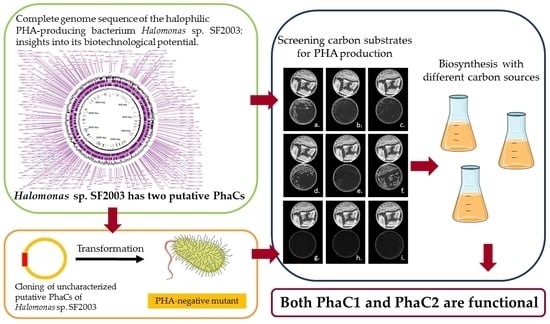
PHA Production and PHA Synthases of the Halophilic Bacterium Halomonas sp. SF2003
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Media
2.1.1. Bacterial Strains
2.1.2. Growth Media
2.1.3. Production Media
2.2. In Silico Study of PHA Synthase of Halomonas sp. SF2003 PhaC1 and PhaC2
2.3. Cloning of Halomonas sp. SF2003 phaC1 and phaC2 Genes
2.4. Screening for Carbon Sources
2.5. PHA Production
2.6. PHA Extraction
3. Results
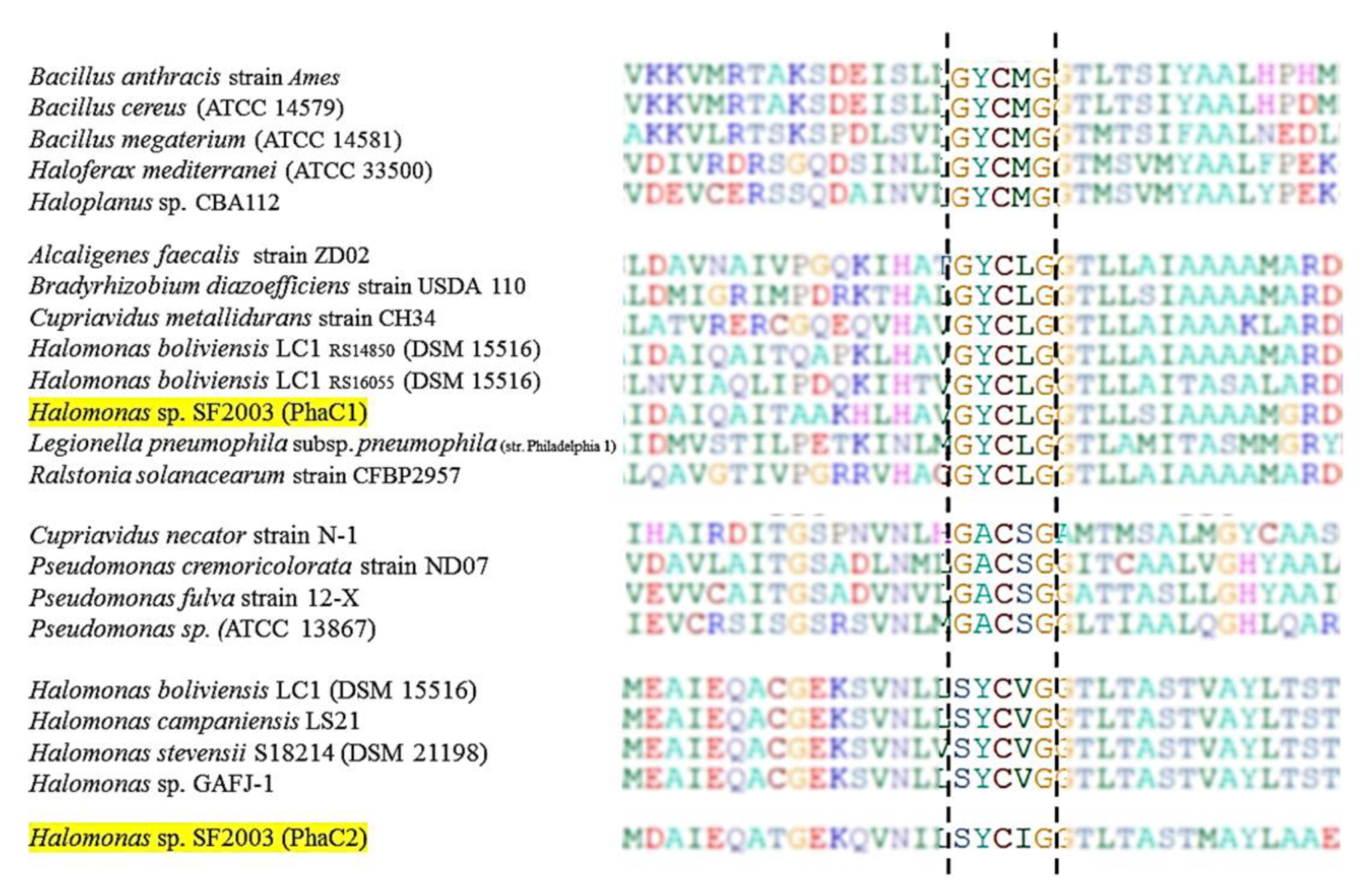
3.1. In Silico Study of PHA Synthases PhaC1 and PhaC2 of Halomonas sp. SF2003
3.2. Screening of Carbon Substrates for PHA Production by Halomonas sp. SF2003
3.3. Study of PHA Synthases
3.3.1. Cloning of PHA Synthases phaC1 and phaC2 of Halomonas sp. SF2003
3.3.2. Characterization of PHA Production by Transformant Strains PHB¯4/pBBR1-ProCn-phaC1 and PHB¯4/pBBR1-ProCn-phaC2
3.3.3. Polyhydroxyalkanoates Production in Shake Flasks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rehm, B.H.A. Bacterial polymers: Biosynthesis, modifications and applications. Nat. Rev. Microbiol. 2010, 8, 578–592. [Google Scholar] [CrossRef] [PubMed]
- Możejko-Ciesielska, J.; Kiewisz, R. Bacterial polyhydroxyalkanoates: Still fabulous? Microbiol. Res. 2016, 192, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz, T.; Roy, I. Polyhydroxyalkanoates: Bioplastics with a green agenda. Curr. Opin. Microbiol. 2010, 13, 321–326. [Google Scholar] [CrossRef]
- Valappil, S.P.; Misra, S.K.; Boccaccini, A.R.; Roy, I. Biomedical applications of polyhydroxyalkanoates, an overview of animal testing and in vivo responses. Expert Rev. Med. Devices 2006, 3, 853–868. [Google Scholar] [CrossRef]
- Williams, H.; Patricia, K. Polyhydroxyalkanoates. Biosynthesis, Chemical Structures and Applications. Materials Science and Technologies; NOVA: Annandale, VA, USA, 2018; p. 332. [Google Scholar]
- Jiang, G.; Hill, D.; Kowalczuk, M.; Johnston, B.; Adamus, G.; Irorere, V.; Radecka, I. Carbon Sources for Polyhydroxyalkanoates and an Integrated Biorefinery. Int. J. Mol. Sci. 2016, 17, 1157. [Google Scholar] [CrossRef]
- Gumel, A.M.; Annuar, M.S.M.; Heidelberg, T. Effects of carbon substrates on biodegradable polymer composition and stability produced by Delftia tsuruhatensis Bet002 isolated from palm oil mill effluent. Polym. Degrad. Stab. 2012, 97, 1224–1231. [Google Scholar] [CrossRef]
- Reddy, C.S.K.; Ghai, R.; Kalia, V. Polyhydroxyalkanoates: An overview. Bioresour. Technol. 2003, 87, 137–146. [Google Scholar] [CrossRef]
- Koller, M.; Maršálek, L.; de Sousa Dias, M.M.; Braunegg, G. Producing microbial polyhydroxyalkanoate (PHA) biopolyesters in a sustainable manner. New Biotechnol. 2017, 37, 4–38. [Google Scholar] [CrossRef]
- Balakrishna Pillai, A.; Kumarapillai, H.K. Bacterial Polyhydroxyalkanoates: Recent Trends in Production and Applications. In Recent Advances in Applied, Microbiology; Shukla, P., Ed.; Springer: Singapore, 2017; pp. 19–53. Available online: http://link.springer.com/10.1007/978-981-10-5275-0_2 (accessed on 15 July 2019).
- Elain, A.; Le Fellic, M.; Corre, Y.-M.; Le Grand, A.; Le Tilly, V.; Audic, J.-L.; Bruzaud, S. Rapid and qualitative fluorescence-based method for the assessment of PHA production in marine bacteria during batch culture. World J. Microbiol. Biotechnol. 2015, 31, 1555–1563. [Google Scholar] [CrossRef]
- Elain, A.; Le Grand, A.; Corre, Y.-M.; Le Fellic, M.; Hachet, N.; Le Tilly, V.; Loulergue, P.; Audic, J.-L.; Bruzaud, S. Valorisation of local agro-industrial processing waters as growth media for polyhydroxyalkanoates (PHA) production. Ind. Crops Prod. 2016, 80, 1–5. [Google Scholar] [CrossRef]
- Thomas, T.; Elain, A.; Bazire, A.; Bruzaud, S. Complete genome sequence of the halophilic PHA-producing bacterium Halomonas sp. SF2003: Insights into its biotechnological potential. World J. Microbiol. Biotechnol. 2019, 35, 50. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.; Pühler, A. A Broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Foong, C.P.; Lau, N.S.; Deguchi, S.; Toyofuku, T.; Taylor, T.; Sudesh, K.; Matsui, M. Whole genome amplification approach reveals novel polyhydroxyalkanoate synthases (PhaCs) from Japan Trench and Nankai Trough seawater. BMC Microbiol. 2014, 14, 318. [Google Scholar] [CrossRef]
- Lemechko, P.; Le Fellic, M.; Bruzaud, S. Production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) using agro-industrial effluents with tunable proportion of 3-hydroxyvalerate monomer units. Int. J. Biol. Macromol. 2019, 128, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Verlinden, R.A.J.; Hill, D.J.; Kenward, M.A.; Williams, C.D.; Radecka, I. Bacterial synthesis of biodegradable polyhydroxyalkanoates. J. Appl. Microbiol. 2007, 102, 1437–1449. [Google Scholar] [CrossRef]
- Labuzek, S.; Radecka, I. Biosynthesis of PHB tercopolymer by Bacillus cereus UW85. J. Appl. Microbiol. 2001, 90, 353–357. [Google Scholar]
- Tan, D.; Xue, Y.-S.; Aibaidula, G.; Chen, G.-Q. Unsterile and continuous production of polyhydroxybutyrate by Halomonas TD01. Bioresour. Technol. 2011, 102, 8130–8136. [Google Scholar] [CrossRef]
- Simon-Colin, C.; Raguénès, G.; Cozien, J.; Guezennec, J.G. Halomonas profundus sp. nov., a new PHA-producing bacterium isolated from a deep-sea hydrothermal vent shrimp. J. Appl. Microbiol. 2008, 104, 1425–1432. [Google Scholar] [CrossRef]
- Balakrishna Pillai, A.; Jaya Kumar, A.; Thulasi, K.; Kumarapillai, H. Evaluation of short-chain-length polyhydroxyalkanoate accumulation in Bacillus aryabhattai. Braz. J. Microbiol. 2017, 48, 451–460. [Google Scholar] [CrossRef]
- Tan, D.; Wu, Q.; Chen, J.-C.; Chen, G.-Q. Engineering Halomonas TD01 for the low-cost production of polyhydroxyalkanoates. Metab. Eng. 2014, 26, 34–47. [Google Scholar] [CrossRef]
- Pernicova, I.; Kucera, D.; Novackova, I.; Vodicka, J.; Kovalcik, A.; Obruca, S. Extremophiles platform strains for sustainable production of polyhydroxyalkanoates. Mater. Sci. Forum 2019, 955, 74–79. [Google Scholar] [CrossRef]
- Page, W.J. Production of poly-β-hydroxybutyrate by Azotobacter vinelandii UWD in media containing sugars and complex nitrogen sources. Appl. Microbiol. Biotechnol. 1992, 38, 117–121. [Google Scholar] [CrossRef]
- Miranda De Sousa Dias, M.; Koller, M.; Puppi, D.; Morelli, A.; Chiellini, F.; Braunegg, G. Fed-batch synthesis of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-4-hydroxybutyrate) from sucrose and 4-hydroxybutyrate precursors by Burkholderia sacchari Strain DSM 17165. Bioengineering 2017, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Daniş, Ö.; Ogan, A.; Tatlican, P.; Attar, A.; Çakmakçi, E.; Mertoglu, B.; Birbir, M.; Cakmakcı, E. Preparation of poly(3-hydroxybutyrate-co-hydroxyvalerate) films from halophilic archaea and their potential use in drug delivery. Extremophiles 2015, 19, 515–524. [Google Scholar]
- Kucera, D.; Pernicová, I.; Kovalcik, A.; Koller, M.; Mullerova, L.; Sedlacek, P.; Mravec, F.; Nebesarova, J.; Kalina, M.; Márova, I.; et al. Characterization of the promising poly(3-hydroxybutyrate) producing halophilic bacterium Halomonas halophila. Bioresour. Technol. 2018, 256, 552–556. [Google Scholar] [CrossRef]
- Giedraitytė, G.; Kalėdienė, L. Purification and characterization of polyhydroxybutyrate produced from thermophilic Geobacillus sp. AY 946034 strain. CHEMIJA 2015, 26, 38–45. [Google Scholar]
- Quillaguamán, J.; Hashim, S.; Bento, F.; Mattiasson, B.; Hatti-Kaul, R. Poly(β-hydroxybutyrate) production by a moderate halophile, Halomonas boliviensis LC1 using starch hydrolysate as substrate. J. Appl. Microbiol. 2005, 99, 151–157. [Google Scholar] [CrossRef]
- Joshi, A.A.; Kanekar, P.P.; Kelkar, A.S.; Sarnaik, S.S.; Shouche, Y.; Wani, A. Moderately halophilic, alkalitolerant Halomonas campisalis MCM B-365 from Lonar Lake, India. J. Basic Microbiol. 2007, 47, 213–221. [Google Scholar] [CrossRef]
- Ilham, M.; Nakanomori, S.; Kihara, T.; Hokamura, A.; Matsusaki, H.; Tsuge, T.; Mizuno, K. Characterization of polyhydroxyalkanoate synthases from Halomonas sp. O-1 and Halomonas elongata DSM2581: Site-directed mutagenesis and recombinant expression. Polym. Degrad. Stab. 2014, 109, 416–423. [Google Scholar] [CrossRef]
- Lu, Q.; Han, J.; Zhou, L.; Zhou, J.; Xiang, H. Genetic and biochemical characterization of the poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthase in Haloferax mediterranei. J. Bacteriol. 2008, 190, 4173–4180. [Google Scholar] [CrossRef]
- Ueda, S.; Yabutani, T.; Maehara, A.; Yamane, T. Molecular analysis of the poly(3-hydroxyalkanoate) synthase gene from a methylotrophic bacterium, Paracoccus denitrificans. J. Bacteriol. 1996, 178, 774–779. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mezzolla, V.; D’Urso, O.; Poltronieri, P. Role of PhaC Type I and Type II Enzymes during PHA Biosynthesis. Polymers 2018, 10, 910. [Google Scholar] [CrossRef] [PubMed]
- Rehm, B.H. Polyester synthases: Natural catalysts for plastics. Biochem. J. 2003, 376, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, T.; Hyakutake, M.; Mizuno, K. Class IV polyhydroxyalkanoate (PHA) synthases and PHA-producing Bacillus. Appl. Microbiol. Biotechnol. 2015, 99, 6231–6240. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, H.; Shafie, N.A.H.; Sudesh, K.; Azizan, M.N.; Majid, M.I.A.; Amirul, A.-A.A. Cupriavidus malaysiensis sp. nov., a novel poly(3-hydroxybutyrate-co-4-hydroxybutyrate) accumulating bacterium isolated from the Malaysian environment. Anton Leeuw. Int. J. G. 2018, 111, 361–372. [Google Scholar] [CrossRef] [PubMed]
- García-Torreiro, M.; López-Abelairas, M.; Lu-Chau, T.A.; Lema, J.M. Production of poly(3-hydroxybutyrate) by simultaneous saccharification and fermentation of cereal mash using Halomonas boliviensis. Biochem. Eng. J. 2016, 114, 140–146. [Google Scholar] [CrossRef]
- Ates, Ö.; Oner, E.; Arga, K.Y. Genome-scale reconstruction of metabolic network for a halophilic extremophile, Chromohalobacter salexigens DSM 3043. BMC Syst. Biol. 2011, 5, 12. [Google Scholar] [CrossRef]
- Kawata, Y.; Aiba, S. Poly(3-hydroxybutyrate) Production by isolated Halomonas sp. KM-1 using waste glycerol. Biosci. Biotechnol. Biochem. 2010, 74, 175–177. [Google Scholar] [CrossRef]
- Chek, M.F.; Kim, S.-Y.; Mori, T.; Arsad, H.; Samian, M.R.; Sudesh, K.; Hakoshima, T. Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp. USM2, producing biodegradable plastics. Sci. Rep. 2017, 7, 5312. [Google Scholar] [CrossRef]
- Wittenborn, E.C.; Jost, M.; Wei, Y.; Stubbe, J.; Drennan, C.L. Structure of the Catalytic Domain of the Class I Polyhydroxybutyrate Synthase from Cupriavidus necator. J. Biol. Chem. 2016, 291, 25264–25277. [Google Scholar] [CrossRef]
- Chek, M.F.; Hiroe, A.; Hakoshima, T.; Sudesh, K.; Taguchi, S. PHA synthase (PhaC): Interpreting the functions of bioplastic-producing enzyme from a structural perspective. Appl. Microbiol. Biotechnol. 2019, 103, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Spiekermann, P.; Rehm, B.H.A.; Kalscheuer, R.; Baumeister, D.; Steinbüchel, A. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 1999, 171, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Peters, V.; Rehm, B.H.A. In vivo monitoring of PHA granule formation using GFP-labeled PHA synthases. FEMS Microbiol. Lett. 2005, 248, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Cavalheiro, J.M.B.T.; de Almeida, M.C.M.D.; da Fonseca, M.M.R.; de Carvalho, C.C.C.R. Adaptation of Cupriavidus necator to conditions favoring polyhydroxyalkanoate production. J. Biotechnol. 2013, 164, 309–317. [Google Scholar] [CrossRef]
- Shrivastav, A.; Mishra, S.K.; Shethia, B.; Pancha, I.; Jain, D.; Mishra, S. Isolation of promising bacterial strains from soil and marine environment for polyhydroxyalkanoates (PHAs) production utilizing Jatropha biodiesel byproduct. Int. J. Biol. Macromol. 2010, 47, 283–287. [Google Scholar] [CrossRef]
- Bramer, C.O.; Vandamme, P.; da Silva, L.F.; Gomez, J.; Steinbüchel, A. Burkholderia sacchari sp. nov., a polyhydroxyalkanoate-accumulating bacterium isolated from soil of a sugar-cane plantation in Brazil. Int. J. Syst. Evol. Microbiol. 2001, 51, 1709–1713. [Google Scholar] [CrossRef]
- Mata, J.A.; Martínez-Cánovas, J.; Quesada, E.; Béjar, V. A detailed phenotypic characterisation of the type strains of Halomonas species. Syst. Appl. Microbiol. 2002, 25, 360–375. [Google Scholar] [CrossRef]
- Vreeland, R.H. Halomonas. In Bergey’s Manual of Systematics of Archaea and, Bacteria; Whitman, W.B., Rainey, F., Kämpfer, P., Trujillo, M., Chun, J., DeVos, P., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2015; pp. 1–19. [Google Scholar]
- Füchtenbusch, B.; Wullbrandt, D.; Steinbüchel, A. Production of polyhydroxyalkanoic acids by Ralstonia eutropha and Pseudomonas oleovorans from an oil remaining from biotechnological rhamnose production. Appl. Microbiol. Biotechnol. 2000, 53, 167–172. [Google Scholar] [CrossRef]
- Chaitanya, K.; Nagamani, P.; Mahmood, S.K.; Sunil Kumar, N. Polyhydroxyalkanoate producing novel Bacillus sp., SKM11 isolated from polluted pond water. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 1159–1165. [Google Scholar]
- Sangkharak, K.; Prasertsan, P. Screening and identification of polyhydroxyalkanoates producing bacteria and biochemical characterization of their possible application. J. Gen. Appl. Microbiol. 2012, 58, 173–182. [Google Scholar] [CrossRef]
- de Paula, F.C.; Kakazu, S.; de Paula, C.B.C.; Gomez, J.G.C.; Contiero, J. Polyhydroxyalkanoate production from crude glycerol by newly isolated Pandoraea sp. J. King Saud Univ. Sci. 2017, 29, 166–173. [Google Scholar] [CrossRef]
- Masood, F.; Yasin, T.; Hameed, A. Production and characterization of tailor-made polyhydroxyalkanoates by Bacillus cereus FC1. Pak. J. Zool. 2015, 47, 491–503. [Google Scholar]
- Koller, M.; Hesse, P.; Bona, R.; Kutschera, C.; Atlić, A.; Braunegg, G. Biosynthesis of high quality polyhydroxyalkanoate co- and terpolyesters for potential medical application by the Archaeon Haloferax mediterranei. Macromol. Symp. 2007, 253, 3–9. [Google Scholar] [CrossRef]
- Koller, M.; Atlić, A.; Gonzalez-Garcia, Y.; Kutschera, C.; Braunegg, G. Polyhydroxyalkanoate (PHA) Biosynthesis from Whey Lactose. Macromol. Symp. 2008, 272, 87–92. [Google Scholar] [CrossRef]
- Koller, M.; Bona, R.; Chiellini, E.; Fernandes, E.G.; Horvat, P.; Kutschera, C.; Hesse, P.; Braunegg, G. Polyhydroxyalkanoate production from whey by Pseudomonas hydrogenovora. Bioresour. Technol. 2008, 99, 4854–4863. [Google Scholar] [CrossRef]
- Alkotaini, B.; Koo, H.; Kim, B.S. Production of polyhydroxyalkanoates by batch and fed-batch cultivations of Bacillus megaterium from acid-treated red algae. Korean J. Chem. Eng. 2016, 33, 1669–1673. [Google Scholar] [CrossRef]
- Obruca, S.; Benesova, P.; Petrik, S.; Oborna, J.; Prikryl, R.; Marova, I. Production of polyhydroxyalkanoates using hydrolysate of spent coffee grounds. Process Biochem. 2014, 49, 1409–1414. [Google Scholar] [CrossRef]
- Pan, W.; Perrotta, J.A.; Stipanovic, A.J.; Nomura, C.T.; Nakas, J.P. Production of polyhydroxyalkanoates by Burkholderia cepacia ATCC 17759 using a detoxified sugar maple hemicellulosic hydrolysate. J. Ind. Microbiol. Biotechnol. 2012, 39, 459–469. [Google Scholar] [CrossRef]
- Cerrone, F.; Davis, R.; Kenny, S.T.; Woods, T.; O’Donovan, A.; Gupta, V.K.; Tuohy, M.; Babu, R.P.; O’Kiely, P.; O’Connor, K. Use of a mannitol rich ensiled grass press juice (EGPJ) as a sole carbon source for polyhydroxyalkanoates (PHAs) production through high cell density cultivation. Bioresour. Technol. 2015, 191, 45–52. [Google Scholar] [CrossRef]
- Bera, A.; Dubey, S.; Bhayani, K.; Mondal, D.; Mishra, S.; Ghosh, P.K. Microbial synthesis of polyhydroxyalkanoate using seaweed-derived crude levulinic acid as co-nutrient. Int. J. Biol. Macromol. 2015, 72, 487–494. [Google Scholar] [CrossRef]
- Ashby, R.D.; Solaiman, D.K.Y.; Strahan, G.D.; Zhu, C.; Tappel, R.C.; Nomura, C.T. Glycerine and levulinic acid: Renewable co-substrates for the fermentative synthesis of short-chain poly(hydroxyalkanoate) biopolymers. Bioresour. Technol. 2012, 118, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Keenan, T.M.; Tanenbaum, S.W.; Stipanovic, A.J.; Nakas, J.P. Production and characterization of poly-β-hydroxyalkanoate copolymers from Burkholderia cepacia utilizing xylose and levulinic acid. Biotechnol. Prog. 2004, 20, 1697–1704. [Google Scholar] [CrossRef] [PubMed]
- Jaremko, M.; Yu, J. The initial metabolic conversion of levulinic acid in Cupriavidus necator. J. Biotechnol. 2011, 155, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, T.; Gomez, J.G.C.; Buffoni, E.; Rodriguez, R.S.; Schripsema, J.; Lopes, M.; Silva, L. Exploring the potential of Burkholderia sacchari to produce polyhydroxyalkanoates. J. Appl. Microbiol. 2013, 116, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Follonier, S.; Goyder, M.S.; Silvestri, A.-C.; Crelier, S.; Kálmán, F.; Riesen, R.; Zinn, M. Fruit pomace and waste frying oil as sustainable resources for the bioproduction of medium-chain-length polyhydroxyalkanoates. Int. J. Biol. Macromol. 2014, 71, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Khosravi-Darani, K.; Mokhtari, Z.-B.; Amai, T.; Tanaka, K. Microbial production of poly(hydroxybutyrate) from C1 carbon sources. Appl. Microbiol. Biotechnol. 2013, 97, 1407–1424. [Google Scholar] [CrossRef]
- Cruz, M.V.; Freitas, F.; Paiva, A.; Mano, F.; Dionísio, M.; Ramos, A.M.; Reis, M.A.; Andrade, M.M.D.; Mano, M.F.M. Valorization of fatty acids-containing wastes and byproducts into short- and medium-chain length polyhydroxyalkanoates. New Biotechnol. 2016, 33, 206–215. [Google Scholar] [CrossRef]
- Obruca, S.; Petrik, S.; Benesova, P.; Svoboda, Z.; Eremka, L.; Marova, I. Utilization of oil extracted from spent coffee grounds for sustainable production of polyhydroxyalkanoates. Appl. Microbiol. Biotechnol. 2014, 98, 5883–5890. [Google Scholar] [CrossRef]
- Chee, J.-Y.; Tan, Y.; Samian, M.-R.; Sudesh, K. Isolation and characterization of a Burkholderia sp. USM (JCM15050) capable of producing polyhydroxyalkanoate (PHA) from triglycerides, fatty acids and glycerols. J. Polym. Environ. 2010, 18, 584–592. [Google Scholar] [CrossRef]
- Pernicova, I.; Kucera, D.; Nebesarova, J.; Kalina, M.; Novackova, I.; Koller, M.; Obruca, S. Production of polyhydroxyalkanoates on waste frying oil employing selected Halomonas strains. Bioresour. Technol. 2019, 292, 122028. [Google Scholar] [CrossRef]
- Luengo, J.M.; García, B.; Sandoval, A.; Naharro, G.; Olivera, E.R. Bioplastics from microorganisms. Curr. Opin. Microbiol. 2003, 6, 251–260. [Google Scholar] [CrossRef]
- Argandoña, M.; Vargas, C.; Reina-Bueno, M.; Rodríguez-Moya, J.; Salvador, M.; Nieto, J.J. An Extended Suite of Genetic Tools for Use in Bacteria of the Halomonadaceae: An Overview; Recombinant Gene, Expression; Lorence, A., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 167–201. Available online: http://link.springer.com/10.1007/978-1-61779-433-9_9 (accessed on 8 November 2017).
- Jia, K.; Cao, R.; Hua, D.H.; Li, P. Study of Class I and Class III Polyhydroxyalkanoate (PHA) synthases with substrates containing a modified side chain. Biomacromolecules 2016, 17, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Mifune, J.; Nakamura, S.; Fukui, T. Targeted engineering of Cupriavidus necator chromosome for biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from vegetable oil. Can. J. Chem. 2008, 86, 621–627. [Google Scholar] [CrossRef]
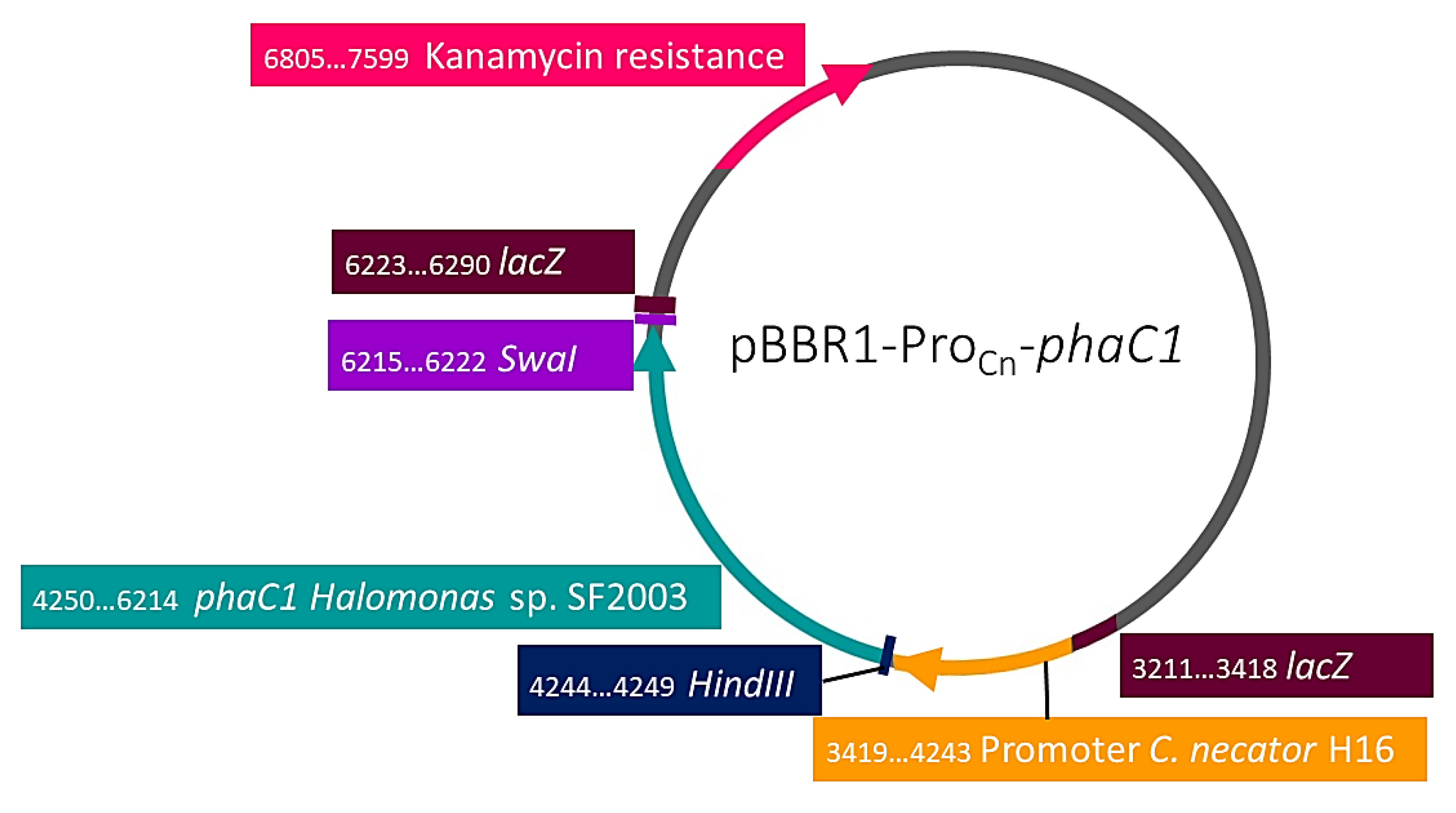
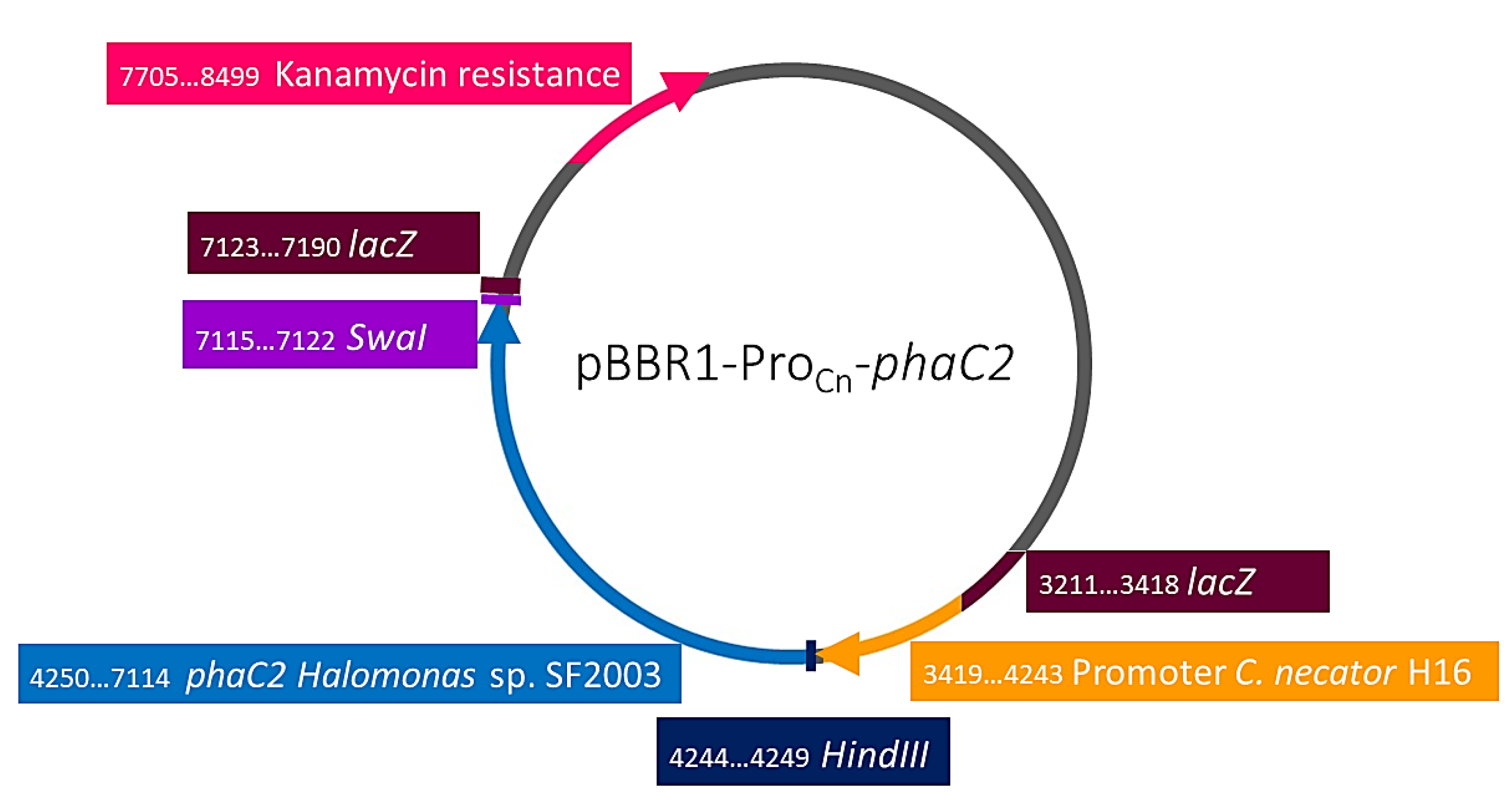
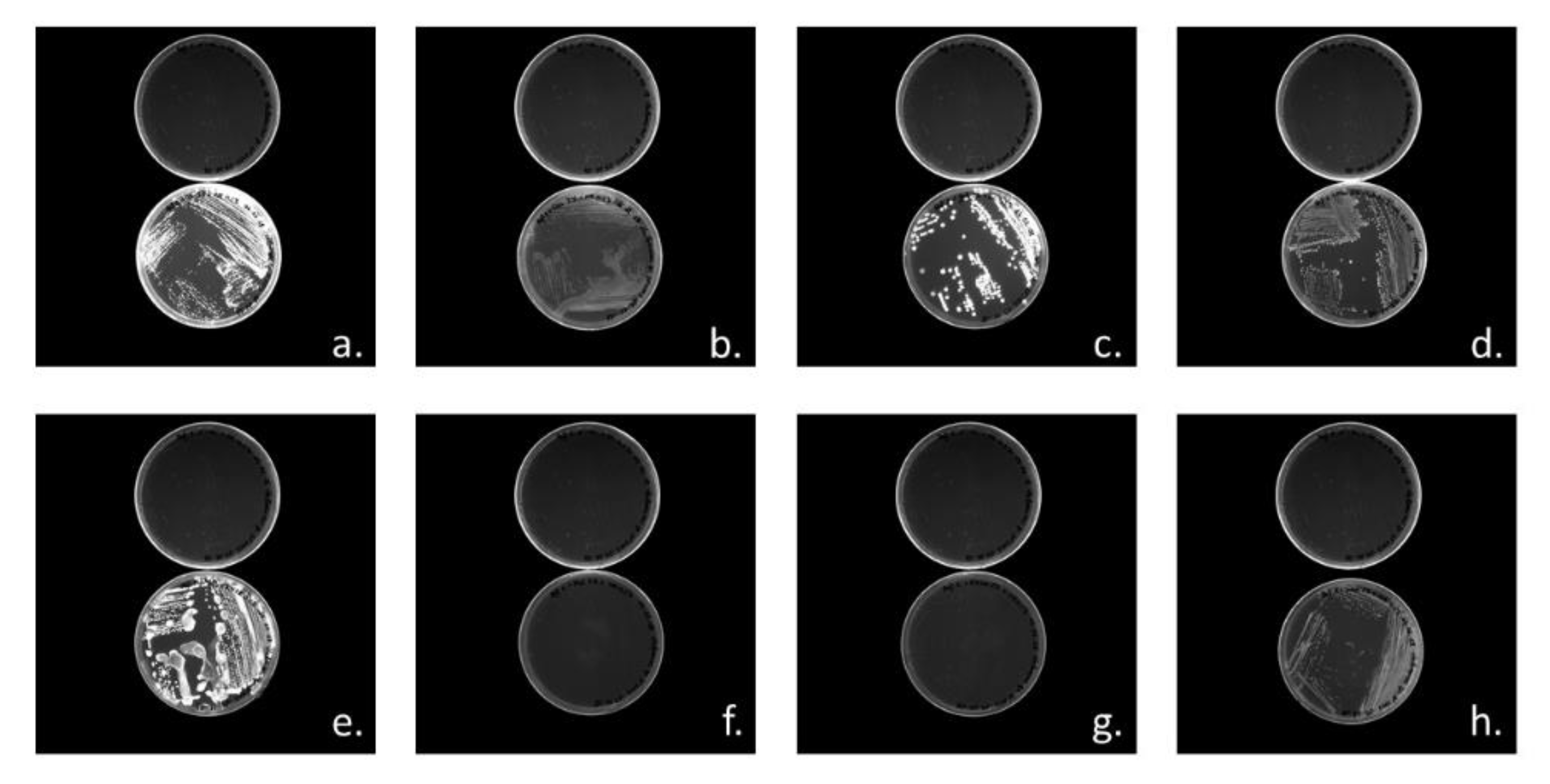
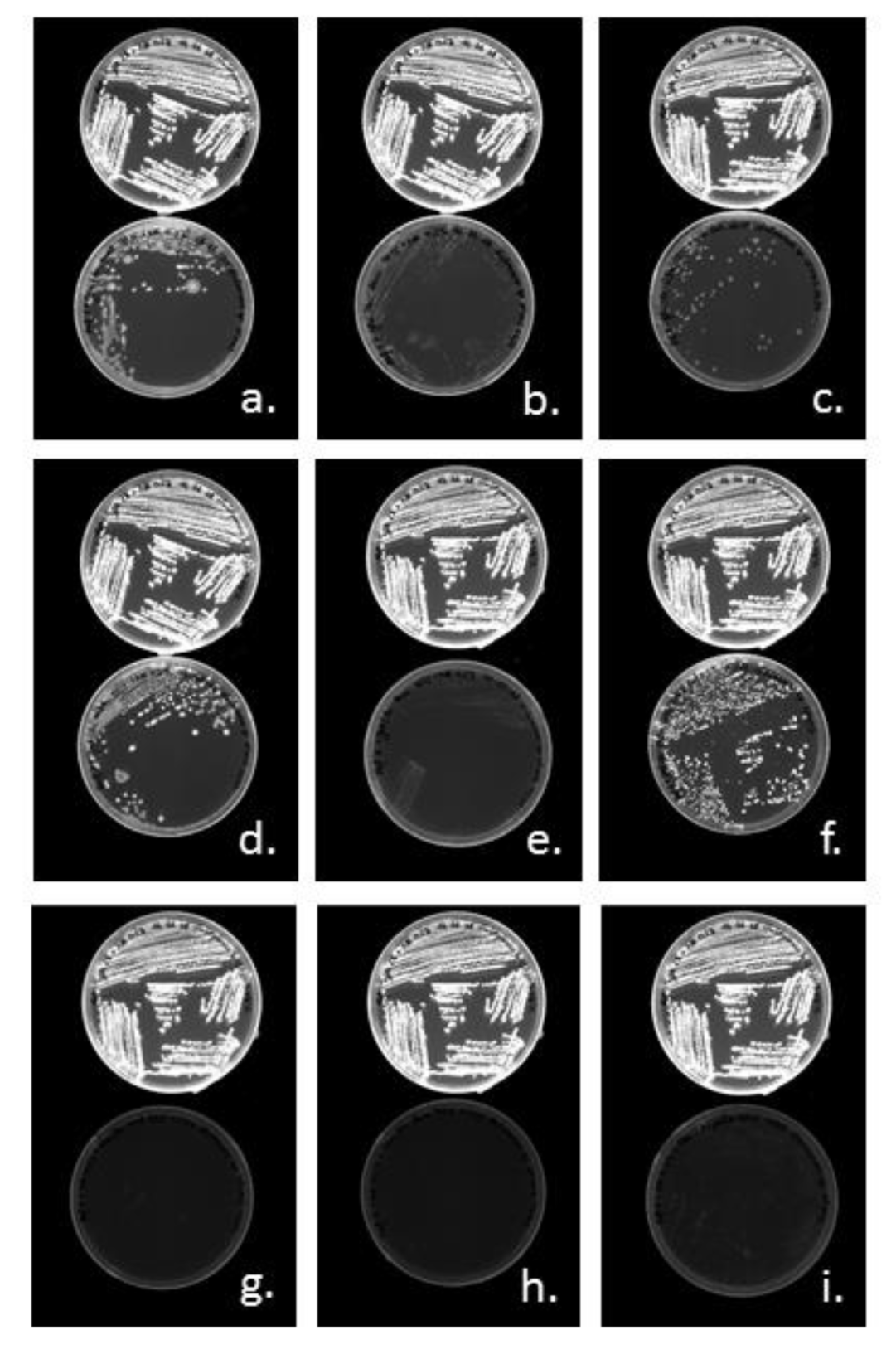
| Strain or Plasmid | Characteristics | Origin |
|---|---|---|
| Halomonas strain | ||
| Halomonas sp. SF2003 ID CNCM-I-4786 | Wild type PHA-producing strain | Sea of Iroise (France), RIDL Collection, UBS |
| Cupriavidus necator strains | ||
| H16 (DSM 428) | Wild type PHA-producing strain | DSMZ Collection |
| PHB¯4 (DSM 541) | Mutant non-PHA-producing strain | DSMZ Collection |
| Escherichia coli strains | ||
| E. cloni® 10G | Competent cells | Lucigen |
| S17-1 | Strain for conjugative transfer of plasmid to C. necator PHB¯4 | Simon et al. 1983 [14] |
| Plasmids | ||
| pBBR1-ProCn | pBBR1MCS-2 derivatives with phaC1 promoter from C. necator. KmR. | Foong et al. 2014 [15] |
| pBBR1-ProCn-phaC1 | pBBR1MCS-2 derivatives with phaC1 promoter from C. necator and phaC1 of Halomonas sp. SF2003. KmR. | This study |
| pBBR1-ProCn-phaC2 | pBBR1MCS-2 derivatives with phaC1 promoter from C. necator and phaC2 of Halomonas sp. SF2003. KmR. | This study |
| Transformants | ||
| PHB¯4/pBBR1-ProCn-phaC1 | Transformant strain with pBBR1MCS-2 plasmid expressing phaC1 of Halomonas sp. SF2003. KmR. | This study |
| PHB¯4/pBBR1-ProCn-phaC2 | Transformant strain with pBBR1MCS-2 plasmid expressing phaC2 of Halomonas sp. SF2003. KmR. | This study |
| Carbon Source | Growth | PHA Accumulation |
|---|---|---|
| (D)-Glucose | + | + |
| (D)-Fructose | + | + |
| (D)-Galactose | + | + |
| (D)-Mannose | + | + |
| (D)-Maltose | + | + |
| (D)-Melibiose | - | - |
| (L)-Rhamnose | - | - |
| (D)-Sucrose | + | + |
| Carbohydrates | Origin | Identification of Pathway for Assimilation |
|---|---|---|
| Fructose * | Fruits, Honey | Total |
| Galactose * | Milk, Honey, Red algae | Partial |
| Glucose * | Food, Metabolism of living organisms | Partial |
| Lactose | Dairy products | Total |
| Maltose * | Starch degradation (barley) | n.i |
| Mannose * | Fruits, Plants, Mannitol | n.i |
| Melibiose * | Plants, Fruits | Total |
| Ribose | RNA | Partial |
| Rhamnose * | Plants | Partial |
| Sucrose * | Plants | Total |
| Xylose | Plants | Partial |
| Carbohydrates | Bacterial Strains/Species | References |
|---|---|---|
| Melibiose | Burkholderia sacchari sp. nov. | [17] |
| Rhamnose | C. necator, P. oleovorans | [17] |
| Glucose | Bacillus cereus UW85, Halomonas sp. TD01, Halomonas profundus, Halomonas sp. SF2003 | [18,19,20] |
| Fructose | Bacillus aryabhattai PHB10, C. necator, Halomonas TD08,Halomonas sp. SF2003, H. halophila, H. organivorans, H. salina | [21,22,23] |
| Sucrose | Azotobacter vinelandii, Burkholderia sacchari DSM 17165, C. necator, Natrinema sp. 5TL6 | [6,24,25,26] |
| Galactose | Halomonas halophila, H. salina, Halomonas sp. SF2003 | [23] |
| Mannose | Halomonas halophila, H. organivorans, H. salina | [23,27] |
| Maltose | B. aryabhattai PHB10, Halomonas sp. TD08, H. boliviensis LC1 and H. campisalis | [17,21,28,29,30] |
| Carbon Source | Growth | PHA Accumulation | |
|---|---|---|---|
| Glucose | Dodecanoic acid | - | - |
| Heptanoic acid | + | - | |
| Hexanoic acid | + | - | |
| Levulinic acid | + | ± | |
| Malic acid | + | + | |
| Palmitic acid | + | - | |
| Trans-2-pentenoic acid | + | - | |
| Galactose | Dodecanoic acid | - | - |
| Heptanoic acid | - | - | |
| Hexanoic acid | - | - | |
| Levulinic acid | + | - | |
| Malic acid | + | + | |
| Palmitic acid | + | ± | |
| Trans-2-pentenoic acid | + | - | |
| Fructose | Dodecanoic acid | ± | ± |
| Heptanoic acid | - | - | |
| Hexanoic acid | - | - | |
| Levulinic acid | - | - | |
| Malic acid | - | - | |
| Palmitic acid | - | - | |
| Trans-2-pentenoic acid | - | - | |
| Carbon Source | PHB¯4/ pBBR1-ProCn-phaC1 | PHB¯4/ pBBR1-ProCn-phaC2 | |||
|---|---|---|---|---|---|
| Growth | PHA Accumulation | Growth | PHA Accumulation | ||
| (D)-Fructose | + | ± | + | + | |
| (D)-Galactose | + | − | + | − | |
| (D)-Glucose | + | − | + | − | |
| (D)-Maltose | + | − | + | − | |
| (D)-Mannose | + | − | + | ± | |
| (D)-Melibiose | + | − | + | − | |
| (L)-Rhamnose | + | − | + | − | |
| (D)-Sucrose | + | − | + | ± | |
| Glucose + | Dodecanoic acid | + | ± | + | + |
| Heptanoic acid | − | − | − | − | |
| Hexanoic acid | − | − | − | − | |
| Levulinic acid | + | − | + | ± | |
| Malic acid | + | − | + | ± | |
| Palmitic acid | + | ± | + | + | |
| Trans-2-pentenoic acid | − | − | − | − | |
| Galactose + | Dodecanoic acid | + | − | + | − |
| Heptanoic acid | − | − | − | − | |
| Hexanoic acid | − | − | − | − | |
| Levulinic acid | ± | − | + | ± | |
| Malic acid | + | − | + | ± | |
| Palmitic acid | − | − | + | − | |
| Trans-2-pentenoic acid | − | − | − | − | |
| Fructose + | Dodecanoic acid | − | − | − | − |
| Heptanoic acid | − | − | − | − | |
| Hexanoic acid | − | − | − | − | |
| Levulinic acid | − | − | − | − | |
| Malic acid | − | − | − | − | |
| Palmitic acid | − | − | − | − | |
| Trans-2-pentenoic acid | − | − | − | − | |
| Strain | Carbon Source | Dry cell Weight (g/L) | PHA (g/L) | PHA Content (wt.%) |
|---|---|---|---|---|
| Halomonas sp. SF2003 | Glucose | 2.63 | 2.25 | 86 |
| C. necator H16 | 2.89 | 2.05 | 71 | |
| PHB¯4/pBBR1-ProCn-phaC1 | 1.05 | 0.32 | 30 | |
| PHB¯4/pBBR1-ProCn-phaC2 | 2.63 | 1.38 | 52 | |
| Halomonas sp. SF2003 | Fructose | 2.63 | 1.02 | 39 |
| C. necator H16 | 3.16 | 2.25 | 71 | |
| PHB¯4/pBBR1-ProCn-phaC1 | 0.79 | 0.26 | 33 | |
| PHB¯4/pBBR1-ProCn-phaC2 | 3.42 | 1.83 | 54 | |
| Halomonas sp. SF2003 | Galactose | 3.16 | 1.23 | 39 |
| C. necator H16 | 0.79 | N.D | N.D | |
| PHB¯4/pBBR1-ProCn-phaC1 | 1.06 | N.D | N.D | |
| PHB¯4/pBBR1-ProCn-phaC2 | 0.79 | N.D | N.D |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, T.; Sudesh, K.; Bazire, A.; Elain, A.; Tan, H.T.; Lim, H.; Bruzaud, S. PHA Production and PHA Synthases of the Halophilic Bacterium Halomonas sp. SF2003. Bioengineering 2020, 7, 29. https://doi.org/10.3390/bioengineering7010029
Thomas T, Sudesh K, Bazire A, Elain A, Tan HT, Lim H, Bruzaud S. PHA Production and PHA Synthases of the Halophilic Bacterium Halomonas sp. SF2003. Bioengineering. 2020; 7(1):29. https://doi.org/10.3390/bioengineering7010029
Chicago/Turabian StyleThomas, Tatiana, Kumar Sudesh, Alexis Bazire, Anne Elain, Hua Tiang Tan, Hui Lim, and Stéphane Bruzaud. 2020. "PHA Production and PHA Synthases of the Halophilic Bacterium Halomonas sp. SF2003" Bioengineering 7, no. 1: 29. https://doi.org/10.3390/bioengineering7010029
APA StyleThomas, T., Sudesh, K., Bazire, A., Elain, A., Tan, H. T., Lim, H., & Bruzaud, S. (2020). PHA Production and PHA Synthases of the Halophilic Bacterium Halomonas sp. SF2003. Bioengineering, 7(1), 29. https://doi.org/10.3390/bioengineering7010029