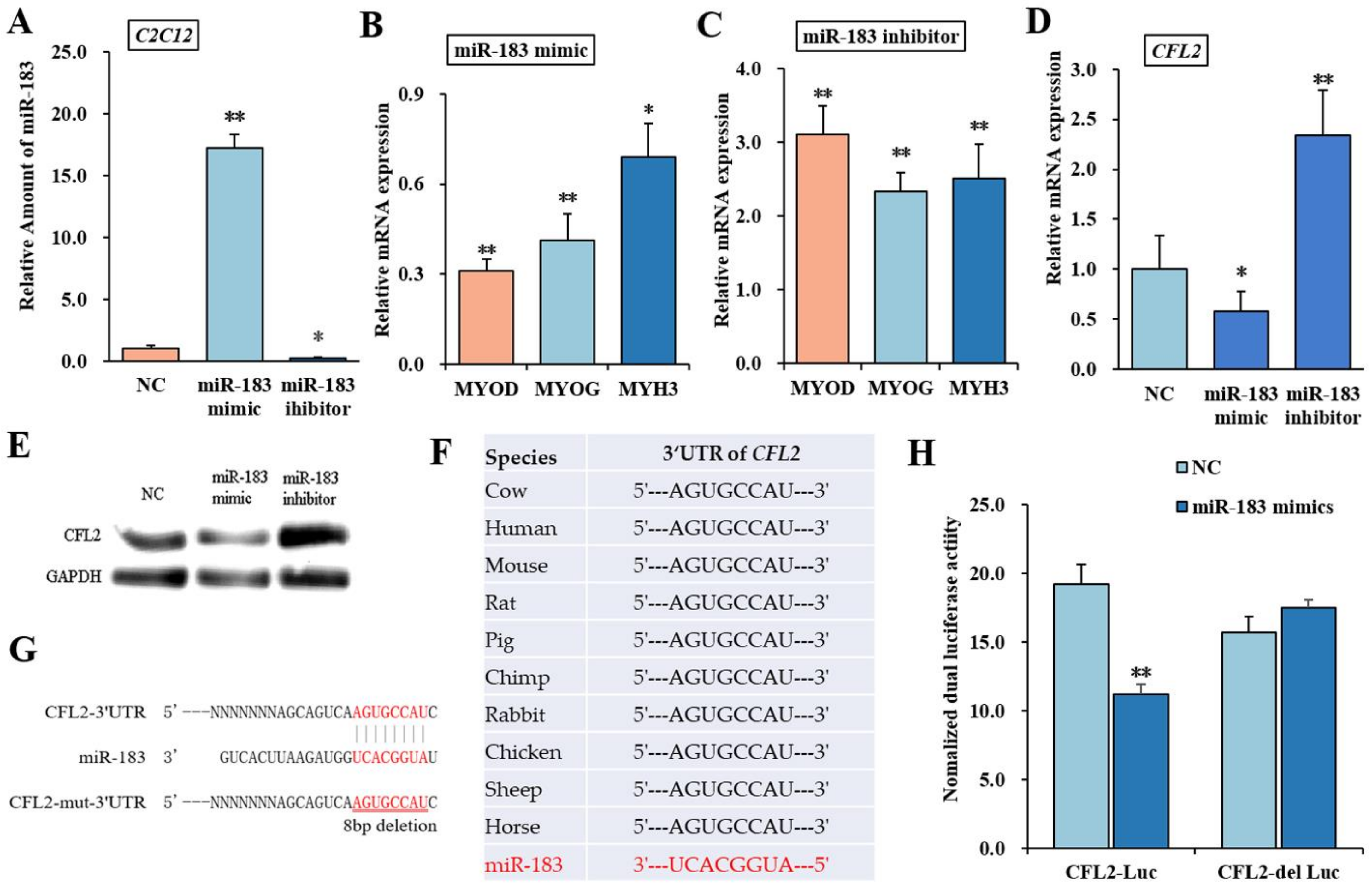
3.3. bta-miR-183 Target Muscle Type CFL2
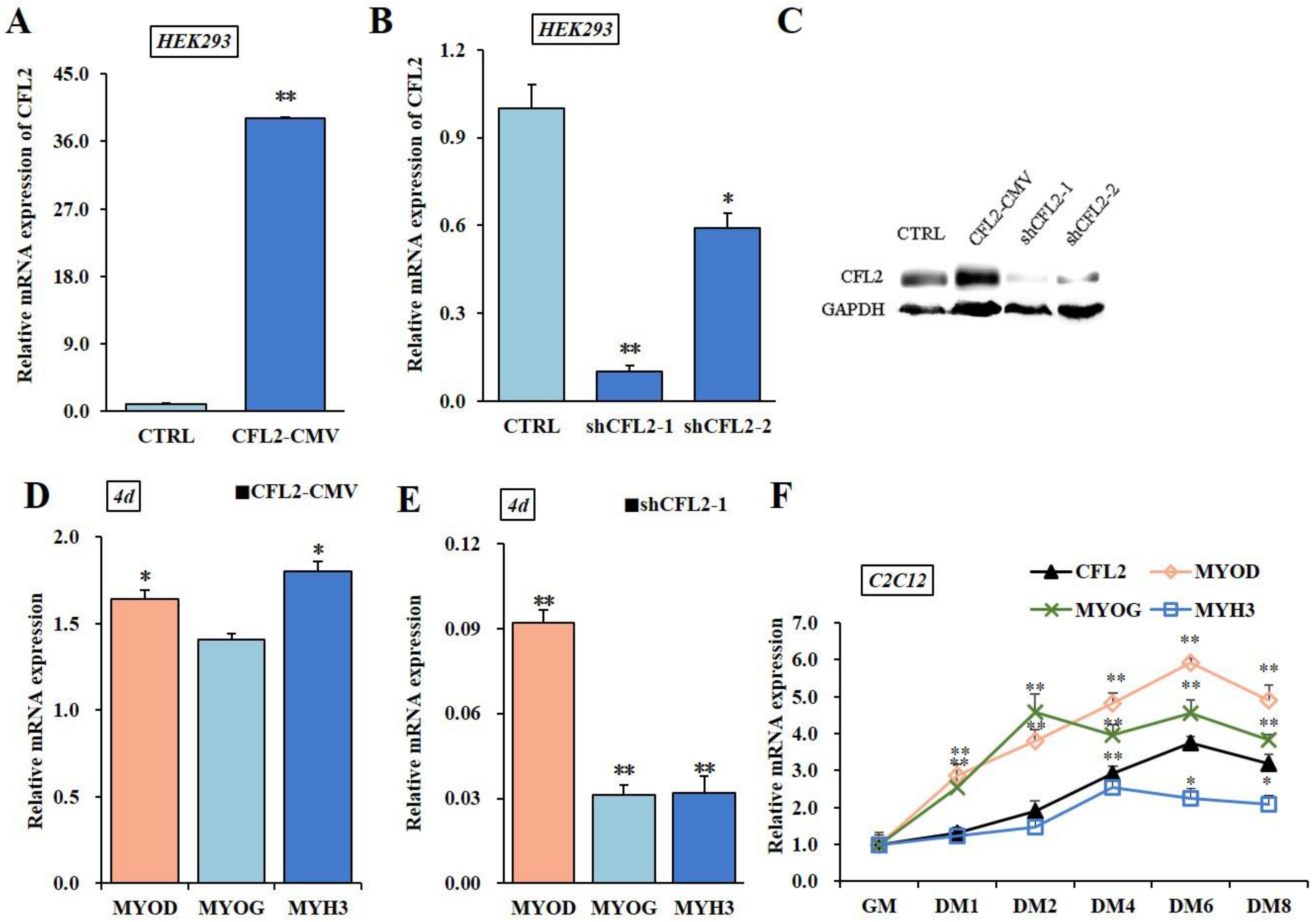
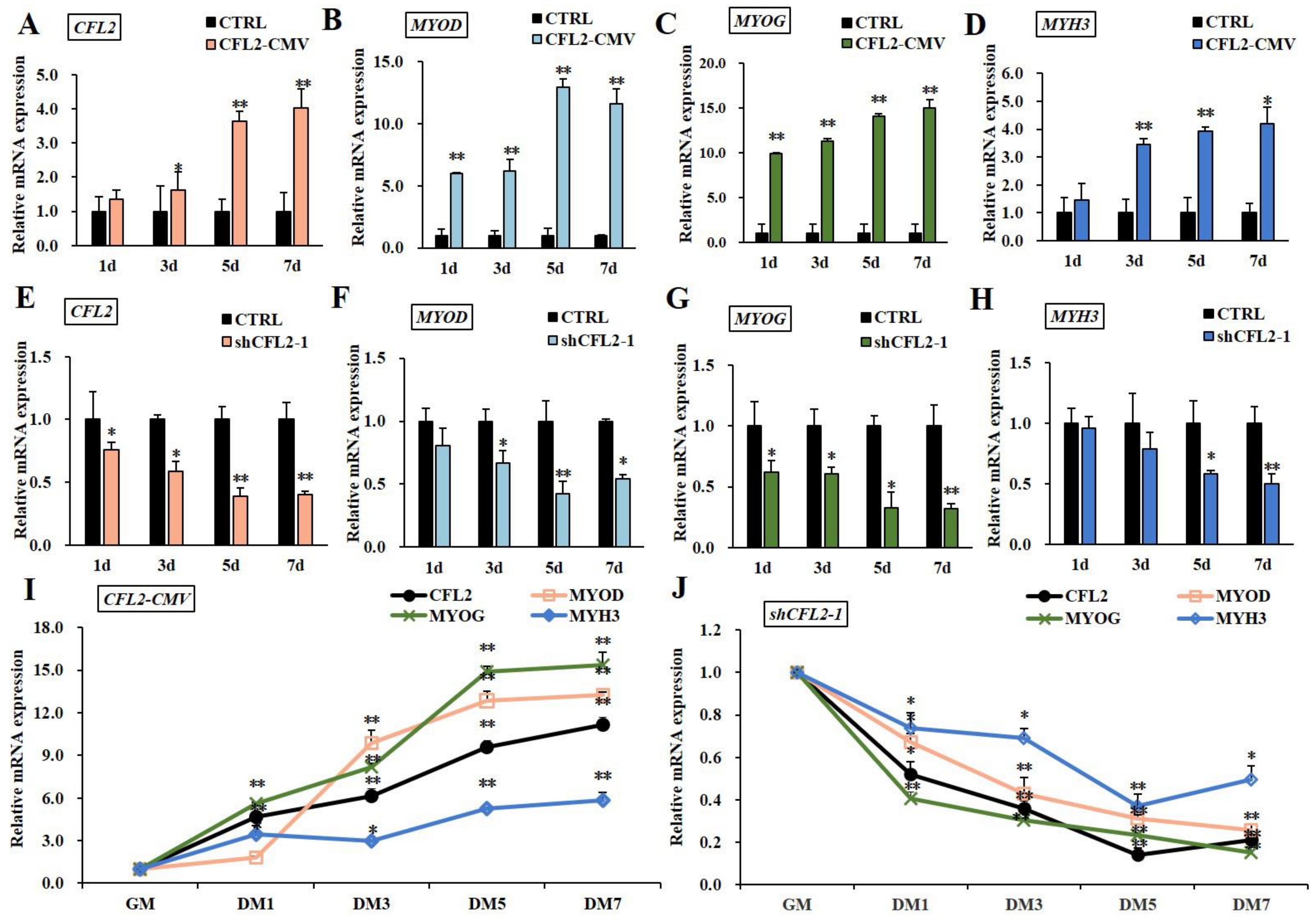
3.4. Effects of CFL2 Gene on Myoblasts Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, C.B.; Xu, X.L.; Zhou, G.H.; Xu, S.Q.; Zhang, J.B. Effects of carcass maturity on meat quality characteristics of beef semitendinosus muscle for chinese native yellow steers. Animal 2007, 1, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Covington, R.C.; Tuma, H.J.; Grant, D.L.; Dayton, A.D. Various chemical and histological characteristics of beef muscle as related to tenderness. J. Anim. Sci. 1970, 30, 191–196. [Google Scholar] [CrossRef]
- Pas, M.T.; Keuning, E.; Hulsegge, B.; Hoving-Bolink, A.H.; Evans, G.; Mulder, H.A. Longissimus muscle transcriptome profiles related to carcass and meat quality traits in fresh meat Pietrain carcasses. J. Anim. Sci. 2010, 88, 4044–4055. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.; Lee, W. Role of MiR-325-3p in the Regulation of CFL2 and Myogenic Differentiation of C2C12 Myoblasts. Cells 2021, 10, 2725. [Google Scholar] [CrossRef] [PubMed]
- Ono, S.; Minami, N.; Abe, H.; Obinata, T. Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J. Biol. Chem. 1994, 269, 15280–15286. [Google Scholar] [CrossRef]
- Sun, Y.; Ma, Y.; Zhao, T.; Li, M.; Mao, Y.; Yang, Z. Epigenetic Regulation Mechanisms of the Cofilin-1 Gene in the Development and Differentiation of Bovine Primary Myoblasts. Genes 2022, 13, 723. [Google Scholar] [CrossRef]
- Kanellos, G.; Frame, M.C. Cellular functions of the ADF/cofilin family at a glance. J. Cell Sci. 2016, 129, 3211. [Google Scholar] [CrossRef]
- Mohri, K.; Takano-Ohmuro, H.; Nakashima, H.; Hayakawa, K.; Endo, T.; Hanaoka, K.; Obinata, T. Expression of cofilin isoforms during development of mouse striated muscles. J. Muscle Res. Cell Motil. 2000, 21, 49–57. [Google Scholar] [CrossRef]
- Ikeda, S.; Cunningham, L.A.; Boggess, D.; Hawes, N.; Hobson, C.D.; Sundberg, J.P.; Naggert, J.K.; Smith, R.S.; Nishina, P.M. Aberrant actin cytoskeleton leads to accelerated proliferation of corneal epithelial cells in mice deficient for destrin (actin depolymerizing factor). Hum. Mol. Genet. 2003, 12, 1029–1037. [Google Scholar] [CrossRef]
- Thirion, C.; Stucka, R.; Mendel, B.; Gruhler, A.; Jaksch, M.; Nowak, K.J.; Binz, N.; Laing, N.G.; Lochmüller, H. Characterization of human muscle type cofilin (CFL2) in normal and regenerating muscle. Eur. J. Biochem. 2001, 268, 3473–3482. [Google Scholar] [CrossRef]
- Mai, T.N.; Min, K.H.; Kim, D.; Park, S.Y.; Wan, L. CFL2 is an essential mediator for myogenic differentiation in C2C12 myoblasts. Biochem. Biophys. Res. Commun. 2020, 533, 710–716. [Google Scholar]
- Agrawal, P.B.; Greenleaf, R.S.; Tomczak, K.K.; Lehtokari, V.-L.; Wallgren-Pettersson, C.; Wallefeld, W.; Laing, N.G.; Darras, B.T.; Maciver, S.K.; Dormitzer, P.R.; et al. Nemaline Myopathy with Minicores Caused by Mutation of the CFL2 Gene Encoding the Skeletal Muscle Actin–Binding Protein, Cofilin-2. Am. J. Hum. Genet. 2007, 80, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Rosen, S.M.; Mugdha, J.; Talia, H.; Beggs, A.H.; Agrawal, P.B. Knockin mouse model of the human CFL2 p.A35T mutation results in a unique splicing defect and severe myopathy phenotype. Hum. Mol. Genet. 2020, 29, 1996–2003. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Lan, X.; Lei, C.; Zhang, C.; Chen, H. Haplotype combination of the bovine CFL2 gene sequence variants and association with growth traits in Qinchuan cattle. Gene 2015, 563, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.D.; Allis, C.; Bernstein, E.J.C. Epigenetics: A Landscape Takes Shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef]
- Luo, J.; Deng, Z.L.; Luo, X.; Tang, N.; Song, W.X.; Chen, J.; Sharff, K.A.; Luu, H.H.; Haydon, R.C.; Kinzler, K.W. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2007, 2, 1236. [Google Scholar] [CrossRef]
- Li, L.C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Kumaki, Y.; Oda, M.; Okano, M. QUMA: Quantification tool for methylation analysis. Nucleic Acids Res. 2008, 36, 170–175. [Google Scholar] [CrossRef]
- Bock, C.; Reither, S.; Mikeska, T.; Paulsen, M.; Walter, J.; Lengauer, T. BiQ Analyzer: Visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 2005, 21, 4067–4068. [Google Scholar] [CrossRef]
- Raza, S.H.A.; Kaster, N.; Khan, R.; Abdelnour, S.A.; El-Hack, M.E.A.; Khafaga, A.F.; Taha, A.; Ohran, H.; Swelum, A.A.; Schreurs, N.M.; et al. The Role of MicroRNAs in Muscle Tissue Development in Beef Cattle. Genes 2020, 11, 295. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, K.; Huang, Y.; Lan, X.; Chen, H. Differential expression of FOXO1 during development and myoblast differentiation of Qinchuan cattle and its association analysis with growth traits. Sci. China Life Sci. 2018, 61, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Gillett, G.T.; Fox, M.F.; Rowe, P.; Casimir, C.M.; Povey, S. Mapping of human non-muscle type cofilin (CFL1) to chromosome 11q13 and muscle-type cofilin (CFL2) to chromosome 14. Ann. Hum. Genet. 2012, 60, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Hocquette, J.F. Endocrine and metabolic regulation of muscle growth and body composition in cattle. Anim. Int. J. Anim. Biosci. 2010, 4, 1797–1809. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, D.; Wen, Y.; Zhang, Z.; Yang, S.; Huang, Y. DNA methylation status of SERPINA3 gene involved in mRNA expression in three cattle breeds. Anim. Biotechnol. 2021, 33, 1289–1295. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Ji, G.; Carrillo, J.A.; Li, Y.; Tian, F.; Baldwin, R.L.; Zan, L.; Song, J. The Profiling of DNA Methylation and Its Regulation on Divergent Tenderness in Angus Beef Cattle. Front. Genet. 2021, 11, 939. [Google Scholar] [CrossRef]
- Huang, Y.Z.; Zhan, Z.Y.; Sun, Y.J.; Cao, X.K.; Li, M.X.; Wang, J.; Lan, X.Y.; Lei, C.Z.; Zhang, C.L.; Chen, H. Intragenic DNA methylation status down-regulates bovine IGF2 gene expression in different developmental stages. Gene 2014, 534, 356–361. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Clifford, R.L.; Singer, C.A.; John, A.E. Epigenetics and miRNA emerge as key regulators of smooth muscle cell phenotype and function. Pulm. Pharmacol. Ther. 2013, 26, 75–85. [Google Scholar] [CrossRef]
- Sannicandro, A.J.; Soriano-Arroquia, A.; Goljanek-Whysall, K. Micro(RNA)-managing muscle wasting. J. Appl. Physiol. 2019, 127, 619–632. [Google Scholar] [CrossRef]
- Hao, D.; Wang, X.; Wang, X.; Thomsen, B.; Yang, Y.; Lan, X.; Huang, Y.; Chen, H. MicroRNA bta-miR-365-3p inhibits proliferation but promotes differentiation of primary bovine myoblasts by targeting the activin A receptor type I. J. Anim. Sci. Biotechnol. 2021, 12, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Amatya, V.J.; Kushitani, K.; Kai, Y.; Kambara, T.; Takeshima, Y. miR-182 and miR-183 Promote Cell Proliferation and Invasion by Targeting FOXO1 in Mesothelioma. Front. Oncol. 2018, 8, 446. [Google Scholar] [CrossRef] [PubMed]
- Shang, A.; Wang, X.; Gu, C.; Liu, W.; Sun, J.; Zeng, B.; Chen, C.; Ji, P.; Wu, J.; Quan, W. Exosomal miR-183-5p promotes angiogenesis in colorectal cancer by regulation of FOXO1. Aging 2020, 12, 8352–8371. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Ma, D.; Wu, Y.; Zhang, Y.; Zhang, L. LncRNA SNHG16 Regulates the Progress of Acute Myeloid Leukemia Through miR183-5p–FOXO1 Axis. OncoTargets Ther. 2020, 13, 12943–12954. [Google Scholar] [CrossRef] [PubMed]
- Ji, A.; Pc, A.; As, A.; Pg, B.; Cc, A.; Yl, C.; Of, C.; Ab, B. Gestational nutrient restriction under extensive grazing conditions: Effects on muscle characteristics and meat quality in heavy lambs. Meat Sci. 2021, 179, 108532. [Google Scholar]
- Wang, J. Checking before changing: Cell cycle checkpoints inhibit muscle differentiation. Cell Cycle 2011, 10, 3234–3235. [Google Scholar] [CrossRef][Green Version]
- Adhikari, A.; Kim, W.; Davie, J. Myogenin is required for assembly of the transcription machinery on muscle genes during skeletal muscle differentiation. PLoS ONE 2021, 16, e0245618. [Google Scholar] [CrossRef]
- Jabre, S.; Hleihel, W.; Coirault, C. Nuclear Mechanotransduction in Skeletal Muscle. Cells 2021, 10, 318. [Google Scholar] [CrossRef]
- Vartiainen, M.K.; Mustonen, T.; Mattila, P.K.; Ojala, P.J.; Thesleff, I.; Partanen, J.; Lappalainen, P. The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol. Biol. Cell 2002, 13, 183–194. [Google Scholar] [CrossRef]
- Bernstein, B.W.; Bamburg, J.R. ADF/cofilin: A functional node in cell biology. Trends Cell Biol. 2010, 20, 187–195. [Google Scholar] [CrossRef]
- Liu, N.; Bassel-Duby, R. Regulation of Skeletal Muscle Development and Disease by microRNAs. Vertebr. Myogenesis 2015, 56, 165–190. [Google Scholar]
- Zhu, H.; Yang, H.; Zhao, S.; Zhang, J.; Liu, D.; Tian, Y.; Shen, Z.; Su, Y. Role of the cofilin 2 gene in regulating the myosin heavy chain genes in mouse myoblast C2C12 cells. Int. J. Mol. Med. 2018, 41, 1096–1102. [Google Scholar] [CrossRef] [PubMed]
| Name | Primer Sequences (5′-3′) |
|---|---|
| CFL2-CMV-F | CGGggtaccATGGCTTCTGGAGTTAC |
| CFL2-CMV-R | CCCaagcttTCAcatcatcaccatcaccatTAAGGGTTTTCCTTC |
| shCFL2-1F | gatccCTGAAAGTGCACCGTTAAATTCAAGAGATTTAACGGTGCACTTTCAGtttttta |
| shCFL2-1R | agcttaaaaaaCTGAAAGTGCACCGTTAAATCTCTTGAATTTAACGGTGCACTTTCAGg |
| shCFL2-2F | gatccGCTCTAAAGATGCCATTAATTCAAGAGATTAATGGCATCTTTAGAGCtttttta |
| shCFL2-2R | agcttaaaaaaGCTCTAAAGATGCCATTAATCTCTTGAATTAATGGCATCTTTAGAGCg |
| shRNA-NC-F | gatccTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAAtttttta |
| shRNA-NC-R | agcttaaaaaaTTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAAg |
| Name | Primer Sequences (5′-3′) |
|---|---|
| CFL2-wild-F | CCGCTCGAGGGAGGCAATGTAGTAGTTTC |
| CFL2-wild-R | ATAAGAATGCGGCCGCCAAGGCAGGTGAGGTGTATG |
| CFL2-mut-F | TATAAAGCAGTCAACTGGATCTTAAGGAG |
| CFL2-mut-R | TCCTTAAGATCCAGTTGACTGCTTTATAAG |
| Name | Primer Sequences (5′-3′) |
|---|---|
| CFL2-F | GGTGACATTGGTGATACTG |
| CFL2-R | CATATCGGCAATCATTCAGA |
| bta-miR-183-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAGTGAAT |
| bta-miR-183-F | ACACTCCAGCTGGGTATGGCACTGGTAGA |
| miR-R | GCAGGGTCCGAGGTATTC |
| U6-F | GCTTCGGCAGCACATATACTAAAAT |
| U6-R | CGCTTCACGAATTTGCGTGTCAT |
| GAPDH-F | AGATAGCCGTAACTTCTGTGC |
| GAPDH-R | ACGATGTCCACTTTGCCAG |
| Name | Primer Sequence (5′-3′) |
|---|---|
| CFL2-DMR-F | GGAAAATTTTTGAAAATGTTTTTT |
| CFL2-DMR-R | CTTTAAAATCATCCTAACCAATACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Zhao, T.; Ma, Y.; Wu, X.; Mao, Y.; Yang, Z.; Chen, H. New Insight into Muscle-Type Cofilin (CFL2) as an Essential Mediator in Promoting Myogenic Differentiation in Cattle. Bioengineering 2022, 9, 729. https://doi.org/10.3390/bioengineering9120729
Sun Y, Zhao T, Ma Y, Wu X, Mao Y, Yang Z, Chen H. New Insight into Muscle-Type Cofilin (CFL2) as an Essential Mediator in Promoting Myogenic Differentiation in Cattle. Bioengineering. 2022; 9(12):729. https://doi.org/10.3390/bioengineering9120729
Chicago/Turabian StyleSun, Yujia, Tianqi Zhao, Yaoyao Ma, Xinyi Wu, Yongjiang Mao, Zhangping Yang, and Hong Chen. 2022. "New Insight into Muscle-Type Cofilin (CFL2) as an Essential Mediator in Promoting Myogenic Differentiation in Cattle" Bioengineering 9, no. 12: 729. https://doi.org/10.3390/bioengineering9120729
APA StyleSun, Y., Zhao, T., Ma, Y., Wu, X., Mao, Y., Yang, Z., & Chen, H. (2022). New Insight into Muscle-Type Cofilin (CFL2) as an Essential Mediator in Promoting Myogenic Differentiation in Cattle. Bioengineering, 9(12), 729. https://doi.org/10.3390/bioengineering9120729