Abstract
Polyhydroxyalkanoates (PHAs) are biodegradable plastic-like materials with versatile properties. Plant oils are excellent carbon sources for a cost-effective PHA production, due to their high carbon content, large availability, and comparatively low prices. Additionally, efficient process development and control is required for competitive PHA production, which can be facilitated by on-line or in-line monitoring devices. To this end, we have evaluated photon density wave (PDW) spectroscopy as a new process analytical technology for Ralstonia eutropha (Cupriavidus necator) H16 plant oil cultivations producing polyhydroxybutyrate (PHB) as an intracellular polymer. PDW spectroscopy was used for in-line recording of the reduced scattering coefficient µs’ and the absorption coefficient µa at 638 nm. A correlation of µs’ with the cell dry weight (CDW) and µa with the residual cell dry weight (RCDW) was observed during growth, PHB accumulation, and PHB degradation phases in batch and pulse feed cultivations. The correlation was used to predict CDW, RCDW, and PHB formation in a high-cell-density fed-batch cultivation with a productivity of 1.65 gPHB·L−1·h−1 and a final biomass of 106 g·L−1 containing 73 wt% PHB. The new method applied in this study allows in-line monitoring of CDW, RCDW, and PHA formation.
1. Introduction
When the US Food and Drug Administration (FDA) announced their process analytical technology (PAT) directives, the investigation of PAT became a key research area in bioprocess development. The main objectives are designing, developing, and operating bioprocesses to guarantee a targeted final product quality [1,2]. The focus of this initiative was predominantly on biopharmaceutical processes, while novel PAT tools could be integrated into any bioprocess. Especially, the implementation of PAT for polyhydroxyalkanoate (PHA) production can provide significant benefits to facilitate a consistent and highly efficient production. Techniques such as FTIR, Raman spectroscopy, fluorescence staining associated with flow cytometry, and enzymatic approaches were reported as novel methods for a rapid characterization of PHA production [3,4,5,6,7]. A comprehensive overview of qualitative and quantitative methods for PHA analysis was published by Koller et al. [8]. However, the reported methods have not been applied for in-line or at-line measurements of the PHA production process so far.
Photon density wave (PDW) spectroscopy is an in-line technique, which has been used as an analytical tool for measurements of various highly turbid chemical processes [9,10,11,12]. The method is based on the theory of photon migration in multiple light scattering material. If intensity-modulated light is introduced into a strongly light scattering but weakly light absorbing material, a PDW is generated. Absorption and scattering properties of the material influence the amplitude and phase of the PDW. By quantifying these shifts as a function of the emitter fiber and detector fiber distance and of the modulation frequency, the absorption coefficient µa and the reduced scattering coefficient µs’ can be determined independently [9,13,14]. The mentioned features make PDW spectroscopy very attractive for the monitoring of high-cell-density bioprocesses.
Currently, PHA production costs are not compatible with the low-priced production of conventional plastics. The main cost driving factors are the feedstocks for PHA accumulation and the recovery process. Thus, alternative low-cost substrates, e.g., biogenic waste streams, are of high interest to reduce the final production price. Other attempts concentrate on finding more sustainable and price efficient purification strategies [15,16,17,18,19,20,21]. Ralstonia eutropha (also known as Cupriavidus necator) is one of the main species studied for polyhydroxybutyrate (PHB) accumulation and the model organism for PHA accumulation [22]. Growth of R. eutropha on oleaginous feedstocks is particularly attractive due to their high carbon contents, high conversion rates to PHA, and low culture dilution in fed-batch processes. Efficient growth on these feedstocks is facilitated by the expression of extracellular lipases, which emulsify the lipids [23,24,25,26,27]. A large biomass accumulation prior to PHA accumulation is very important for a high final product titer. In this context, it has been shown that urea is an inexpensive nitrogen source, which allows excellent growth [24,28]. Despite alternative substrates and downstream approaches, highly efficient bioprocesses are required for an economic feasible PHA production. Recently, high-cell-density cultivations with R. eutropha on various renewable feedstocks have been published presenting the production of over 100 g·L−1 PHA and space time yields from 1 to 2.5 gPHA·L−1·h−1 [21,24,29,30,31]. However, none of the presented studies describe in-line PAT-based monitoring or control strategies for the enhancement of process results.
This work aims to integrate PDW spectroscopy into high-cell-density bioprocesses, for the monitoring of the highly turbid and complex PHB production with R. eutropha in plant oil cultivations. As a result, total cell dry weight (CDW) and residual cell dry weight (RCDW, the difference of CDW and the PHB concentration) accumulation could be distinguished with the PDW spectroscopy probe as a new in-line tool for bioprocesses.
2. Materials and Methods
2.1. Bacterial Strain
All cultivations were performed with the wild type strain R. eutropha H16 (DSM-428, Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Germany).
2.2. Growth Media and Preculture Cultivation Conditions
Tryptic soy broth (TSB) media (17 g·L−1 tryptone, 5 g·L−1 NaCl, 3 g·L−1 peptone) was used for the first precultures and with an additional supply of 2% (w·v−1) agar for culture plates. The second precultures and bioreactor cultivations were conducted in mineral salt media (MSM) containing 4.62 g·L−1 NaH2PO4·H2O, 5.74 g·L−1 Na2HPO4·2H2O, 0.45 g·L−1 K2SO4, 0.04 g·L−1 NaOH, 0.80 g·L−1 MgSO4·7H2O, 0.06 g·L−1 CaCl2·2H2O and 1 mL·L−1 trace element solution consisting of 0.48 g·L−1 CuSO4·5H2O, 2.4 g·L−1 ZnSO4·7H2O, 2.4 g·L−1 MnSO4·H2O, 15 g·L−1 FeSO4·7H2O. All cultivation media and plates contained 10 mg·L−1 sterile filtered gentamycin sulfate. Rapeseed oil (Edeka Zentrale AG & Co. KG, Germany) was used as the sole carbon source and urea as the sole nitrogen source in the MSM. The explicit amounts are described in the text. All chemicals were purchased from Carl Roth GmbH & Co. KG (Germany) unless stated otherwise.
R. eutropha H16 was streaked from a cryoculture on a TSB agar plate and incubated for 3–4 days at 30 °C. A single colony from the plate was used to inoculate the first preculture in 10 mL TSB media in a 125-mL Ultra Yield™ Flask (Thomson Instrument Company, USA) sealed with an AirOtop™ enhanced flask seal (Thomson Instrument Company, USA). After incubating for 16 h, 2.5 mL were used to inoculate the second preculture (250 mL MSM with 3% (w·v−1) rapeseed oil and 4.5 g·L−1 urea) in a 1-L DURAN® baffled glass flask with a GL45 thread (DWK Life Sciences GmbH, Germany) sealed with an AirOtop membrane. After 24 h of incubation, the complete second preculture was used to inoculate the main bioreactor culture. The precultures were incubated at 30 °C and shaken at 200 rpm (first preculture) or 180 rpm (second preculture) in an orbital shaker (Kühner LT-X incubator, Adolf Kühner AG, Switzerland, 50 mm amplitude).
2.3. Bioreactor Cultivation Conditions
Mineral salts dissolved in deionized (DI) water and rapeseed oil were added prior autoclavation in a 6.6-L stirred tank bioreactor with two six-blade Rushton impellers (BIOSTAT® Aplus, Sartorius AG, Germany). MgSO4, CaCl2, trace elements, gentamycin, and urea were added into the medium after autoclavation from sterile stock solutions. The temperature was maintained at 30 °C and the pH was kept constant at 6.8 ± 0.2 using 2 M NaOH and 1 M H3PO4 for pH control. The dissolved oxygen concentration (DO) was kept above 40% using a stirrer cascade ranging from 400 to 1350 rpm. The cultures were aerated with a constant aeration rate of 0.5 vvm throughout the cultivations. Five pairs of cable ties were mounted on the upper part of the stirrer shaft in order to break the foam mechanically and thus preventing overfoaming of the reactor.
2.3.1. Batch Cultivations
For a first evaluation of the PDW spectroscopy signal, three batch cultivations were performed in which the carbon and nitrogen content was varied. The concentrations of rapeseed oil were 3, 4, and 4% (w·v−1), and 2.25 (corresponding to 75 mM nitrogen), 4.5, and 2.25 g·L−1 for urea, respectively.
2.3.2. Pulse-Based Fed-Batch Cultivation
A cultivation strategy with a pulse feeding was performed in biological duplicates. The cultures initially contained 0.5% (w·v−1) rapeseed oil and 4.5 g·L−1 urea. Pulses were given whenever the PDW spectroscopy in-line signal (µs’ at 638 nm) indicated a decreased cell activity. After 8.2 h, the first pulse (15 g rapeseed oil) was added, followed by two more rapeseed oil pulses at 14.3 h (30 g) and at 21.1 h (60 g). At 31.7 h a pulse consisting of 110 mL urea solution (122 g·L−1), 15.6 mL 0.5 M K2SO4, 30 mL 0.042 M CaCl2, 30 mL 0.32 M MgSO4, and 3 mL trace element solution was added to restore the initial media concentrations of the components. The last pulse (120 g rapeseed oil) was added after 48.4 h.
2.3.3. Fed-Batch High-Cell-Density Cultivation
The culture initially contained 4% (w·v−1) rapeseed oil and 4.5 g·L−1 urea (150 mM nitrogen). Continuous feeding of pure rapeseed oil and a 30% (w·v−1) urea solution was started 7 h after inoculation with initial feeding rates of 3.5 g·h−1 and 0.39 mL·h−1, respectively. Both feeding rates were linearly increased up to 6.58 mL·h−1 (urea) at 16 h, after which the urea feed was stopped to cause nitrogen starvation, and 23 g·h−1 (rapeseed oil) at 35 h to final concentrations of 480 mM nitrogen and 170 g·L−1 rapeseed oil. A single injection of MgSO4, CaCl2, K2SO4, and trace elements was performed after 20 h with the amounts as described above (see Section 2.3.2.) to restore the initial concentrations and prevent nutrient depletion. An additional pulse of CaCl2 with the same concentration was added after 32 h.
2.4. Photon Density Wave Spectroscopy
A PDW spectrometer built by the University of Potsdam was used for the measurement of the absorption coefficient µa and the reduced scattering coefficient µs’. Identical devices are commercially available at PDW Analytics GmbH (Potsdam, Germany). The general set-up of the PDW spectrometer was described by Bressel et al., as follows: “A schematic set-up of the spectrometer is shown in Figure 1. Light from a laser diode with wavelength λ [m] (typ. 400–1000 nm) is sinusoidally intensity modulated by a vector network analyzer (typ. f = ω/(2π) = 10–1300 MHz). The light is then coupled into the material via an optical fiber, acting as a point-like light source. A second optical fiber, positioned at a distance r to the emission fiber (typ. r = 5–30 mm), collects light of the PDW and guides it onto an avalanche photodiode (APD) as detector.” [13].
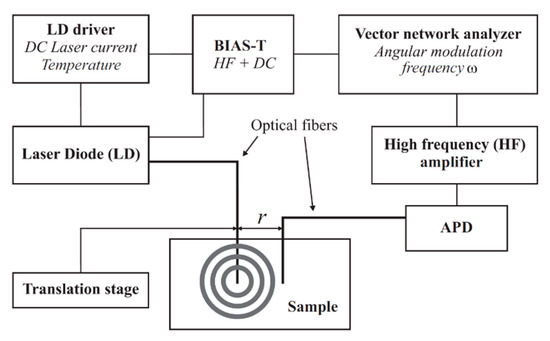
Figure 1.
Schematic experimental set-up of a photon density wave (PDW) spectrometer [13].
To integrate the multifiber PDW spectroscopy in-line probe into the system, a DN25 safety Ingold socket (elpotech GmbH & Co. KG, Germany) was welded onto the lid of the bioreactor. The probe was mounted before autoclaving and sterilized with the bioreactor inside the autoclave. The optical fibers of the probe were connected to the PDW spectrometer after autoclaving. µs’ and µa were analyzed at 638 nm with a temporal resolution of 0.8 min−1. A 10-point moving average was used to reduce the signal noise.
2.5. Analytical Methods
For each off-line reference analysis time point, two aliquots of 10 mL were sampled in preweighed 15-mL polypropylene test tubes. The samples were centrifuged for 15 min at 6000× g and pellets were washed either with a mixture of 5 mL cold deionized (DI) water and 2 mL cold hexane or with 7 mL cold DI water to remove residual lipids. The washed pellets were resuspended in 2–4 mL ice cold DI water, frozen at −80 °C, and dried for 24 h by lyophilization (Gamma 1–20, Martin Christ Gefriertrocknungsanlagen GmbH, Germany). Then the CDW was determined by weighing the test tubes.
The PHB content was determined off-line by high-performance liquid chromatography with a diode array detector (HPLC-DAD 1200 series, Agilent Technologies, USA). The method was adapted from Karr et al. [32]. Pure PHB (Sigma-Aldrich Corporation, USA) or 8–15 mg freeze-dried cells were depolymerized by boiling samples with 1 mL concentrated H2SO4 to yield crotonic acid. Dilution series of the depolymerized PHB were prepared to yield standards in the range of 0.1–10 mg·mL−1. Samples were diluted with 4 mL 5 mM H2SO4, filtered through a 0.2 µm cellulose acetate syringe filter, and subsequently 100 µL were transferred to a HPLC vial containing 900 µL of 5 mM H2SO4. HPLC analysis was performed with an injection volume of 20 µL using 5 mM H2SO4 as an eluent with an isocratic flow rate of 0.4 mL·min−1 for 60 min on a NUCLEOGEL® ION 300 OA column (Macherey-Nagel, Germany). Crotonic acid was detected at 210 nm. RCDW was determined by subtracting the PHB from the CDW concentration.
The nitrogen content was indirectly determined off-line by measuring ammonia, resulting from urea cleavage, in the supernatant using a pipetting robot (Cedex Bio HT Analyzer, Roche Diagnostics International AG, Switzerland) with the NH3 Bio HT test kit (Roche Diagnostics International AG, Switzerland).
During the fed-batch cultivation the in-line PDW spectroscopy signals were used for an on-line determination of the CDW, RCDW, and PHB content (see Section 3.4).
3. Results
A reduction of the PHA production price is crucial for commercialization [33], which can be facilitated by maximization of the biotechnological process performance using PAT. In this context, our group is interested in developing biotechnological processes for PHA production from renewable resources [24] and biogenic waste streams [17,18].
In this study, PDW spectroscopy was evaluated as a novel in-line tool to monitor the PHA production process with R. eutropha from plant oil.
3.1. Batch Cultivations
For an initial evaluation of the PDW spectroscopy signals, batch cultivations with different C/N ratios were performed to trigger different PHB and CDW accumulation. The results of the three batch cultivations are shown in Figure 2. Detailed graphs of the cultivations can be found in the in Supplementary Materials in Figures S1–S3. The first batch cultivation was performed as a reference batch containing 3% (w·v−1) rapeseed oil and 75 mM nitrogen (2.25 g·L−1 urea). Nitrogen limitation was indirectly detected by ammonia quantification, which is released from urea cleavage prior nitrogen uptake [24,28]. Nitrogen was depleted between 9.2 and 14.2 h, which triggered PHB accumulation. Within this period, the CDW increased from 6 to 15 g·L−1 and the PHB content increased from 7 to 35 wt% (0.4 to 5.3 gPHB·L−1). The PDW spectroscopy signals µa and µs’ did not show any analyzable signals until 6.5 h. Subsequently, both signals exponentially increased until 10.5 h. After this time point, µa linearly increased until 16 h and did not further rise afterwards. An increase of the µs’ signal was detected from 10.5 to 20 h and remained constant afterwards. The maximum CDW (25.5 g·L−1) was achieved after 33.2 h containing 65 wt% PHB (16.8 gPHB·L−1), which represents an overall yield of 0.56 gPHB·gOil−1. Over the entire cultivation, the urea consumption for biomass accumulation was 0.26 gUrea·gRCDW−1.
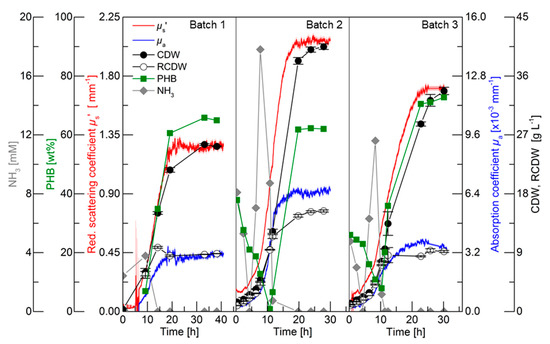
Figure 2.
Batch cultivations for polyhydroxybutyrate (PHB) production by R. eutropha H16. Batch 1 contained 3% (w·v−1) rapeseed oil and 2.25 g·L−1 urea (75 mM nitrogen), Batch 2 contained 4% (w·v−1) rapeseed oil and 4.5 g·L−1 urea, and Batch 3 contained 4% (w·v−1) rapeseed oil and 2.25 g·L−1 urea. Ammonia content (grey diamonds, mM), PHB content (green squares, wt%), cell dry weight (CDW) (filled circles, g·L−1), residual cell dry weight (RCDW) (empty circles, g·L−1), reduced scattering coefficient µs’ (red line, mm−1), and absorption coefficient µa (blue line, ×10−3 mm−1) at 638 nm are shown. Error bars indicate minimum and maximum values of technical duplicates.
The purpose of Batch 2 was to decrease the C/N ratio compared to the reference batch for an increased accumulation of active biomass (RCDW) and decreased PHB content. Nitrogen was depleted between 11.6 and 19.8 h. Within this period, the CDW and PHB content increased from 12.3 to 38.3 g·L−1 and 7 to 62 wt% (0.8 to 23.7 gPHB·L−1), respectively. The PDW spectroscopy signals µa and µs’ simultaneously increased from the beginning of the cultivation until 12 h in an exponential manner. No significant changes of µa were detected after that time point, whereas µs’ increased until 18 h and subsequently remained constant. After 23.8 h the maximum CDW of 40 g·L−1 containing 63 wt% (26.2 gPHB·L−1) was achieved. Overall, a PHB yield of 0.66 gPHB·gOil−1 and urea consumption of 0.29 gUrea·gRCDW−1 was achieved.
The C/N ratio in the third batch was increased compared to the reference batch by keeping the initial nitrogen concentration constant (75 mM) but increasing the rapeseed oil content to 4% (w·v−1). Depletion of nitrogen occurred between 10.3 and 11.3 h. Within this period the CDW increased from 7.7 g·L−1 with 8 wt% PHB (0.6 gPHB·L−1) to 9.6 g·L−1 with 20 wt% PHB (1.9 gPHB·L−1). PDW signals were detectable after 2 h. A comparable increase of µa and µs’ was detected until 10 h. An attenuated increase of µa until 17.5 h was detected, whereas a diminished increase of µs’ was detected from 10 to 22.5 h. The CDW further increased to 33.7 g·L−1 containing 73 wt% PHB (24.5 gPHB·L−1). Overall, a PHB yield of 0.61 gPHB·gOil−1 and urea consumption of 0.25 gUrea·gRCDW−1 was achieved.
To summarize, the C/N ratio influenced the yield coefficients for PHB accumulation and urea usage within these three batch cultivations. During the batch cultivations, µs’ and µa simultaneously increased until nitrogen depletion. Subsequently, µa leveled off and µs’ increased further until maximum PHB accumulation.
3.2. Pulse-Based Fed-Batch Cultivation
While the batch cultivations aimed to initially evaluate the relationship of the PDW spectroscopy signals with process relevant characteristics, a pulse-based fed-batch cultivation was conducted to: (i) show the feasibility to control the process by monitoring the process with PDW spectroscopy; (ii) confirm signal relationships; and (iii) validate the reproducibility during biological duplicate cultivations with independent seed trains. The process intention was implemented without difficulty: a pulse of either rapeseed oil or a nutrient bolus (see dashed lines in Figure 3 and details in the legend), respectively, was added to the culture when the µs’ signal showed no further changes and indicated either carbon or nitrogen limitation. It is worth mentioning, that no signal deflections were observed whenever oil pulses were added to the bioreactor, which would account for an effect of the added oil on µs’ or µa.
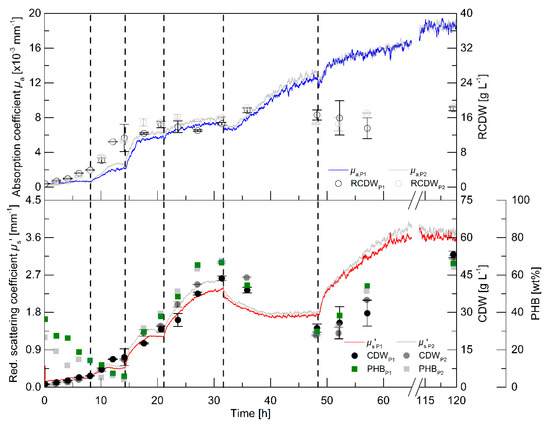
Figure 3.
Pulse feeding cultivations for PHB production by R. eutropha H16. The cultures were initially started with 0.5% (w·v−1) rapeseed oil and 4.5 g·L−1 urea (150 mM nitrogen). The dashed vertical lines represent time points of pulse additions: 0.5% (w·v−1) rapeseed oil at 8.2 h, 1% (w·v−1) rapeseed oil at 14.3 h, and 2% (w·v−1) rapeseed oil at 21.1 h. At 31.7 h a bolus consisting of 110 mL urea solution (122 g·L−1), 15.6 mL 0.5 M K2SO4, 30 mL 0.32 M MgSO4, 30 mL 0.042 mM CaCl2, and 3 mL trace element solution was added to the bioreactor. A final bolus of 4% (w·v−1) rapeseed oil was added at 48.4 h. Data from the reference experiment is shown in color (indexed P1) and the biological duplicate (with an independent seed train) in transparent grey (indexed P2). Absorption coefficient µa (upper graph, solid line, ×10−3 mm−1), RCDW (upper graph, empty circles, g·L−1), PHB content (bottom graph, squares, wt%), CDW (bottom graph, filled circles, g·L−1), and reduced scattering coefficient µs’ (bottom graph, solid line, mm−1) are shown. Error bars indicate minimum and maximum values of technical duplicates.
Within the first 8 h, the intracellular PHB content decreased from 36 to 14 wt% (Figure 3). An increase of the CDW, RCDW, µs’ and µa was observed in the first 6 h of the cultivation. Subsequently, only minor increases of the CDW and RCDW of about 0.7 g·L−1 were detected until 8 h, whereas the PDW spectroscopy signals did not further increase during this period. After addition of the first pulse (0.5% (w·v−1) rapeseed oil), µs’ and µa resumed to increase until reaching constant levels at 11.5 h until the next pulse addition (1% (w·v−1) rapeseed oil). The increase of µa stopped at 16.5 h, whereas µs’ increased further until 18 h. Nitrogen depletion was detected at 17.5 h (Figure S4, Supplementary Materials). The PHB content had already increased to 30 wt% at this time point and further increased to 38 wt% before addition of the next pulse. The CDW increased up to 23 g·L−1, whereas the RCDW stopped at a value of 14 g·L−1 at 20.5 h. Addition of the next rapeseed oil pulse (2% (w·v−1)) at 21 h did not trigger a significant change of the µs’ signal and of the RCDW. In contrast, a sharp increase of µs’ resulted from the rapeseed oil addition and the CDW increased up to 43 g·L−1 at 31 h containing 66 wt% PHB. At 31.6 h, a bolus containing urea as a nitrogen source was supplemented to the culture. The addition resulted in a dilution of the culture, which was seen in a step decrease of both PDW spectroscopy signals at this time point. Subsequently, µs’ decreased until 42 h and stayed constant until the next pulse addition. In contrast, µa resumed to increase until 45 h. The CDW decreased during this period to 24 g·L−1, resulting from intracellular PHB degradation. The PHB content decreased to 30 wt%. At the same time, the RCDW increased to 17 g·L−1. The addition of the next pulse (4% (w·v−1) rapeseed oil) resulted in resumed growth on rapeseed oil as the primary carbon source instead of degrading the intracellular carbon storage. The PDW spectroscopy signals increased after the rapeseed oil supplementation. The scattering signal µs’ increased until 70 h, whereas the absorption coefficient µa only slightly increased after 52 h. At 52 h, nitrogen was depleted again. The cells accumulated 66 wt% PHB until the end of the cultivation and the CDW increased to 53 g·L−1. When the whole cultivation was repeated, the PDW spectroscopy signals showed an equivalent course and a final CDW of 52 g·L−1 CDW containing 64 wt% PHB, indicating a high robustness, i.e., repeatability, of the process.
The pulse-based fed-batch experiment showed the possibility to control the rapeseed oil-based cultivation for PHB production using the in-line PDW spectroscopy probe. The highly reproducible course of the PDW spectroscopy signals strongly imply the connection of biological events with this measurement technique. The coefficients µs’ and µa show the same trend as the CDW and RCDW, respectively. Nevertheless, a more significant change was observed for the µa signal after addition of the nitrogen pulse than the off-line determined RCDW. Regardless of this discrepancy, the hypothesis was that the µs’ and µa correspond and can be correlated with the CDW and RCDW, respectively.
3.3. PDW Spectroscopy Signal Correlation
For an analysis of the correlation between the PDW spectroscopy signals µs’ and µa at 638 nm, respectively, with process relevant characteristics, the experiments described above were analyzed. The reduced scattering coefficient µs’ followed the course of the CDW, while the absorption coefficient µa increased with the rise of RCDW. The correlation of the respective values from all five cultivations (i.e., batch and pulse-based fed-batch cultures) are shown in Figure 4. A root mean squared error of 0.96 for the linear correlation of µs’ and the CDW was obtained, whereas the linear correlation of µa and the RCDW resulted in a R2 of 0.90. Equations (A1)–(A3) in Appendix A show the obtained formulas for calculating the CDW, RCDW, and subsequently the PHB concentration using the linear relationships of the in-line PDW spectroscopy signals with the corresponding off-line values. Due to disproportionally large PDW spectroscopy signal intensification after 48 h of the pulse feeding experiments (Figure 3), the last four data points of each replicate were not included in the correlation analysis.
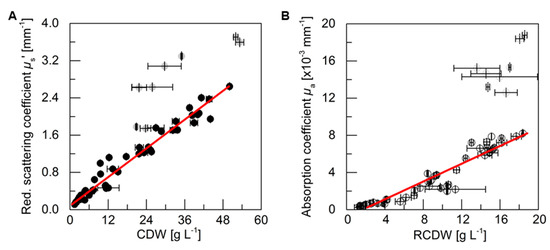
Figure 4.
Correlation analysis of PDW spectroscopy signals with off-line values. (A) Reduced scattering coefficient µs’ at 638 nm is correlated with CDW. (B) Absorption coefficient µa at 638 nm is correlated with RCDW. Data points are from five cultivations with different rapeseed oil and urea contents (cf. Figure 2 and Figure 3). The gray data points comprise the last four samples of the two pulse-based fed-batch experiments (cf. Figure 3), which differ significantly from the other samples and were therefore not considered for the linear fit with the experimental data. A squared correlation coefficient R2 of 0.96 was obtained for µs’ and CDW and R2 of 0.90 for µa and RCDW. Error bars indicate minimum and maximum values of technical duplicates (CDW, RCDW). STDEV of the 10-point average is shown for µs’ and µa.
3.4. High-Cell-Density Fed-Batch Cultivation
High-cell-density cultivations are essential for a competitive production of PHA biopolymers. Therefore, it was aimed to perform such a cultivation to evaluate the PDW spectroscopy probe performance at industrial relevant biomass concentrations. The correlation factors obtained from the first five cultivations (Equations (A1)–(A3) in Appendix A) were used to calculate the CDW, RCDW, and PHB content from the in-line PDW spectroscopy signals (Figure 5).
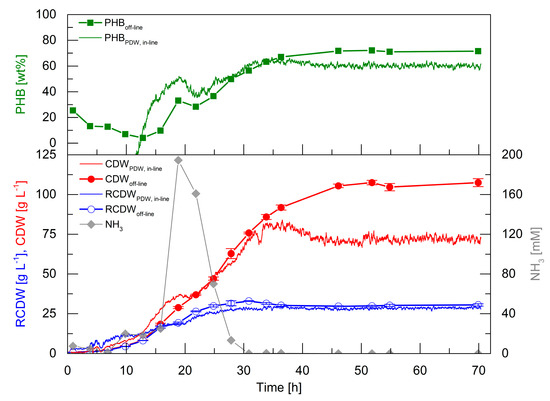
Figure 5.
Fed-batch cultivation of R. eutropha H16 for PHB production. The culture was started with 4% (w·v−1) rapeseed oil and 4.5 g·L−1 urea (150 mM nitrogen). Rapeseed oil feeding linearly increased from 7 h with an initial feeding rate of 3.5 g·h−1 to 35 h with a final feeding rate of 23 g·h−1 up to a total concentration of 17% (w·v−1). Urea (30% (w·v−1)) feeding was linearly increased from 7 h with an initial feeding rate of 0.39 mL·h−1 up to a final feeding rate of 6.58 mL·h−1 at 16 h to a total concentration of 14.4 g·L−1. In-line PHB content (upper graph, green line, wt%) by PDW spectroscopy and off-line PHB content (upper graph, green squares, wt%), estimated in-line CDW by PDW spectroscopy (bottom graph, red line, g·L−1), off-line CDW (bottom graph, red filled circles, g·L−1), estimated in-line RCDW by PDW spectroscopy (bottom graph, blue line, g·L−1), off-line RCDW (bottom graph, blue empty circles, g·L−1), and ammonia content (bottom graph, grey diamonds, mM) are shown. Error bars indicate minimum and maximum values of technical duplicates.
In the first 13 h, the off-line CDW increased to 8.5 g·L−1 and the RCDW to 8.2 g·L−1. In the same period, the PHB content decreased to 4 wt%. Within this time frame, the in-line signals overestimated CDW and RCDW to 13.9 and 12.3 g·L−1 at 13 h, respectively. The estimated RCDW was higher than the estimated CDW until 12 h, which yielded a negative calculated PHB content. An accumulation of ammonia up to 195 mM at 19 h was detected, which subsequently decreased and was depleted after 28 h. The CDW and RCDW increased up to 63 and 32 g·L−1, respectively, during this period. Contrary to expectation, PHB accumulation was detected from 13 to 19 h up to 33 wt%. Subsequently, the PHB content decreased again to 28 wt% during ammonia consumption and further accumulation started after 25 h. Subsequently, the PHB content increased to 72 wt% at 46 h. The preliminary accumulation and degradation of PHB until 25 h was also detected by the calculated in-line PHB signal. Subsequently, the in-line PHB content increased to 61 wt% at 46 h and ceased afterwards. At 46 h, the off-line CDW and RCDW had increased to 106 and 30 g·L−1, respectively. In total, 76 g·L−1 PHB accumulated in 46 h, which represents a space time yield of 1.65 gPHB·L−1·h−1 and a yield coefficient of 0.43 gPHB·gOil−1. The overall urea usage was 0.48 gUrea·gRCDW−1. The in-line RCDW signal indicated an accumulation until 29 h to a final RCDW of 29 g·L−1. The in-line CDW signal increased simultaneously with the off-line CDW until 36 h to 82 g·L−1. Unexpectedly, the in-line CDW signal decreased afterwards until 46 h and stayed constant at a value of 71 g·L−1.
The high-cell-density cultivation showed that PDW spectroscopy is capable of a qualitative tracking of the CDW, RCDW, and PHB content. Nevertheless, the quantitative accuracy was not precise during the first 12 h of the cultivation at low cell densities. However, the estimated in-line CDW showed a good representation until 36 h. Also, the in-line RCDW was estimated from the µa with a very good accuracy for the rest of the cultivation. In contrast, a drop of the µs’ signal after 36 h did not correlate with CDW during PHB production.
4. Discussion
The purpose of this study was to evaluate the potential of PDW spectroscopy for monitoring plant oil-based R. eutropha cultivations. The batch (Figure 2) and pulse-based fed-batch (Figure 3) cultivations showed that the reduced scattering coefficient µs’ correlates strongly with the CDW and the absorption coefficient µa with the RCDW (Figure 4). These results demonstrate that PDW spectroscopy is a valuable tool for in-line monitoring of the CDW, RCDW, and PHB accumulation. To the best of our knowledge, the results of this study are the first data showing in-line quantification of PHB. The lack of such in-line or on-line monitoring devices for an adaptive control of the production process was recently emphasized by Koller et al. [3]. Previously, Cruz et al. reported the possibility to use a NIR transflectance probe for an in-line quantification of PHB. However, the authors showed only at-line data quantifying the PHB and CDW concentration (up to 9.3 g·L−1 and 13.7 g·L−1, respectively) during batch cultivations [34].
During the initial batch cultivations, PHB yields of 0.56–0.66 gPHB·gOil−1 were obtained (Figure 2), which are similar to previously reported yields for R. eutropha H16 growth on palm oil [35]. During the high-cell-density fed-batch cultivation 106 g·L−1 CDW (72 wt% PHB) with a space time yield of 1.65 gPHB·L−1·h−1 were reached, which is comparable to other published high-cell-density plant oil cultivations [21,24,25,29,36,37].
In the pulse experiment, a nitrogen bolus was added (32 h) after the PHB production phase to trigger PHB degradation, as described previously [38]. The PDW spectroscopy signal µs‘ decreased with the declining CDW while µa increased with an increasing RCDW (Figure 3). Currently, we do not understand why the strength of the signal was not proportional with the determined off-line value changes after that time point (32 h). For this reason, these measurement points were not used for the linear correlation. A potential hypothesis could be an unequal distribution of PHA granules during PHA mobilization, as it was reported for Pseudomonas putida [39], which might have an effect on scattering and absorption coefficients during PHA degradation.
Atypical PHB formation before nitrogen depletion was detected during the high-cell-density cultivation (Figure 5). This preliminary formation of PHB could explain the low yield coefficient of 0.43 gPHB·gOil−1, which is significant lower than the typical yield of PHB in R. eutropha plant oil cultivations [37]. The formation of PHB without nutrient starvation could indicate a stress response triggered by the high urea levels. Stress responses typically involve the formation of the alarmone (p)ppGpp. For R. eutropha it is known that formation of this alarmone triggers PHB formation [38,40], but (p)ppGpp formation due to excess urea or ammonia availability was not studied so far. Additionally, it was reported that controlled induction of stress could also been used for an enhanced PHB formation [41]. Such stress responses should be thoroughly considered during the scale-up of a R. eutropha PHA production process, as zones of high or low substrate availability occur in large scale bioreactors. The high impact of such substrate gradients on a reduction of biomass and product yields were intensively studied for Escherichia coli [42]. Adapting the feeding strategy during the PHA production process by using in-line monitoring devices could be a potential scenario for avoiding such negative impacts on the process.
A reduction of the µs’ signal after 35 h was observed during the high-cell-density fed-batch cultivation, whereas µa stayed constant during that period (Figure 5). The decrease of µs’ instead of a leveling off of the signal contradicts that a signal saturation effect was observed. Additionally, scattering coefficients in suspensions with particle contents of up to 40% (v·v−1) were measured successfully with this technology [9,10]. Heavy foaming, which occurred after 35 h, could be the reason for the observed signal reductions. The surplus of foam was constantly forced into the liquid phase, which increased the overall gas hold-up and total reaction volume in the system. This additional gas volume results in a dilution of the system, which could explain the µs’ decrease (35–45 h) even though the culture continued to accumulate PHB (Figure 5). The foaming occurred after the end of the continuous rapeseed oil feed at 35 h. Before 35 h, the added oil functioned as a natural antifoam agent by decreasing the surface tension of the culture broth. During plant oil cultivations foaming occurs through the emulsification process. R. eutropha emulsifies plant oils before uptake, which is catalyzed by extracellular lipases. The lipases cleave the triacylglycerols in diacylglycerols, monoacylglycerols, glycerol, and free fatty acids (FFAs) [23,43,44], which causes heavy foaming during aerated bioreactor cultivations. Nevertheless, µs’ stayed constant after the CDW did not further increase at 45 h, which could indicate the perfect time point for harvesting in an industrial process. A reliable in-line quantification of CDW, RCDW, and subsequently PHA concentration was reached until a CDW of 84 g·L−1. To increase the robustness of the method further at higher cell densities, the calculated gas-hold up in the bioreactor [45] could be integrated in the correlation of the PDW spectroscopy signals. In order to quantify the direct impact of the PHA concentration on the optical coefficients, further studies referencing cell counts and sizes by flow cytometry and microscopy need to be conducted.
A wavelength of 638 nm was used to evaluate the PDW spectroscopy signals during this study, which did not show any correlations with the oil addition or emulsification (Figure 3). The emulsification process is very important for an efficient growth. It was previously shown that an overexpression of lipases results in a reduced lag phase and subsequently a more efficient process [43]. In previous studies, it was shown that PDW spectroscopy was used to measure emulsions [46,47]. An in-line determination of the oil content or the emulsion formation would be a very valuable additional information for bioprocess development and process control. This information might result from integration of additional wavelengths into the PDW spectroscopy set-up.
To summarize, PDW spectroscopy allows in-line estimation of the CDW, RCDW, and PHB content in real-time. In contrast, off-line analysis is typically carried out to determine the PHB content, which includes drying the cells and polymer derivatization before time-consuming HPLC or GC analysis. By using PDW spectroscopy, process development and scale-up could be accelerated. In addition, this technology could be used at a large scale for process monitoring and control of R. eutropha cultivations. Specifically, the real-time adjustment of feeding strategies according to the PHA production rates—determined by PDW spectroscopy signals—holds great potential.
5. Conclusions
Here, we show that in-line PDW spectroscopy is a powerful PAT tool for monitoring R. eutropha-based PHB production. The reduced scattering coefficient µs’ and absorption coefficient µa showed very reproducible signals during different biological cultivations. The new method described in this study allows in-line monitoring of CDW, RCDW, and PHB concentrations in R. eutropha cultivations up to a CDW of 84 g·L−1. PDW spectroscopy could contribute to improving the scaling-up process and thus to performing PHA production processes in an economical efficient way with the ultimate goal to commercialize a green sustainable plastic.
Supplementary Materials
The following are available online at https://www.mdpi.com/2306-5354/6/3/85/s1, Figure S1: Detailed figure of Batch 1, Figure S2: Detailed figure of Batch 2, Figure S3: Detailed figure of Batch 3, Figure S4: Ammonia contents from the pulse feed cultivations.
Author Contributions
Conceptualization, B.G., T.S., R.H., S.L.R.; Methodology, B.G., T.S., R.H., S.L.R.; Validation, B.G., T.S.; Formal analysis, B.G., T.S.; Investigation, B.G., T.S., M.T.H.W.; Resources, P.N., R.H., S.L.R.; Data curation, B.G., T.S.; Writing—original draft preparation, B.G., S.L.R.; Writing—review and editing, B.G., T.S., M.T.H.W., P.N., R.H., S.L.R.; Visualization, B.G.; Supervision, R.H., S.L.R.; Project administration, S.L.R.; Funding acquisition, R.H., S.L.R., P.N.
Funding
This research was funded by the German Federal Ministry of Education and Research, grant number 03Z22AN12.
Acknowledgments
We thank Roche CustomBiotech (Mannheim, Germany) for the supply of the Cedex Bio HT Analyzer. We thank Thomas Högl for helping with the installation of the PDW spectroscopy probe. We acknowledge support by the Open Access Publication Funds of TU Berlin.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
CDW [g·L−1] = 19.012 [mm·g·L−1] µs’ [mm−1] − 1.2404 [g·L−1] R2 = 0.96
RCDW [g·L−1] = 1872 [mm·g·L−1] µa [mm−1] + 2.5722 [g·L−1] R2 = 0.90
PHB [wt%] = 100 [wt%] (CDW [g·L−1] – RCDW [g·L−1])/CDW [g·L−1]
References
- FDA Guidance for Industry PAT: A Framework for Innovative Pharmaceutical Development, Manufacuring, and Quality Assurance. FDA Off. Doc.; 2004. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/pat-framework-innovative-pharmaceutical-development-manufacturing-and-quality-assurance (accessed on 18 September 2019).
- Gomes, J.; Chopda, V.R.; Rathore, A.S. Integrating systems analysis and control for implementing process analytical technology in bioprocess development. J. Chem. Technol. Biotechnol. 2015, 90, 583–589. [Google Scholar] [CrossRef]
- Karmann, S.; Follonier, S.; Bassas-Galia, M.; Panke, S.; Zinn, M. Robust at-line quantification of poly(3-hydroxyalkanoate) biosynthesis by flow cytometry using a BODIPY 493/503-SYTO 62 double-staining. J. Microbiol. Methods 2016, 131, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, S.H.; Yim, S.S.; Kang, K.; Lee, S.Y.; Park, S.J.; Jeong, K.J. Quantified High-Throughput Screening of Escherichia coli Producing Poly (3-hydroxybutyrate ) Based on FACS. Appl. Biochem. Biotechnol. 2013, 170, 1767–1779. [Google Scholar] [CrossRef] [PubMed]
- Samek, O.; Obruča, S.; Šiler, M.; Sedláček, P.; Benešová, P.; Kučera, D.; Márova, I.; Ježek, J.; Bernatová, S.; Zemánek, P. Quantitative Raman Spectroscopy Analysis of Polyhydroxyalkanoates Produced by Cupriavidus necator H16. Sensors 2016, 16, 1808. [Google Scholar] [CrossRef] [PubMed]
- Porras, M.A.; Cubitto, M.A.; Villar, M.A. A new way of quantifying the production of poly(hydroxyalkanoate)s using FTIR. J. Chem. Technol. Biotechnol. 2016, 91, 1240–1249. [Google Scholar] [CrossRef]
- Hesselmann, R.P.X.; Fleischmann, T.; Hany, R.; Zehnder, A.J.B. Determination of polyhydroxyalkanoates in activated sludge by ion chromatographic and enzymatic methods. J. Microbiol. Methods 1999, 35, 111–119. [Google Scholar] [CrossRef]
- Koller, M.; Rodríguez-Contreras, A. Techniques for tracing PHA-producing organisms and for qualitative and quantitative analysis of intra- and extracellular PHA. Eng. Life Sci. 2015, 15, 558–581. [Google Scholar] [CrossRef]
- Hass, R.; Munzke, D.; Vargas Ruiz, S.; Tippmann, J.; Reich, O. Optical monitoring of chemical processes in turbid biogenic liquid dispersions by Photon Density Wave spectroscopy. Anal. Bioanal. Chem. 2015, 407, 2791–2802. [Google Scholar] [CrossRef]
- Münzberg, M.; Hass, R.; Dinh Duc Khanh, N.; Reich, O. Limitations of turbidity process probes and formazine as their calibration standard. Anal. Bioanal. Chem. 2017, 409, 719–728. [Google Scholar] [CrossRef]
- Häne, J.; Brühwiler, D.; Ecker, A.; Hass, R. Real-time inline monitoring of zeolite synthesis by Photon Density Wave spectroscopy. Microporous Mesoporous Mater. 2019, 288, 109580. [Google Scholar] [CrossRef]
- Hass, R.; Münzberg, M.; Bressel, L.; Reich, O. Industrial applications of photon density wave spectroscopy for in-line particle sizing. Appl. Opt. 2013, 52, 1429–1431. [Google Scholar] [CrossRef] [PubMed]
- Bressel, L.; Hass, R.; Reich, O. Particle sizing in highly turbid dispersions by Photon Density Wave spectroscopy. J. Quant. Spectrosc. Radiat. Transf. 2013, 126, 122–129. [Google Scholar] [CrossRef]
- Hass, R.; Reich, O. Photon density wave spectroscopy for dilution-free sizing of highly concentrated nanoparticles during starved-feed polymerization. ChemPhysChem 2011, 12, 2572–2575. [Google Scholar] [CrossRef] [PubMed]
- Koller, M.; Shahzad, K.; Braunegg, G. Waste streams of the animal-processing industry as feedstocks to produce polyhydroxyalkanoate biopolyesters. Appl. Food Biotechnol. 2018, 5, 193–203. [Google Scholar]
- Kourmentza, C.; Plácido, J.; Venetsaneas, N.; Burniol-Figols, A.; Varrone, C.; Gavala, H.N.; Reis, M.A.M. Recent Advances and Challenges towards Sustainable Polyhydroxyalkanoate (PHA) Production. Bioengineering 2017, 4, 55. [Google Scholar] [CrossRef] [PubMed]
- Riedel, S.L.; Jahns, S.; Koenig, S.; Bock, M.C.E.; Brigham, C.J.; Bader, J.; Stahl, U. Polyhydroxyalkanoates production with Ralstonia eutropha from low quality waste animal fats. J. Biotechnol. 2015, 214, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Brigham, C.J.; Riedel, S.L. The Potential of Polyhydroxyalkanoate Production from Food Wastes. Appl. Food Biotechnol. 2018, 6, 7–18. [Google Scholar]
- Ong, S.Y.; Kho, H.P.; Riedel, S.L.; Kim, S.W.; Gan, C.Y.; Taylor, T.D.; Sudesh, K. An integrative study on biologically recovered polyhydroxyalkanoates (PHAs) and simultaneous assessment of gut microbiome in yellow mealworm. J. Biotechnol. 2018, 265, 31–39. [Google Scholar] [CrossRef]
- Riedel, S.L.; Brigham, C.J.; Budde, C.F.; Bader, J.; Rha, C.; Stahl, U.; Sinskey, A.J. Recovery of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from Ralstonia eutropha cultures with non-halogenated solvents. Biotechnol. Bioeng. 2013, 110, 461–470. [Google Scholar] [CrossRef]
- Obruca, S.; Marova, I.; Snajdar, O.; Mravcova, L.; Svoboda, Z. Production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by Cupriavidus necator from waste rapeseed oil using propanol as a precursor of 3-hydroxyvalerate. Biotechnol. Lett. 2010, 32, 1925–1932. [Google Scholar] [CrossRef]
- Reinecke, F.; Steinbüchel, A. Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J. Mol. Microbiol. Biotechnol. 2008, 16, 91–108. [Google Scholar] [CrossRef] [PubMed]
- Brigham, C.J.; Budde, C.F.; Holder, J.W.; Zeng, Q.; Mahan, A.E.; Rha, C.K.; Sinskey, A.J. Elucidation of β-oxidation pathways in Ralstonia eutropha H16 by examination of global gene expression. J. Bacteriol. 2010, 192, 5454–5464. [Google Scholar] [CrossRef] [PubMed]
- Riedel, S.L.; Bader, J.; Brigham, C.J.; Budde, C.F.; Yusof, Z.A.M.; Rha, C.; Sinskey, A.J. Production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) by Ralstonia eutropha in high cell density palm oil fermentations. Biotechnol. Bioeng. 2012, 109, 74–83. [Google Scholar] [CrossRef]
- Kahar, P.; Tsuge, T.; Taguchi, K.; Doi, Y. High yield production of polyhydroxyalkanoates from soybean oil by Ralstonia eutropha and its recombinant strain. Polym. Degrad. Stab. 2004, 83, 79–86. [Google Scholar] [CrossRef]
- Rehm, B.H.A. Polyester synthases: Natural catalysts for plastics. Biochem. J. 2003, 376, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Pohlmann, A.; Fricke, W.F.; Reinecke, F.; Kusian, B.; Liesegang, H.; Cramm, R.; Eitinger, T.; Ewering, C.; Pötter, M.; Schwartz, E.; et al. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 2006, 24, 1257–1262. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.S.; Ooi, W.Y.; Goh, L.K.; Shenbagarathai, R.; Sudesh, K. Evaluation of jatropha oil to produce poly(3-hydroxybutyrate) by Cupriavidus necator H16. Polym. Degrad. Stab. 2010, 95, 1365–1369. [Google Scholar] [CrossRef]
- Arikawa, H.; Matsumoto, K. Evaluation of gene expression cassettes and production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) with a fine modulated monomer composition by using it in Cupriavidus necator. Microb. Cell Fact. 2016, 15, 1–11. [Google Scholar] [CrossRef]
- Arikawa, H.; Matsumoto, K.; Fujiki, T. Polyhydroxyalkanoate production from sucrose by Cupriavidus necator strains harboring csc genes from Escherichia coli W. Appl. Microbiol. Biotechnol. 2017, 101, 7497–7507. [Google Scholar] [CrossRef]
- Sato, S.; Maruyama, H.; Fujiki, T.; Matsumoto, K. Regulation of 3-hydroxyhexanoate composition in PHBH synthesized by recombinant Cupriavidus necator H16 from plant oil by using butyrate as a co-substrate. J. Biosci. Bioeng. 2015, 120, 246–251. [Google Scholar] [CrossRef]
- Karr, D.B.; Waters, J.K.; Emerich, D.W. Analysis of poly-β-hydroxybutyrate in Rhizobium japonicum bacteroids by ion-exclusion high-pressure liquid chromatography and UV detection. Appl. Environ. Microbiol. 1983, 46, 1339–1344. [Google Scholar] [PubMed]
- Koller, M. Advances in Polyhydroxyalkanoate (PHA) Production. Bioengineering 2017, 4, 88. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.V.; Sarraguça, M.C.; Freitas, F.; Lopes, J.A.; Reis, M.A.M. Online monitoring of P(3HB) produced from used cooking oil with near-infrared spectroscopy. J. Biotechnol. 2015, 194, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Budde, C.F.; Riedel, S.L.; Hübner, F.; Risch, S.; Popović, M.K.; Rha, C.; Sinskey, A.J. Growth and polyhydroxybutyrate production by Ralstonia eutropha in emulsified plant oil medium. Appl. Microbiol. Biotechnol. 2011, 89, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Fujiki, T.; Matsumoto, K. Construction of a stable plasmid vector for industrial production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) by a recombinant Cupriavidus necator H16 strain. J. Biosci. Bioeng. 2013, 116, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Fadzil, F.I.B.M.; Tsuge, T. Bioproduction of polyhydroxyalkanoate from plant oils. In Microbial Applications; Springer: Berlin, Germany, 2017; Volume 2, pp. 231–260. ISBN 9783319526690. [Google Scholar]
- Juengert, J.R.; Borisova, M.; Mayer, C.; Wolz, C.; Brigham, C.J.; Sinskey, A.J.; Jendrossek, D. Absence of ppGpp leads to increased mobilization of intermediately accumulated poly(3-hydroxybutyrate) in Ralstonia eutropha H16. Appl. Environ. Microbiol. 2017, 83, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Karmann, S.; Panke, S.; Zinn, M. The Bistable Behaviour of Pseudomonas putida KT2440 during PHA Depolymerization under Carbon Limitation. Bioengineering 2017, 4, 58. [Google Scholar] [CrossRef] [PubMed]
- Brigham, C.J.; Speth, D.R.; Rha, C.K.; Sinskey, A.J. Whole-genome microarray and gene deletion studies reveal regulation of the polyhydroxyalkanoate production cycle by the stringent response in Ralstonia eutropha H16. Appl. Environ. Microbiol. 2012, 78, 8033–8044. [Google Scholar] [CrossRef] [PubMed]
- Obruca, S.; Marova, I.; Svoboda, Z.; Mikulikova, R. Use of controlled exogenous stress for improvement of poly(3-hydroxybutyrate) production in Cupriavidus necator. Folia Microbiol. 2010, 55, 17–22. [Google Scholar] [CrossRef]
- Enfors, S.O.; Jahic, M.; Rozkov, A.; Xu, B.; Hecker, M.; Jürgen, B.; Krüger, E.; Schweder, T.; Hamer, G.; O’Beirne, D.; et al. Physiological responses to mixing in large scale bioreactors. J. Biotechnol. 2001, 85, 175–185. [Google Scholar] [CrossRef]
- Lu, J.; Brigham, C.J.; Rha, C.; Sinskey, A.J. Characterization of an extracellular lipase and its chaperone from Ralstonia eutropha H16. Appl. Microbiol. Biotechnol. 2013, 97, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Riedel, S.L.; Lu, J.; Stahl, U.; Brigham, C.J. Lipid and fatty acid metabolism in Ralstonia eutropha: Relevance for the biotechnological production of value-added products. Appl. Microbiol. Biotechnol. 2014, 98, 1469–1483. [Google Scholar] [CrossRef] [PubMed]
- Sieblist, C.; Lübbert, A. Gas Holdup in Bioreactors. In Encyclopedia of Industrial Biotechnology; John Wiley & Sons: New York, NY, USA, 2010; pp. 1–8. [Google Scholar]
- Reich, O.; Bressel, L.; Hass, R. Sensing emulsification processes by Photon Density Wave spectroscopy. In the 21st International Conference on Optical Fiber Sensors; The Institute of Electrical and Electronics Engineers: New York, NY, USA, 2011; Volume 7753, p. 77532J. [Google Scholar]
- Münzberg, M.; Hass, R.; Reich, O. In-line characterization of phase inversion temperature emulsification by photon density wave spectroscopy. SOFW J. 2013, 4, 38–46. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).