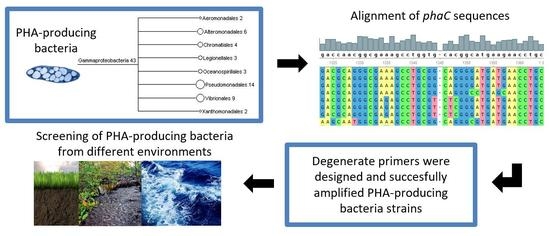
Molecular Diagnostic for Prospecting Polyhydroxyalkanoate-Producing Bacteria
Abstract
:1. Introduction
2. Experimental Procedures
2.1. Bacteria Strains and Media
2.2. Design and Evaluation of Primers for the Amplification of the Gene phaC
2.3. Amplification of phaC Gene by PCR
2.4. DNA Sequencing
3. Results
3.1. Design of Primers for Gene phaC Amplification
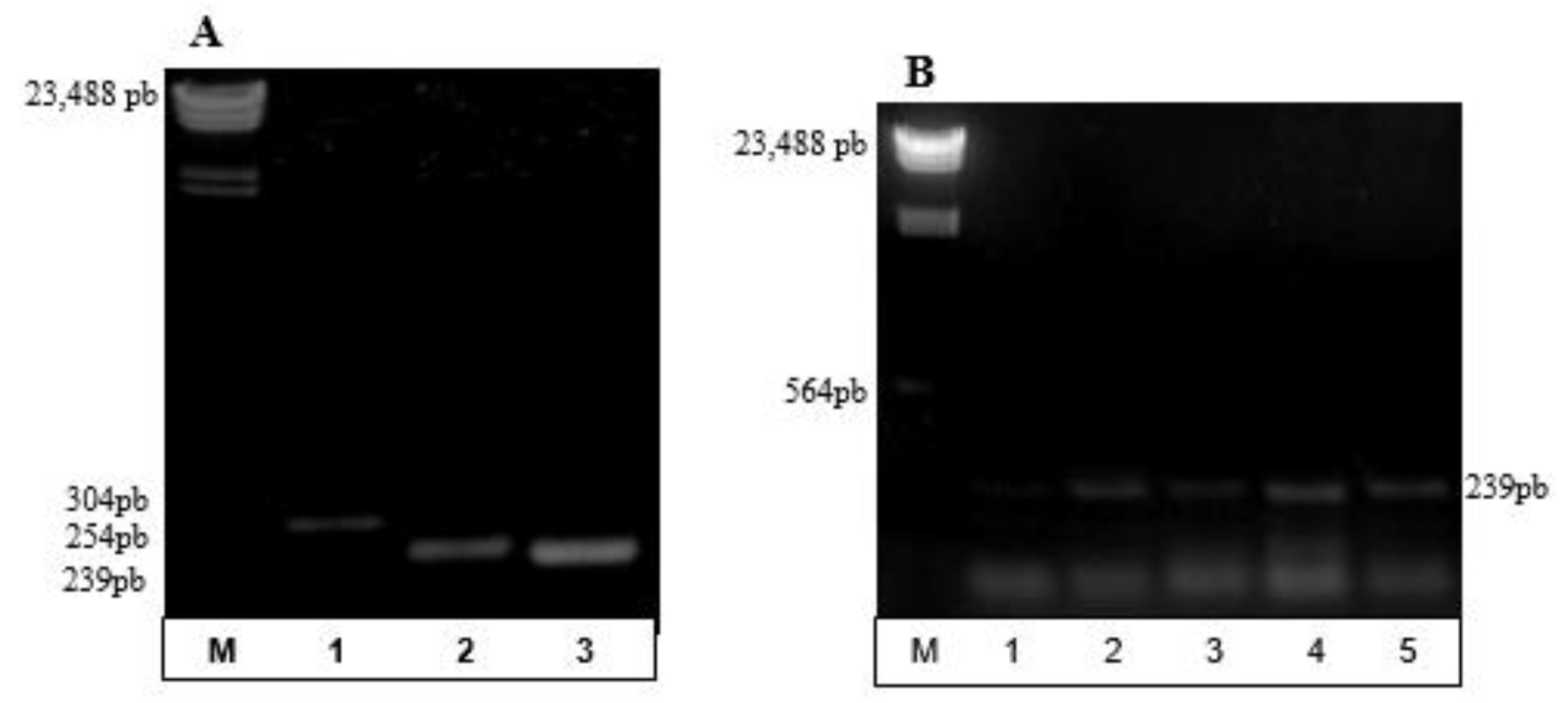
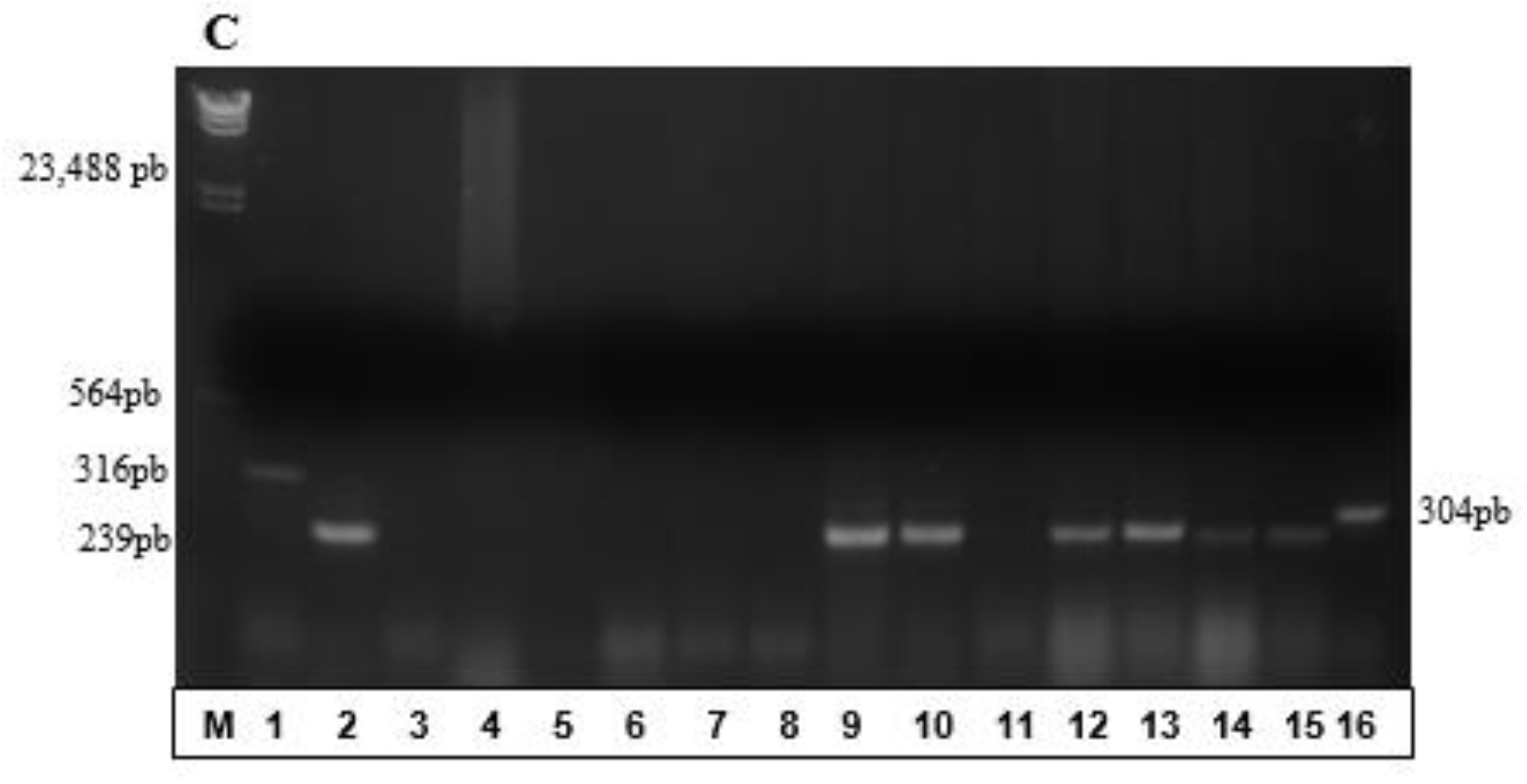
3.2. Partial Amplification of phaC Gene
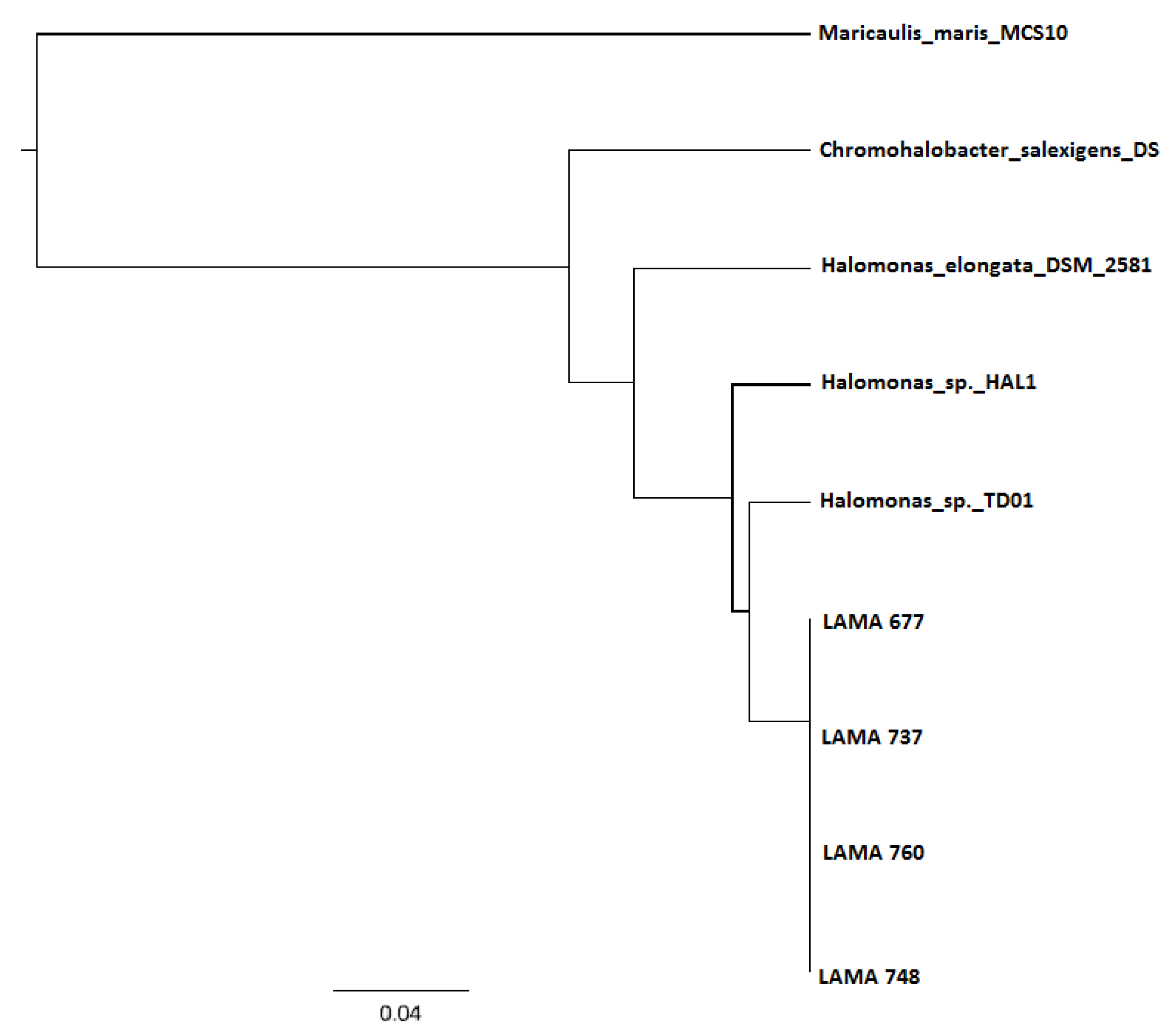
3.3. DNA Sequencing and phaC Gene Identification
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Song, J.H.; Murphy, R.J.; Narayan, R.; Davies, G.B.H. Biodegradable and compostable alternatives to conventional plastics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 2127–2139. [Google Scholar] [CrossRef] [PubMed]
- Barnes, D.K.A.; Galgani, F.; Thompson, R.C.; Barlaz, M. Accumulation and fragmentation of plastic debris in global environments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Steinbüchel, A.; Füchtenbush, B. Bacterial and other biological systems for polyester production. Trends Biotechnol. 1998, 16, 419–427. [Google Scholar] [CrossRef]
- Rehm, B.H. Biogenesis of microbial polyhydroxyalkanoate granules: A platform technology for the production of tailor-made bioparticles. Curr. Issues Mol. Biol. 2007, 9, 41–62. [Google Scholar] [PubMed]
- Bugnicourt, E.; Cinelli, P.; Lazzeri, A.; Alvarez, V.A. Polyhydroxyalkanoate (PHA): Review of synthesis, characteristics, processing and potential applications in packaging. Express Polym. Lett. 2014, 8, 791–808. [Google Scholar] [CrossRef]
- Godbole, S. Methods for identification, quantification and characterization of polyhydroxyalkanoates. Int. J. Bioassays 2016, 5, 4977–4983. [Google Scholar] [CrossRef]
- Khardenavis, A.A.; Kumar, M.S.; Mudliar, S.N.; Chakrabarti, T. Biotechnological conversion of agro industrial wastewaters into biodegradable plastic, poly-β-hydroxybutyrate. Biosour. Technol. 2007, 98, 3579–3584. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.Y.A.; Chen, C.L.; Li, L.; Ge, L.; Wang, L.; Razaad, I.M.N.; Li, Y.; Zhao, L.; Mo, Y.; Wang, J.-Y. Start are search on biopolymer polyhydroxyalkanoate (PHA): A review. Polymers 2014, 6, 706–754. [Google Scholar] [CrossRef]
- Lee, S.Y.; Choi, J.; Wong, H.H. Recent advances in polyhydroxyalkanoate production by bacterial fermentation: Mini-review. Int. J. Biol. Macromol. 1999, 25, 31–36. [Google Scholar] [CrossRef]
- Khanna, S.; Srivastava, A.K. Recent advances in microbial polyhydroxyalkanoates. Process Biochem. 2005, 40, 607–619. [Google Scholar] [CrossRef]
- Silva, A.L.; dos Santosa, E.C.; dos Santosa, Í.A.; Lópeza, A.M. Seleção polifásica de microrganismos produtores de polihidroxialcanoatos. Quim. Nova. 2016, 39, 782–788. [Google Scholar]
- Rehm, B.H. Polyester synthases: Natural catalysts for plastics. Biochem. J. 2003, 376, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Solaiman, D.K.; Ashby, R.D. Rapid genetic characterization of poly(hydroxyalkanoate) synthase and its applications. Biomacromolecules 2005, 6, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Hein, S.; Paletta, J.R.; Steinbüchel, A. Cloning, characterization and comparison of the Pseudomonas mendocina polyhydroxyalkanoate synthases PhaC1 and PhaC2. Appl. Microbiol. Biotechnol. 2002, 58, 229–236. [Google Scholar] [PubMed]
- Yuan, W.; Jia, Y.; Tian, J.; Snell, K.D.; Muh, U.; Sinskey, A.J.; Lambalot, R.H.; Walsh, C.T.; Stubbe, J. Class I and III polyhydroxyalkanoate synthases from Ralstonia eutropha and Allochromatium vinosum: Characterization and substrate specificity studies. Arch. Biochem. Biophys. 2001, 394, 87–98. [Google Scholar] [CrossRef] [PubMed]
- McCooL, G.J.; Cannon, M.C. PhaC and PhaR are required for polyhydroxyalkanoic acid synthase activity in Bacillus megaterium. J. Bacteriol. 2001, 183, 4235–4243. [Google Scholar] [CrossRef] [PubMed]
- Koller, M.; Rodríguez-Contreras, A. Techniques for tracing PHA-producing organisms and for qualitative and quantitative analysis of intra-and extracellular PHA. Eng. Life Sci. 2015, 15, 558–581. [Google Scholar] [CrossRef]
- Murray, R.G.E.; Doetsch, R.N.; Robinow, C.F. Determinative and cytological light microscopy. Am. Soc. Microbiol. 1994, 1, 21–41. [Google Scholar]
- Ostle, A.G.; Holt, J.G. Nile Blue A as a fluorescent stain for polybeta-hydroxybutyrate. Appl. Environ. Microbiol. 1982, 44, 238–241. [Google Scholar] [PubMed]
- Spiekermann, P.; Rehm, B.H.; Kalscheuer, R.; Baumeister, D.; Steinbüchel, A. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 1999, 171, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Shamala, T.R.; Chandrashekar, A.; Vijayendra, S.V.; Kshama, L. Identification of polyhydroxyalkanoate (PHA)-producing Bacillus spp. using the polymerase chain reaction (PCR). J. Appl. Microbiol. 2003, 94, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.S.; Bhat, S.G.; Chandrasekaran, M. Amplification and sequence analysis of phaC gene of polyhydroxybutyrate producing Vibrio azureus BTKB33 isolated from marine sediments. Ann. Microbiol. 2016, 66, 299–306. [Google Scholar] [CrossRef]
- Lane, C.E.; Benton, M.G. Detection of the enzymatically-active polyhydroxyalkanoate synthase subunit gene, phaC, in cyanobacteria via colony PCR. Mol. Cell. Probes 2015, 29, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Sheu, D.S.; Wang, Y.T.; Lee, C.Y. Rapid detection of polyhydroxyalkanoate accumulating bacteria isolated from the environment by colony PCR. Microbiology 2000, 146, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Solaiman, D.K.; Ashby, R.D.; Foglia, T.A. Rapid and specific identification of medium-chain-length polyhydroxyalkanoate synthase gene by polymerase chain reaction. Appl. Microbiol. Biotechnol. 2000, 53, 690–694. [Google Scholar] [CrossRef] [PubMed]
- Kung, S.S.; Chuang, Y.C.; Chen, C.H.; Chien, C.C. Isolation of polyhydroxyalkanoates-producing bacteria using a combination of phenotypic and genotypic approach. Lett. Appl. Microbiol. 2007, 44, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Desetty, R.D.; Mahajan, V.S.; Khan, B.M.; Rawal, S.K. Isolation and heterologous expression of PHA synthesising genes from Bacillus thuringiensis R1. World J. Microbiol. Biotechnol. 2008, 24, 1769–1774. [Google Scholar] [CrossRef]
- Lima, A.O.S.; Garcês, S.P.S. Intragenic Primer Design: Bringing Bioinformatics Tools to the Class. Biochem. Mol. Biol. Educ. 2006, 34, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Biocyc Database Collection. Available online: http://biocyc.org (accessed on 20 October 2016).
- Huson, D.H.; Auch, A.; Qi, J.; Schuster, S.C. Megan Analysis of Metagenome Data. Genome Res. 2011, 17, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Nacional Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 11 November 2016).
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. ClustalW and ClustalX version 2. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Fursov, M.Y.; Oshchepkov, D.Y; Novikova, O.S. UGENE: Interactive computational schemes for genome analysis. In Proceedings of the Fifth Moscow International Congress on Biotechnology, Moscow, Russia, 16–20 March 2009; Volume 3, pp. 14–15. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Tzu, H.Y.; Semblante, G.U. Detection of polyhydroxyalkanoate-accumulating bacteria from domestic wastewater treatment plant using highly sensitive PCR primers. J. Microbiol. Biotechnol. 2012, 22, 1141–1147. [Google Scholar]
- Castroverde, C.D.M.; San Luis, B.B.; Monsalud, R.G.; Hedreyda, C.T. Differential detection of vibrios pathogenic to shrimp by multiplex PCR. J. Gen. Appl. Microbiol. 2006, 52, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Sujatha, K.; Mahalakshmi, A.; Shenbagarathai, R. Molecular characterization of Pseudomonas sp. LDC-5 involved in accumulation of poly 3 hydroxybutyrate and medium-chain-length poly 3-hydroxyalkanoates. Arch. Microbiol. 2007, 188, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Aneja, K.K.; Ashby, R.D.; Solaiman, D.K.Y. Altered composition of Ralstonia eutropha poly (hydroxyalkanoate) through expression of PHA synthase from Allochromatium vinosum ATCC 35206. Biotechnol. Lett. 2009, 31, 1601–1612. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C.; Lal, S.; Cheema, S. Insight in to the phylogeny of polyhydroxyalkanoate biosynthesis: Horizontal gene transfer. Gene 2007, 389, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Gao, M.; Wang, M.; Liu, H.; Sun, J.; Gao, J. Subtilase genes diversity in the biogas digester microbiota. Curr. Microbiol. 2011, 62, 1542–1547. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.T.; Foong, C.P.; Najimudin, N.; Sudesh, K. Discovery of a new polyhydroxyalkanoate synthase from limestone soil through metagenomic approach. J. Biosci. Bioeng. 2016, 121, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wu, H.; Wang, A.; Du, P.; Pei, X.; Li, H.; Yin, X.; Huang, L.; Xiong, X. Prospecting Metagenomic Enzyme Subfamily Genes for DNA Family Shuffling by a Novel PCR-based Approach. J. Biol. Chem. 2010, 285, 41509–41516. [Google Scholar] [CrossRef] [PubMed]


| Primer ID | Sequence | Tm | Consensus Position | Aligned Sequences * |
|---|---|---|---|---|
| phaCF1 | 5′TGATSSAGCTGATCCAGTAC3′ | 53.9° | 489–508 | 18 |
| phaCF2 | 5′CCGCTGCTGATCGTBCCGCC3′ | 65.5° | 539–558 | 41 |
| phaCF3 | 5′CCGCCSTGGATCAACAAGT3′ | 58.0° | 554–572 | 61 |
| phaCF4 | 5′CTACATCCTCGACCTGMAGCCGGA3′ | 63.1° | 574–597 | 24 |
| phaCF5 | 5′GGCTACTGCRTCGGCGGCAC3′ | 65.1° | 773–792 | 47 |
| phaCF6 | 5′TGGAACDSCGACDCCACCAAC3′ | 61.6° | 1078–1098 | 0 |
| phaCF7 | 5′CGACRCCACCAACMTGCCGGG3′ | 65.8° | 1086–1106 | 4 |
| phaCR1 | 5′GTGCCGCCGAYGCAGTAGCC3′ | 65.1° | 773–792 | 47 |
| phaCR2 | 5′CCCGGCAKGTTGGTGGYGTCG3′ | 65.8° | 1086–1106 | 4 |
| phaCR3 | 5′CAGTSCGGCCACCAGSWGCC3′ | 66.3° | 1432–1451 | 0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montenegro, E.M.d.S.; Delabary, G.S.; Silva, M.A.C.d.; Andreote, F.D.; Lima, A.O.d.S. Molecular Diagnostic for Prospecting Polyhydroxyalkanoate-Producing Bacteria. Bioengineering 2017, 4, 52. https://doi.org/10.3390/bioengineering4020052
Montenegro EMdS, Delabary GS, Silva MACd, Andreote FD, Lima AOdS. Molecular Diagnostic for Prospecting Polyhydroxyalkanoate-Producing Bacteria. Bioengineering. 2017; 4(2):52. https://doi.org/10.3390/bioengineering4020052
Chicago/Turabian StyleMontenegro, Eduarda Morgana da Silva, Gabriela Scholante Delabary, Marcus Adonai Castro da Silva, Fernando Dini Andreote, and André Oliveira de Souza Lima. 2017. "Molecular Diagnostic for Prospecting Polyhydroxyalkanoate-Producing Bacteria" Bioengineering 4, no. 2: 52. https://doi.org/10.3390/bioengineering4020052
APA StyleMontenegro, E. M. d. S., Delabary, G. S., Silva, M. A. C. d., Andreote, F. D., & Lima, A. O. d. S. (2017). Molecular Diagnostic for Prospecting Polyhydroxyalkanoate-Producing Bacteria. Bioengineering, 4(2), 52. https://doi.org/10.3390/bioengineering4020052