Assessment of Skeletal Tumor Load in Metastasized Castration-Resistant Prostate Cancer Patients: A Review of Available Methods and an Overview on Future Perspectives
Abstract
1. Introduction
2. Methods
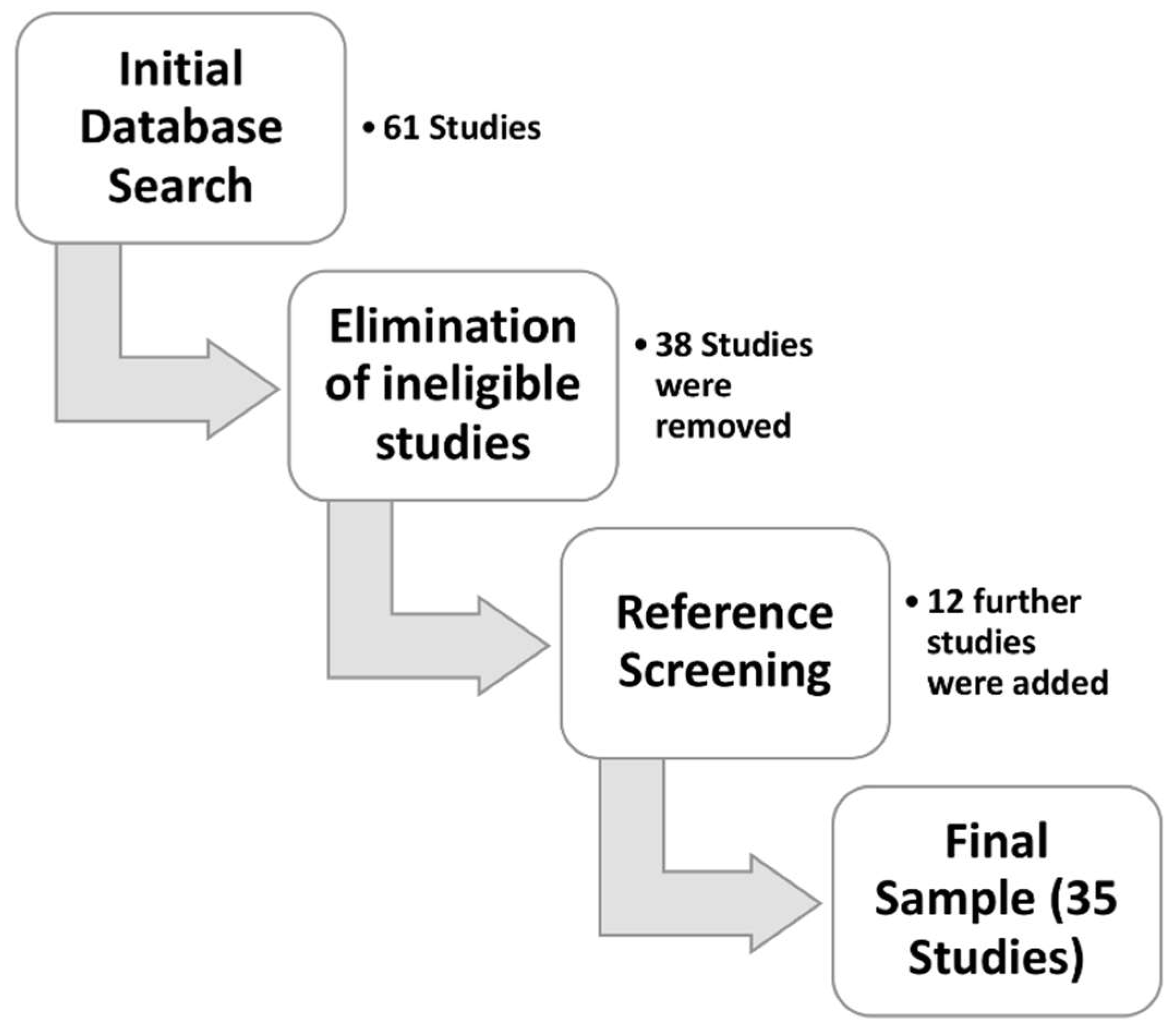
2.1. Search Strategy
2.2. Study Selection
2.3. Article Categorization
3. Results and Discussion
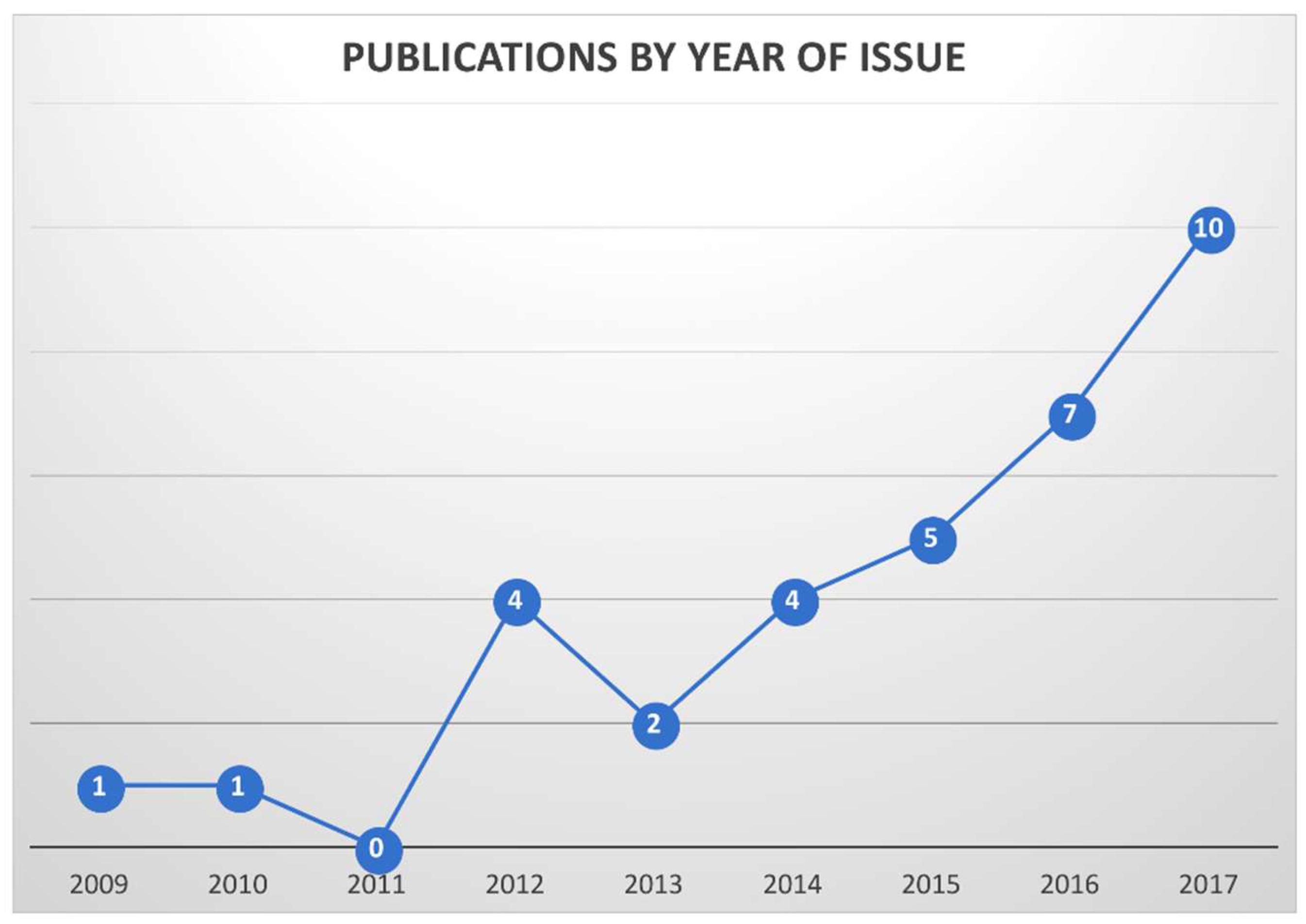
3.1. General Parameter
3.2. Types of Computational Approaches
3.2.1. Automated 2D Analysis of Bone Scans
3.2.2. SUV-Based Thresholding
3.2.3. Hybrid CT- and SUV-Based Thresholding
3.2.4. MR-Based and other Non-Isotopic Methods
4. Conclusions
Funding
Conflicts of Interest
References
- Jemal, A.; Fedewa, S.A.; Ma, J.; Siegel, R.; Lin, C.C.; Brawley, O.; Ward, E.M. Prostate Cancer Incidence and PSA Testing Patterns in Relation to USPSTF Screening Recommendations. JAMA 2015, 314, 2054–2061. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, A.; Bastian, P.J.; Bellmunt, J.; Bolla, M.; Joniau, S.; van der Kwast, T.; Mason, M.; Matveev, V.; Wiegel, T.; Zattoni, F.; et al. EAU guidelines on prostate cancer. Part II: Treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur. Urol. 2014, 65, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Quayle, L.; Ottewell, P.D.; Holen, I. Bone Metastasis: Molecular Mechanisms Implicated in Tumour Cell Dormancy in Breast and Prostate Cancer. Curr. Cancer Drug Targets 2015, 15, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Loriot, Y.; Bianchini, D.; Ileana, E.; Sandhu, S.; Patrikidou, A.; Pezaro, C.; Albiges, L.; Attard, G.; Fizazi, K.; De Bono, J.S.; et al. Antitumour activity of abiraterone acetate against metastatic castration-resistant prostate cancer progressing after docetaxel and enzalutamide (MDV3100). Ann. Oncol. 2013, 24, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Kantoff, P.W.; Higano, C.S.; Shore, N.D.; Berger, E.R.; Small, E.J.; Penson, D.F.; Redfern, C.H.; Ferrari, A.C.; Dreicer, R.; Sims, R.B.; et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 2010, 363, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Berthold, D.R.; Pond, G.R.; Soban, F.; de Wit, R.; Eisenberger, M.; Tannock, I.F. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer: Updated survival in the TAX 327 study. J. Clin. Oncol. 2008, 26, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.; Nilsson, S.; Heinrich, D.; Helle, S.I.; O’Sullivan, J.M.; Fossa, S.D.; Chodacki, A.; Wiechno, P.; Logue, J.; Seke, M.; et al. Alpha emitter radium-223 and survival in metastatic prostate cancer. N. Engl. J. Med. 2013, 369, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Omlin, A.; Pezaro, C.; Mukherji, D.; Mulick Cassidy, A.; Sandhu, S.; Bianchini, D.; Olmos, D.; Ferraldeschi, R.; Maier, G.; Thompson, E.; et al. Improved survival in a cohort of trial participants with metastatic castration-resistant prostate cancer demonstrates the need for updated prognostic nomograms. Eur. Urol. 2013, 64, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Fizazi, K.; Massard, C.; Smith, M.; Rader, M.; Brown, J.; Milecki, P.; Shore, N.; Oudard, S.; Karsh, L.; Carducci, M.; et al. Bone-related Parameters are the Main Prognostic Factors for Overall Survival in Men with Bone Metastases from Castration-resistant Prostate Cancer. Eur. Urol. 2015, 68, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Even-Sapir, E.; Metser, U.; Mishani, E.; Lievshitz, G.; Lerman, H.; Leibovitch, I. The detection of bone metastases in patients with high-risk prostate cancer: 99mTc-MDP Planar bone scintigraphy, single- and multi-field-of-view SPECT, 18F-fluoride PET, and 18F-fluoride PET/CT. J. Nucl. Med. 2006, 47, 287–297. [Google Scholar] [PubMed]
- Nakajima, K.; Edenbrandt, L.; Mizokami, A. Bone scan index: A new biomarker of bone metastasis in patients with prostate cancer. Int. J. Urol. 2017, 24, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Kwee, S.A.; Lim, J.; Watanabe, A.; Kromer-Baker, K.; Coel, M.N. Prognosis Related to Metastatic Burden Measured by (1)(8)F-Fluorocholine PET/CT in Castration-Resistant Prostate Cancer. J. Nucl. Med. 2014, 55, 905–910. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.; Jeraj, R. Use of articulated registration for response assessment of individual metastatic bone lesions. Phys. Med. Biol. 2014, 59, 1501–1514. [Google Scholar] [CrossRef] [PubMed]
- Etchebehere, E.C.; Araujo, J.C.; Fox, P.S.; Swanston, N.M.; Macapinlac, H.A.; Rohren, E.M. Prognostic Factors in Patients Treated with 223Ra: The Role of Skeletal Tumor Burden on Baseline 18F-Fluoride PET/CT in Predicting Overall Survival. J. Nucl. Med. 2015, 56, 1177–1184. [Google Scholar] [CrossRef] [PubMed]
- Rohren, E.M.; Etchebehere, E.C.; Araujo, J.C.; Hobbs, B.P.; Swanston, N.M.; Everding, M.; Moody, T.; Macapinlac, H.A. Determination of Skeletal Tumor Burden on 18F-Fluoride PET/CT. J. Nucl. Med. 2015, 56, 1507–1512. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Bradshaw, T.; Perk, T.; Harmon, S.; Eickhoff, J.; Jallow, N.; Choyke, P.L.; Dahut, W.L.; Larson, S.; Humm, J.L.; et al. Repeatability of Quantitative 18F-NaF PET: A Multicenter Study. J. Nucl. Med. 2016, 57, 1872–1879. [Google Scholar] [CrossRef] [PubMed]
- Harmon, S.A.; Perk, T.; Lin, C.; Eickhoff, J.; Choyke, P.L.; Dahut, W.L.; Apolo, A.B.; Humm, J.L.; Larson, S.M.; Morris, M.J.; et al. Quantitative Assessment of Early [18F]Sodium Fluoride Positron Emission Tomography/Computed Tomography Response to Treatment in Men With Metastatic Prostate Cancer to Bone. J. Clin. Oncol. 2017, 35, 2829–2837. [Google Scholar] [CrossRef] [PubMed]
- Etchebehere, E.C.; Araujo, J.C.; Milton, D.R.; Erwin, W.D.; Wendt, R.E., 3rd; Swanston, N.M.; Fox, P.; Macapinlac, H.A.; Rohren, E.M. Skeletal Tumor Burden on Baseline 18F-Fluoride PET/CT Predicts Bone Marrow Failure After 223Ra Therapy. Clin. Nucl. Med. 2016, 41, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Sato, M.M.; Coel, M.N.; Lee, K.H.; Kwee, S.A. Prediction of PSA Progression in Castration-Resistant Prostate Cancer Based on Treatment-Associated Change in Tumor Burden Quantified by 18F-Fluorocholine PET/CT. J. Nucl. Med. 2016, 57, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Alva, A.; Nordquist, L.; Daignault, S.; George, S.; Ramos, J.; Albany, C.; Isharwal, S.; McDonald, M.; Campbell, G.; Danchaivijitr, P.; et al. Clinical Correlates of Benefit From Radium-223 Therapy in Metastatic Castration Resistant Prostate Cancer. Prostate 2017, 77, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Morris, M.J.; Larson, S.M.; Minarik, D.; Josefsson, A.; Helgstrand, J.T.; Oturai, P.S.; Edenbrandt, L.; Roder, M.A.; Bjartell, A. Automated Bone Scan Index as a quantitative imaging biomarker in metastatic castration-resistant prostate cancer patients being treated with enzalutamide. EJNMMI Res. 2016, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.J.; Kaboteh, R.; Carducci, M.A.; Damber, J.E.; Stadler, W.M.; Hansen, M.; Edenbrandt, L.; Forsberg, G.; Nordle, O.; Pili, R.; et al. Assessment of the bone scan index in a randomized placebo-controlled trial of tasquinimod in men with metastatic castration-resistant prostate cancer (mCRPC). Urol. Oncol. 2014, 32, 1308–1316. [Google Scholar] [CrossRef] [PubMed]
- Bieth, M.; Kronke, M.; Tauber, R.; Dahlbender, M.; Retz, M.; Nekolla, S.G.; Menze, B.; Maurer, T.; Eiber, M.; Schwaiger, M. Exploring New Multimodal Quantitative Imaging Indices for the Assessment of Osseous Tumor Burden in Prostate Cancer Using (68)Ga-PSMA PET/CT. J. Nucl. Med. 2017, 58, 1632–1637. [Google Scholar] [CrossRef] [PubMed]
- Schmuck, S.; von Klot, C.A.; Henkenberens, C.; Sohns, J.M.; Christiansen, H.; Wester, H.J.; Ross, T.L.; Bengel, F.M.; Derlin, T. Initial Experience with Volumetric 68Ga-PSMA I&T PET/CT for Assessment of Whole-Body Tumor Burden as a Quantitative Imaging Biomarker in Patients with Prostate Cancer. J. Nucl. Med. 2017, 58, 1962–1968. [Google Scholar] [PubMed]
- Thomas, L.; Balmus, C.; Ahmadzadehfar, H.; Essler, M.; Strunk, H.; Bundschuh, R.A. Assessment of Bone Metastases in Patients with Prostate Cancer-A Comparison between 99mTc-Bone-Scintigraphy and [68Ga]Ga-PSMA PET/CT. Pharmaceuticals 2017, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Fiz, F.; Sahbai, S.; Campi, C.; Weissinger, M.; Dittmann, H.; Marini, C.; Piana, M.; Sambuceti, G.; la Fougere, C. Tumor Burden and Intraosseous Metabolic Activity as Predictors of Bone Marrow Failure during Radioisotope Therapy in Metastasized Prostate Cancer Patients. Biomed. Res. Int. 2017, 2017, 3905216. [Google Scholar] [CrossRef] [PubMed]
- Miederer, M.; Thomas, C.; Beck, J.; Hampel, C.; Krieger, C.; Baque, P.E.; Helisch, A.; Schreckenberger, M. Haematopoietic toxicity of radium-223 in patients with high skeletal tumour burden. Nuklearmedizin 2015, 54, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Sadik, M.; Suurkula, M.; Hoglund, P.; Jarund, A.; Edenbrandt, L. Improved classifications of planar whole-body bone scans using a computer-assisted diagnosis system: A multicenter, multiple-reader, multiple-case study. J. Nucl. Med. 2009, 50, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Lindgren Belal, S.; Sadik, M.; Kaboteh, R.; Hasani, N.; Enqvist, O.; Svarm, L.; Kahl, F.; Simonsen, J.; Poulsen, M.H.; Ohlsson, M.; et al. 3D skeletal uptake of 18F sodium fluoride in PET/CT images is associated with overall survival in patients with prostate cancer. EJNMMI Res. 2017, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Wassberg, C.; Lubberink, M.; Sorensen, J.; Johansson, S. Repeatability of quantitative parameters of 18F-fluoride PET/CT and biochemical tumour and specific bone remodelling markers in prostate cancer bone metastases. EJNMMI Res. 2017, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Kaboteh, R.; Gjertsson, P.; Leek, H.; Lomsky, M.; Ohlsson, M.; Sjostrand, K.; Edenbrandt, L. Progression of bone metastases in patients with prostate cancer—Automated detection of new lesions and calculation of bone scan index. EJNMMI Res. 2013, 3, 64. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Yoshimura, M.; Suzuki, K.; Hashimoto, T.; Hirose, H.; Uchida, K.; Inoue, S.; Koizumi, K.; Tokuuye, K. Assessment of bone scans in advanced prostate carcinoma using fully automated and semi-automated bone scan index methods. Ann. Nucl. Med. 2012, 26, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, H.; Nakajima, K.; Mizokami, A.; Namiki, M.; Inaki, A.; Taki, J.; Kinuya, S. Bone scintigraphy as a new imaging biomarker: The relationship between bone scan index and bone metabolic markers in prostate cancer patients with bone metastases. Ann. Nucl. Med. 2013, 27, 802–807. [Google Scholar] [CrossRef] [PubMed]
- Shintawati, R.; Achmad, A.; Higuchi, T.; Shimada, H.; Hirasawa, H.; Arisaka, Y.; Takahashi, A.; Nakajima, T.; Tsushima, Y. Evaluation of bone scan index change over time on automated calculation in bone scintigraphy. Ann. Nucl. Med. 2015, 29, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, Y.; Shiina, H.; Yamamoto, Y.; Haramoto, M.; Arichi, N.; Yasumoto, H.; Kitagaki, H.; Igawa, M. Prediction of survival benefit using an automated bone scan index in patients with castration-resistant prostate cancer. BJU Int. 2012, 110, E628–E634. [Google Scholar] [CrossRef] [PubMed]
- Uemura, K.; Miyoshi, Y.; Kawahara, T.; Yoneyama, S.; Hattori, Y.; Teranishi, J.; Kondo, K.; Moriyama, M.; Takebayashi, S.; Yokomizo, Y.; et al. Prognostic value of a computer-aided diagnosis system involving bone scans among men treated with docetaxel for metastatic castration-resistant prostate cancer. BMC Cancer 2016, 16, 109. [Google Scholar] [CrossRef] [PubMed]
- Umeda, T.; Koizumi, M.; Fukai, S.; Miyaji, N.; Motegi, K.; Nakazawa, S.; Takiguchi, T. Evaluation of bone metastatic burden by bone SPECT/CT in metastatic prostate cancer patients: Defining threshold value for total bone uptake and assessment in radium-223 treated patients. Ann. Nucl. Med. 2018, 32, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Chu, G.H.; Kim, H.J.; Allen-Auerbach, M.; Poon, C.; Bridges, J.; Vidovic, A.; Ramakrishna, B.; Ho, J.; Morris, M.J.; et al. Computer-aided quantitative bone scan assessment of prostate cancer treatment response. Nucl. Med. Commun. 2012, 33, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Meirelles, G.S.; Schoder, H.; Ravizzini, G.C.; Gonen, M.; Fox, J.J.; Humm, J.; Morris, M.J.; Scher, H.I.; Larson, S.M. Prognostic value of baseline [18F] fluorodeoxyglucose positron emission tomography and 99mTc-MDP bone scan in progressing metastatic prostate cancer. Clin. Cancer Res. 2010, 16, 6093–6099. [Google Scholar] [CrossRef] [PubMed]
- Dennis, E.R.; Jia, X.; Mezheritskiy, I.S.; Stephenson, R.D.; Schoder, H.; Fox, J.J.; Heller, G.; Scher, H.I.; Larson, S.M.; Morris, M.J. Bone scan index: A quantitative treatment response biomarker for castration-resistant metastatic prostate cancer. J. Clin. Oncol. 2012, 30, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Reza, M.; Jones, R.; Aspegren, J.; Massard, C.; Mattila, L.; Mustonen, M.; Wollmer, P.; Tragardh, E.; Bondesson, E.; Edenbrandt, L.; et al. Bone Scan Index and Progression-free Survival Data for Progressive Metastatic Castration-resistant Prostate Cancer Patients Who Received ODM-201 in the ARADES Multicentre Study. Eur. Urol. Focus 2016, 2, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Fosbol, M.O.; Petersen, P.M.; Kjaer, A.; Mortensen, J. 223Ra Therapy of Advanced Metastatic Castration-Resistant Prostate Cancer: Quantitative Assessment of Skeletal Tumor Burden for Prognostication of Clinical Outcome and Hematologic Toxicity. J. Nucl. Med. 2018, 59, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, M.D.; Collins, D.J.; Tunariu, N.; Orton, M.R.; Padhani, A.R.; Leach, M.O.; Koh, D.M. Assessment of treatment response by total tumor volume and global apparent diffusion coefficient using diffusion-weighted MRI in patients with metastatic bone disease: A feasibility study. PLoS ONE 2014, 9, e91779. [Google Scholar] [CrossRef] [PubMed]
- Perez-Lopez, R.; Lorente, D.; Blackledge, M.D.; Collins, D.J.; Mateo, J.; Bianchini, D.; Omlin, A.; Zivi, A.; Leach, M.O.; de Bono, J.S.; et al. Volume of Bone Metastasis Assessed with Whole-Body Diffusion-weighted Imaging Is Associated with Overall Survival in Metastatic Castration-resistant Prostate Cancer. Radiology 2016, 280, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Brisset, J.C.; Hoff, B.A.; Chenevert, T.L.; Jacobson, J.A.; Boes, J.L.; Galban, S.; Rehemtulla, A.; Johnson, T.D.; Pienta, K.J.; Galban, C.J.; et al. Integrated multimodal imaging of dynamic bone-tumor alterations associated with metastatic prostate cancer. PLoS ONE 2015, 10, e0123877. [Google Scholar] [CrossRef] [PubMed]
- Bastawrous, S.; Bhargava, P.; Behnia, F.; Djang, D.S.; Haseley, D.R. Newer PET application with an old tracer: Role of 18F-NaF skeletal PET/CT in oncologic practice. Radiographics 2014, 34, 1295–1316. [Google Scholar] [CrossRef] [PubMed]
- Eder, M.; Schafer, M.; Bauder-Wust, U.; Hull, W.E.; Wangler, C.; Mier, W.; Haberkorn, U.; Eisenhut, M. 68Ga-complex lipophilicity and the targeting property of a urea-based PSMA inhibitor for PET imaging. Bioconjug. Chem. 2012, 23, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Ryan, C.J.; Shah, S.; Efstathiou, E.; Smith, M.R.; Taplin, M.E.; Bubley, G.J.; Logothetis, C.J.; Kheoh, T.; Kilian, C.; Haqq, C.M.; et al. Phase II study of abiraterone acetate in chemotherapy-naive metastatic castration-resistant prostate cancer displaying bone flare discordant with serologic response. Clin. Cancer Res. 2011, 17, 4854–4861. [Google Scholar] [CrossRef] [PubMed]
- Matheoud, R.; Goertzen, A.L.; Vigna, L.; Ducharme, J.; Sacchetti, G.; Brambilla, M. Five-year experience of quality control for a 3D LSO-based whole-body PET scanner: Results and considerations. Phys. Med. 2012, 28, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Oldan, J.D.; Hawkins, A.S.; Chin, B.B. 18F Sodium Fluoride PET/CT in Patients with Prostate Cancer: Quantification of Normal Tissues, Benign Degenerative Lesions, and Malignant Lesions. World J. Nucl. Med. 2016, 15, 102–108. [Google Scholar] [PubMed]
- Sambuceti, G.; Brignone, M.; Marini, C.; Massollo, M.; Fiz, F.; Morbelli, S.; Buschiazzo, A.; Campi, C.; Piva, R.; Massone, A.M.; et al. Estimating the whole bone-marrow asset in humans by a computational approach to integrated PET/CT imaging. Eur. J. Nucl. Med. Mol. Imaging 2012, 39, 1326–1338. [Google Scholar] [CrossRef] [PubMed]
- Fiz, F.; Marini, C.; Piva, R.; Miglino, M.; Massollo, M.; Bongioanni, F.; Morbelli, S.; Bottoni, G.; Campi, C.; Bacigalupo, A.; et al. Adult advanced chronic lymphocytic leukemia: Computational analysis of whole-body CT documents a bone structure alteration. Radiology 2014, 271, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Fiz, F.; Marini, C.; Campi, C.; Massone, A.M.; Podesta, M.; Bottoni, G.; Piva, R.; Bongioanni, F.; Bacigalupo, A.; Piana, M.; et al. Allogeneic cell transplant expands bone marrow distribution by colonizing previously abandoned areas: An FDG PET/CT analysis. Blood 2015, 125, 4095–4102. [Google Scholar] [CrossRef] [PubMed]
- Marini, C.; Bruno, S.; Fiz, F.; Campi, C.; Piva, R.; Cutrona, G.; Matis, S.; Nieri, A.; Miglino, M.; Ibatici, A.; et al. Functional Activation of Osteoclast Commitment in Chronic Lymphocytic Leukaemia: A Possible Role for RANK/RANKL Pathway. Sci. Rep. 2017, 7, 14159. [Google Scholar] [CrossRef] [PubMed]
| First Author | Year | Reference | Country | Type of Study | Pts. Number | Techinique | Tracer | Analysis | Mean Age | Mean Gleason | Mean PSA (ng/mL) | High Risk Ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAKAJIMA | 2017 | [11] | JAPAN/SWEDEN | RW | - | - | - | REVIEW | - | - | - | - |
| KWEE | 2014 | [12] | USA | P | 30 | PET/CT | 18F-CHOLINE | PET-BASED SEGMENTATION | 73 | N/A | 35.1 | N/A |
| YIP | 2014 | [13] | USA | NS | 16 | PET/CT | 18F-FLUORIDE | HYBRID CT- AND PET-BASED SEGMENTATION | 69 | N/A | N/A | N/A |
| ETCHEBEHERE | 2015 | [14] | USA | R | 42 | PET/CT | 18F-FLUORIDE | PET-BASED SEGMENTATION | 71.7 | N/A | 54 | 64.3% |
| ROHREN | 2015 | [15] | USA | R | 68 | PET/CT | 18F-FLUORIDE | PET-BASED SEGMENTATION | 65.7 | N/A | N/A | N/A |
| LIN | 2016 | [16] | USA | P | 35 | PET/CT | 18F-FLUORIDE | HYBRID CT- AND PET-BASED SEGMENTATION | 71.5 | 7.8 | 49 | 41% |
| HARMON | 2017 | [17] | USA | P | 58 | PET/CT | 18F-FLUORIDE | HYBRID CT- AND PET-BASED SEGMENTATION | 71 | N/A | N/A | 48% |
| ETCHEBEHERE | 2016 | [18] | USA/BRASIL | R | 41 | PET/CT | 18F-FLUORIDE | PET-BASED SEGMENTATION | 71 | N/A | 150 | 61.9% |
| LEE | 2016 | [19] | SOUTH KOREA/USA | P | 42 | PET/CT | 18F-CHOLINE | PET-BASED SEGMENTATION | 73 | N/A | 329 | N/A |
| ALVA | 2017 | [20] | USA/SWEDEN | R | 145 | BONE SCAN | 99mTc-DPD | EXINI BONE SCAN ANN | 71.8 | 9 | 188.7 | 70% |
| ANAND | 2016 | [21] | USA/SWEDEN | R | 80 | BONE SCAN | 99mTc-MDP | EXINI BONE SCAN ANN | 71 | N/A | 157.5 | N/A |
| ARMSTRONG | 2014 | [22] | USA/SWEDEN | R | 85 | BONE SCAN | NOT SPECIFIED * | EXINI BONE SCAN ANN | N/A | N/A | N/A | N/A |
| BIETH | 2017 | [23] | GERMANY | R | 45 | PET/CT | 68-Ga-PSMA | HYBRID CT- AND PET-BASED SEGMENTATION | 71 | N/A | 43 | N/A |
| SCHMUCK | 2017 | [24] | GERMANY | R | 101 | PET/CT | 68-Ga-PSMA | PET-BASED SEGMENTATION | 69.1 | 7 *** | 4.1 | N/A |
| THOMAS | 2017 | [25] | GERMANY | R | 30 | BONE SCAN AND PET/CT | 99mTc-MPD AND 68-Ga-PSMA | EXINI BONE SCAN ANN; VISUAL ANALYSIS | N/A | N/A | N/A | N/A |
| FIZ | 2017 | [26] | GERMANY/ITALY | R | 47 | BONE SPECT/CT | 99mTc-DPD | CT-BASED SEGMENTATION | 69.5 | 8 | 788 | 68% |
| MIEDERER | 2015 | [27] | GERMANY | R | 14 ** | BONE SCAN | 99mTc-DPD | EXINI BONE SCAN ANN | 71 | N/A | N/A | N/A |
| SADIK | 2009 | [28] | SWEDEN | R | 41 | BONE SCAN | 99mTc-MPD | EXINI BONE SCAN ANN | 65 | N/A | N/A | N/A |
| LINDGREN BELAI | 2017 | [29] | SWEDEN | R | 48 | BONE SCAN AND PET/CT | 99mTc-HPD AND 18-F-FLUORIDE | HYBRID CT- AND PET-BASED SEGMENTATION; EXINI BONE SCAN ANN | 73 | 7.7 | 374 | N/A |
| WASSBERG | 2017 | [30] | SWEDEN | P | 10 | PET/CT | 18F-FLUORIDE | PET-BASED SEGMENTATION | 74.6 | 8.1 | 208.5 | 50% |
| KABOTEH | 2013 | [31] | SWEDEN | R | 266 | BONE SCAN | 99mTc-MDP | EXINI BONE SCAN ANN | 76 | N/A | N/A | N/A |
| TAKAHASHI | 2012 | [32] | JAPAN | R | 158 | BONE SCAN | 99mTc-MPD | BONENAVI BONE SCAN ANN | 69.5 | N/A | 148 | N/A |
| WAKABAYASHI | 2013 | [33] | JAPAN | R | 52 | BONE SCAN | 99mTc-MPD | BONENAVI BONE SCAN ANN | 71 | 9 | N/A **** | N/A |
| SHINTAWATI | 2015 | [34] | JAPAN | P | 20 | BONE SCAN | 99mTc-MPD | BONENAVI BONE SCAN ANN | N/A | N/A | N/A | N/A |
| MITSUI | 2012 | [35] | JAPAN | R | 42 | BONE SCAN | 99mTc-MDP | BONENAVI BONE SCAN ANN | 73 | 8 | 65.3 | N/A |
| UEMURA | 2016 | [36] | JAPAN | R | 41 | BONE SCAN | NOT SPECIFIED * | BONENAVI BONE SCAN ANN | 73 | N/A | 56.8 | N/A |
| UMEDA | 2018 | [37] | JAPAN | R | 47 | BONE SPECT/CT | 99mTc-MDP | SPECT-BASED SEGMENTATION; BONENAVI BONE SCAN ANN | 74 | N/A | N/A | N/A |
| BROWN | 2012 | [38] | USA | R | 20 | BONE SCAN | 99mTc-MDP | CAD ANALYSIS | N/A | N/A | N/A | N/A |
| MEIRELLES | 2010 | [39] | USA | P | 39 | BONE SCAN AND PET/CT | 99mTc-HPD AND 18-F-FDG | EXINI BONE SCAN ANN | 68 | N/A | N/A | N/A |
| DENNIS | 2012 | [40] | USA | R | 88 | BONE SCAN | NOT SPECIFIED * | EXINI BONE SCAN ANN | 67.7 | 8 | 95.95 | N/A |
| REZA | 2016 | [41] | SWEDEN/UK/FINLAND/FRANCE | R | 47 | BONE SCAN | NOT SPECIFIED * | EXINI BONE SCAN ANN | 68 | N/A | 83.1 | N/A |
| FOSBØL | 2018 | [42] | DENMARK | R | 88 | BONE SCAN | NOT SPECIFIED * | EXINI BONE SCAN ANN | 71 | N/A | 212 | N/A |
| BLACKLEDGE | 2014 | [43] | UK | P | 7 | MRI | NONE | MARKOV RANDOM FIELD MODEL | N/A | N/A | N/A | N/A |
| PEREZ-LOPEZ | 2016 | [44] | UK | R | 43 | MRI AND BONE SCAN | NOT SPECIFIED * | MR SEGMENTATION AND EXINI BONE SCAN ANN | N/A | N/A | 43 | N/A |
| BRISSET | 2015 | [45] | USA/HOLLAND | P | 12 | CT AND MR | NONE | VOXEL-BASED ANALYSIS | N/A | N/A | N/A | N/A |
| Method | Advantages | Disadvantages | Relative Frequency |
|---|---|---|---|
| Neural network analysis applied to planar bone scan |
|
| Common (prevalent diffusion of bone scan) |
| PET-based thresholding |
|
| Uncommon |
| Hybrid CT- and PET/SPECT-based thresholding |
|
| Rare (presently only research application) |
| MR-based and other non-isotopic methods |
|
| Rare (presently only research application) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiz, F.; Dittman, H.; Campi, C.; Morbelli, S.; Marini, C.; Brignone, M.; Bauckneht, M.; Piva, R.; Massone, A.M.; Piana, M.; et al. Assessment of Skeletal Tumor Load in Metastasized Castration-Resistant Prostate Cancer Patients: A Review of Available Methods and an Overview on Future Perspectives. Bioengineering 2018, 5, 58. https://doi.org/10.3390/bioengineering5030058
Fiz F, Dittman H, Campi C, Morbelli S, Marini C, Brignone M, Bauckneht M, Piva R, Massone AM, Piana M, et al. Assessment of Skeletal Tumor Load in Metastasized Castration-Resistant Prostate Cancer Patients: A Review of Available Methods and an Overview on Future Perspectives. Bioengineering. 2018; 5(3):58. https://doi.org/10.3390/bioengineering5030058
Chicago/Turabian StyleFiz, Francesco, Helmut Dittman, Cristina Campi, Silvia Morbelli, Cecilia Marini, Massimo Brignone, Matteo Bauckneht, Roberta Piva, Anna Maria Massone, Michele Piana, and et al. 2018. "Assessment of Skeletal Tumor Load in Metastasized Castration-Resistant Prostate Cancer Patients: A Review of Available Methods and an Overview on Future Perspectives" Bioengineering 5, no. 3: 58. https://doi.org/10.3390/bioengineering5030058
APA StyleFiz, F., Dittman, H., Campi, C., Morbelli, S., Marini, C., Brignone, M., Bauckneht, M., Piva, R., Massone, A. M., Piana, M., Sambuceti, G., & La Fougère, C. (2018). Assessment of Skeletal Tumor Load in Metastasized Castration-Resistant Prostate Cancer Patients: A Review of Available Methods and an Overview on Future Perspectives. Bioengineering, 5(3), 58. https://doi.org/10.3390/bioengineering5030058










