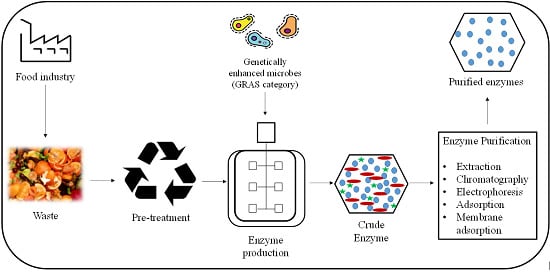
Microbial Enzyme Production Using Lignocellulosic Food Industry Wastes as Feedstock: A Review
Abstract
:1. Introduction
2. Market Potential
3. Types of Food Waste
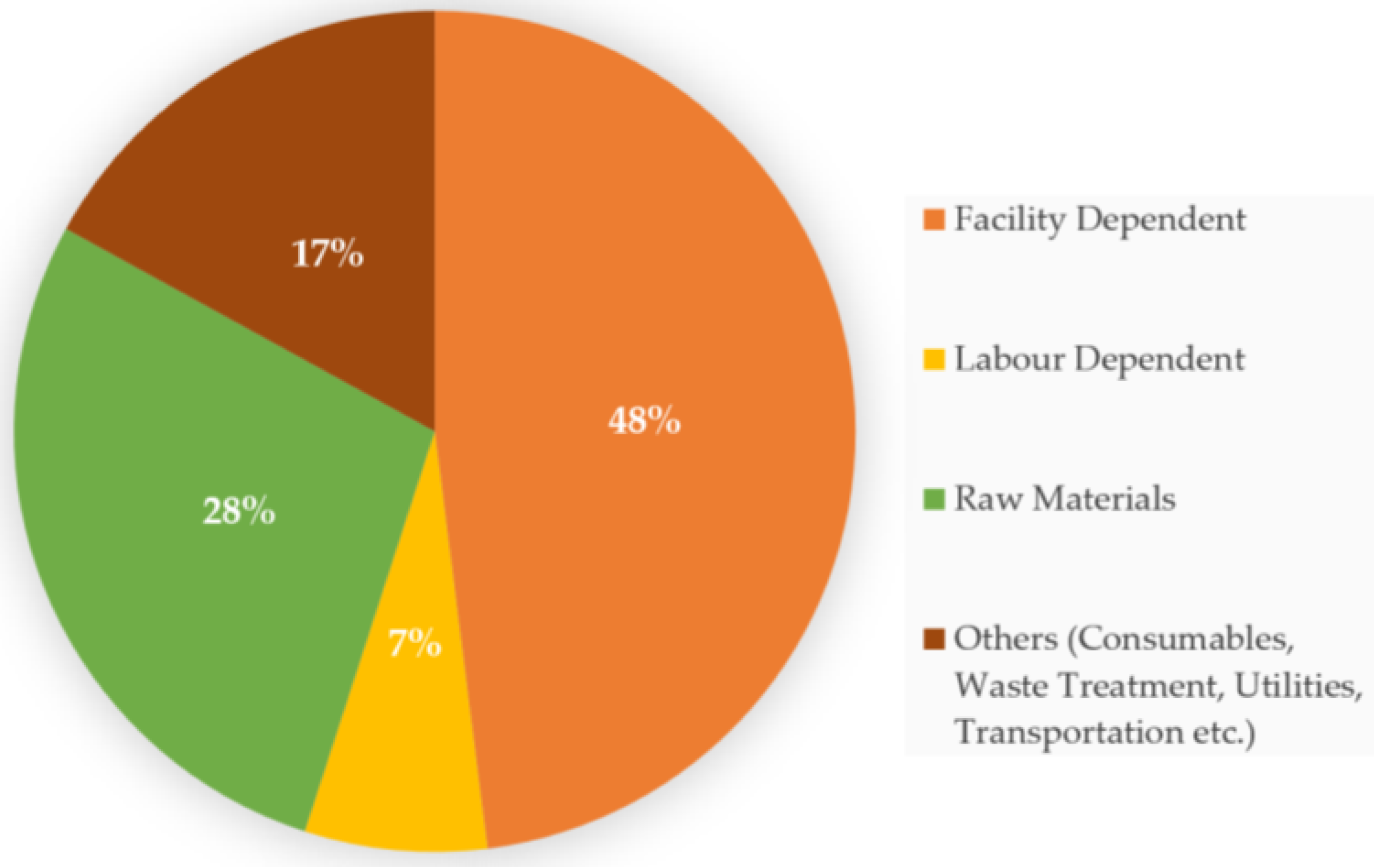
4. Economic Impact of Food Waste Valorization
5. Food Industry Waste (Global Status)
6. Challenges in Enzyme Production Using Lignocellulosic Food Waste
6.1. Lignocellulose as a Raw Material
6.2. Pretreatment of Lignocellulose
6.3. Choice of Microorganism
6.4. Fermentation Strategies
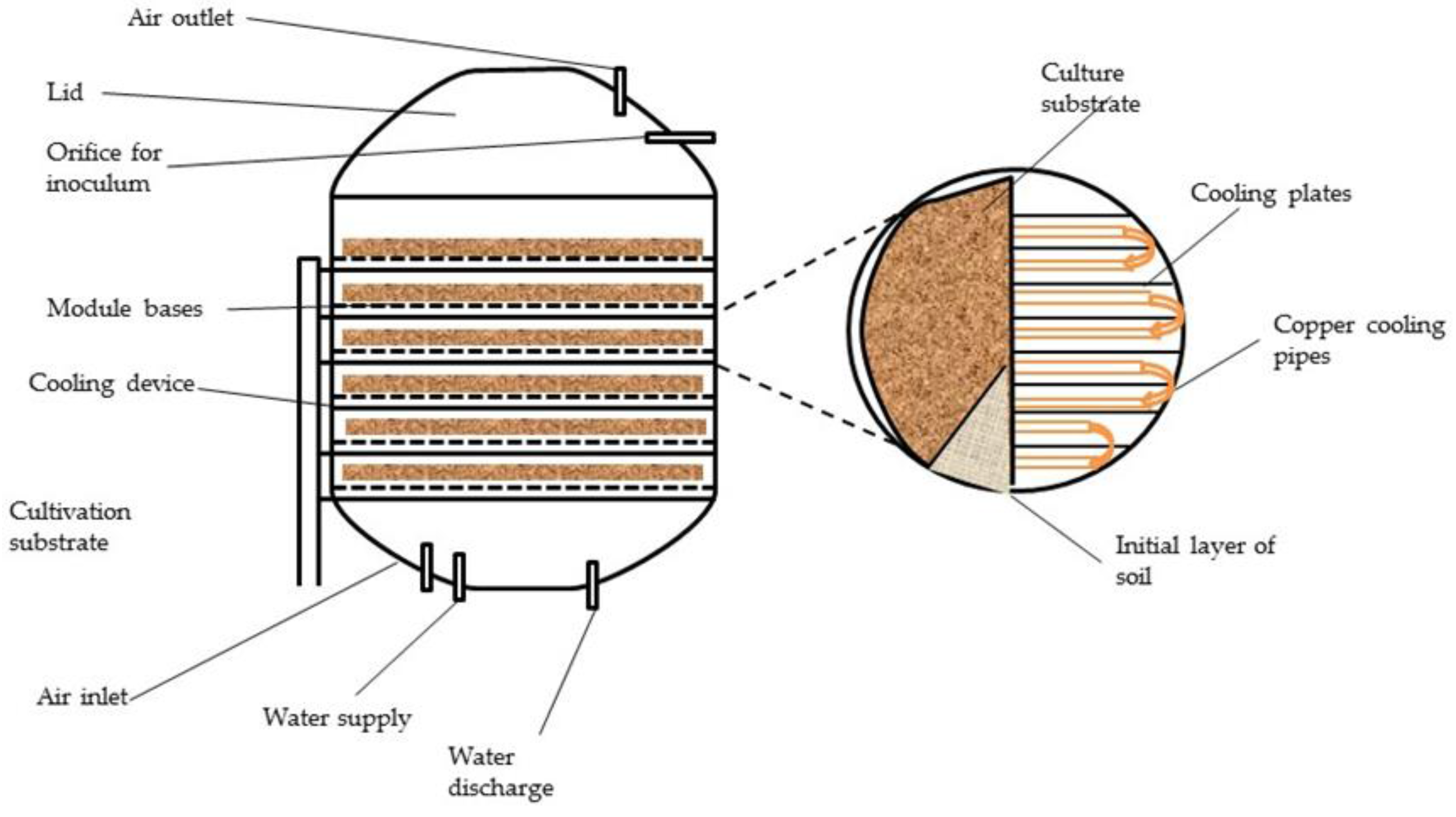
6.4.1. Solid State Fermentation
6.4.2. Submerged Fermentation
6.4.3. Alternative Fermentation Modes
7. Isolation, Purification and Recovery of Enzymes
7.1. Source of Enzyme
7.2. Isolation of Enzymes
7.3. Fractionation
7.3.1. Precipitation, Centrifugation and Ultrafiltration
7.3.2. Liquid–Liquid Extraction
7.3.3. Chromatography
7.3.4. Electrophoresis
7.3.5. Expansion Bed Adsorption
7.3.6. Membrane Adsorption
7.4. One-Step Immobilization and Purification
- (i)
- immobilization via one point;
- (ii)
- introduction of different domains; and
- (iii)
- immobilization using heterofunctional supports.
8. Technical Problems
9. Conclusions
Acknowledgments
Conflicts of Interest
References
- Adrio, J.L.; Demain, A.L. Microbial enzymes: Tools for biotechnological processes. Biomolecules 2014, 4, 117–139. [Google Scholar] [CrossRef] [PubMed]
- Klein-Marcuschamer, D.; Oleskowicz-Popiel, P.; Simmons, B.A.; Blanch, H.W. The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol. Bioeng. 2012, 109, 1083–1087. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, R.; Jaiswal, A.K. Exploitation of food industry waste for high value products. Trends Biotechnol. 2016, 34, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Shukla, A.; Tiwari, S.; Srivastava, M. A review on delignification of lignocellulosic biomass for enhancement of ethanol production potential. Renew. Sustain. Energy Rev. 2014, 32, 713–728. [Google Scholar] [CrossRef]
- Maitan-Alfenas, G.P.; Visser, E.M.; Guimarães, V.M. Enzymatic hydrolysis of lignocellulosic biomass: Converting food waste in valuable products. Curr. Opin. Food Sci. 2015, 1, 44–49. [Google Scholar] [CrossRef]
- Meng, X.; Ragauskas, A.J. Recent advances in understanding the role of cellulose accessibility in enzymatic hydrolysis of lignocellulosic substrates. Curr. Opin. Biotechnol. 2014, 27, 150–158. [Google Scholar] [CrossRef] [PubMed]
- BCC Research. Global Market for Industrial Enzymes to Reach Nearly $7.1 Billion by 2018; Detergent Enzyme Market to Record Maximum Growth; BCC Research: Wellesley, MA, USA, 2014. [Google Scholar]
- Jegannathan, K.R.; Nielsen, P.H. Environmental assessment of enzyme use in industrial production—A literature review. J. Clean. Prod. 2013, 42, 228–240. [Google Scholar] [CrossRef]
- Phitsuwan, P.; Laohakunjit, N.; Kerdchoechuen, O.; Kyu, K.L.; Ratanakhanokchai, K. Present and potential applications of cellulases in agriculture, biotechnology, and bioenergy. Folia Microbiol. 2013, 58, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Laboratory NRE. Reducing Enzyme Cost Increases Market Potential of Biofuels; Laboratory NRE: Golden, CO, USA, 2010. [Google Scholar]
- Mussatto, S.I.; Dragone, G.; Roberto, I.C. Brewers’ spent grain: Generation, characteristics and potential applications. J. Cereal Sci. 2006, 43, 1–14. [Google Scholar] [CrossRef]
- Sun, X.; Liu, Z.; Qu, Y.; Li, X. The effects of wheat bran composition on the production of biomass-hydrolyzing enzymes by Penicillium decumbens. Appl. Microbiol. Biotechnol. 2008, 146, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Saunders, R.M. Rice bran: Composition and potential food uses. Food Rev. Int. 1985, 1, 465–495. [Google Scholar] [CrossRef]
- Pirmohammadi, R.; Rouzbehan, Y.; Rezayazdi, K.; Zahedifar, M. Chemical composition, digestibility and in situ degradability of dried and ensiled apple pomace and maize silage. Small Rumin. Res. 2006, 66, 150–155. [Google Scholar] [CrossRef]
- Bhushan, S.; Kalia, K.; Sharma, M.; Singh, B.; Ahuja, P.S. Processing of Apple Pomace for Bioactive Molecules. Crit. Rev. Biotechnol. 2008, 28, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Freestone, P.P.E.; Haigh, R.D.; Lyte, M. Bacterial Growth Enhancer. U.S. Patent Application 12/092,956, 2 April 2009. [Google Scholar]
- European Commission. Towards a Circular Economy: A Zero Waste Programme for Europe; European Commission: Brussels, Belgium, 2014. [Google Scholar]
- Lin, C.S.K.; Pfaltzgraff, L.A.; Herrero-Davila, L.; Mubofu, E.B.; Abderrahim, S.; Clark, J.H. Food waste as a valuable resource for the production of chemicals, materials and fuels: Current situation and global perspective. Energy Environ. Sci. 2013, 6, 426–464. [Google Scholar] [CrossRef]
- Pfaltzgraff, L.A.; Cooper, E.C.; Budarin, V.; Clark, J.H. Food waste biomass: A resource for high-value chemicals. Green Chem. 2013, 15, 307–314. [Google Scholar] [CrossRef]
- Yasin, N.H.M.; Mumtaz, T.; Hassan, M.A. Food waste and food processing waste for biohydrogen production: A review. J. Environ. Manag. 2013, 130, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Couto, S.R.; Herrera, J.L.T. Industrial and biotechnological applications of laccases: A review. Biotechnol. Adv. 2006, 24, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Fan, X.; Mei, X.; Wei, Z.; Raza, W.; Shen, Q.; Xu, Y. Production and characterization of cellulolytic enzyme from Penicillium oxalicum GZ-2 and its application in lignocellulose saccharification. Biomass Bioenery 2015, 74, 122–134. [Google Scholar] [CrossRef]
- Ragauskas, A.J.; Beckham, G.T.; Biddy, M.J.; Chandra, R.; Chen, F.; Davis, M.F. Lignin valorization: Improving lignin processing in the biorefinery. Science 2014, 344, 1246843. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zhao, S.; Yang, S.; Ding, S.Y. Lignin plays a negative role in the biochemical process for producing lignocellulosic biofuels. Curr. Opin. Biotechnol. 2014, 27, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Chang, V.; Holtzapple, M. Fundamental factors affecting biomass enzymatic reactivity. In Twenty-First Symposium on Biotechnology for Fuels and Chemicals; Finkelstein, M., Davison, B., Eds.; Humana Press: Totowa, NJ, USA, 2000; pp. 5–37. [Google Scholar]
- Ghaffar, S.H.; Fan, M. Structural analysis for lignin characteristics in biomass straw. Biomass Bioenergy 2013, 57, 264–279. [Google Scholar] [CrossRef]
- Bansal, P.; Hall, M.; Realff, M.J.; Lee, J.H.; Bommarius, A.S. Multivariate statistical analysis of X-ray data from cellulose: A new method to determine degree of crystallinity and predict hydrolysis rates. Bioresour. Technol. 2010, 101, 4461–4471. [Google Scholar] [CrossRef] [PubMed]
- Gurgel, L.V.A.; Marabezi, K.; Ramos, L.A.; Curvelo, A.A.D. Characterization of depolymerized residues from extremely low acid hydrolysis (ELA) of sugarcane bagasse cellulose: Effects of degree of polymerization, crystallinity and crystallite size on thermal decomposition. Ind. Crops Prod. 2012, 36, 560–571. [Google Scholar] [CrossRef]
- Wang, K.; Kim, K.H.; Brown, R.C. Catalytic pyrolysis of individual components of lignocellulosic biomass. Green Chem. 2014, 16, 727–735. [Google Scholar] [CrossRef]
- DeMartini, J.D.; Pattathil, S.; Miller, J.S.; Li, H.; Hahn, M.G.; Wyman, C.E. Investigating plant cell wall components that affect biomass recalcitrance in poplar and switchgrass. Energy Environ. Sci. 2013, 6, 898–909. [Google Scholar] [CrossRef]
- Motta, F.; Andrade, C.; Santana, M. A review of xylanase production by the fermentation of xylan: Classification, characterization and applications. In Sustainable Degradation of Lignocellulosic Biomass-Techniques, Applications and Commercialization; InTech: Rijeka, Croatia, 2013. [Google Scholar]
- Chesini, M.; Neila, L.P.; de la Parra, D.F.; Rojas, L.; Esquivel, J.C.C.; Cavalitto, S.F. Aspergillus kawachii produces an inulinase in cultures with yacon (Smallanthus sonchifolius) as substrate. Electron. J. Biotechnol. 2013, 16, 8. [Google Scholar]
- Xiao, C.; Anderson, C.T. Roles of pectin in biomass yield and processing for biofuels. Front. Plant Sci. 2013, 4, 67. [Google Scholar] [CrossRef] [PubMed]
- Olsen, H.S. Biogas Production Process with Enzymatic Pre-Treatment. U.S. Patent Application 13/521,463, 14 February 2013. [Google Scholar]
- Singh, J.; Suhag, M.; Dhaka, A. Augmented digestion of lignocellulose by steam explosion, acid and alkaline pretreatment methods: A review. Carbohydr. Polym. 2015, 117, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Mai, N.L.; Ha, S.H.; Koo, Y.M. Efficient pretreatment of lignocellulose in ionic liquids/co-solvent for enzymatic hydrolysis enhancement into fermentable sugars. Process Biochem. 2014, 49, 1144–1151. [Google Scholar] [CrossRef]
- Bellido, C.; Bolado, S.; Coca, M.; Lucas, S.; González-Benito, G.; García-Cubero, M.T. Effect of inhibitors formed during wheat straw pretreatment on ethanol fermentation by Pichia stipitis. Bioresour. Technol. 2011, 102, 10868–10874. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, R.; Jaiswal, A.K. A comprehensive review on pre-treatment strategy for lignocellulosic food industry waste: Challenges and opportunities. Bioresour. Technol. 2016, 199, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Vandenberghe, L.P.; Carvalho, J.C.; Libardi, N.; Rodrigues, C.; Soccol, C.R. Chapter 1—Microbial Enzyme Factories: Current Trends in Production Processes and Commercial Aspects. In Agro-Industrial Wastes as Feedstock for Enzyme Production; Dhillon, G.S., Kaur, S., Eds.; Academic Press: San Diego, CA, USA, 2016; pp. 1–22. [Google Scholar]
- Arslan, S.; Eyi, A.; Küçüksarı, R. Toxigenic genes, spoilage potential, and antimicrobial resistance of Bacillus cereus group strains from ice cream. Anaerobe 2014, 25, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Pariza, M.W.; Johnson, E.A. Evaluating the Safety of Microbial Enzyme Preparations Used in Food Processing: Update for a New Century. Regul. Toxicol. Pharm. 2001, 33, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Olempska-Beer, Z.S.; Merker, R.I.; Ditto, M.D.; DiNovi, M.J. Food-processing enzymes from recombinant microorganisms—A review. Regul. Toxicol. Pharm. 2006, 45, 144–158. [Google Scholar] [CrossRef] [PubMed]
- Youssef, N.H.; Couger, M.B.; Struchtemeyer, C.G.; Liggenstoffer, A.S.; Prade, R.A.; Najar, F.Z. The Genome of the Anaerobic Fungus Orpinomyces sp. Strain C1A Reveals the Unique Evolutionary History of a Remarkable Plant Biomass Degrader. Appl. Environ. Microbiol. 2013, 79, 4620–4634. [Google Scholar] [PubMed]
- Nevoigt, E. Progress in Metabolic Engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2008, 73, 379–412. [Google Scholar] [CrossRef] [PubMed]
- Keasling, J.D. Synthetic biology and the development of tools for metabolic engineering. Metab. Eng. 2012, 14, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Agrimi, G.; Pisano, I.; Palmieri, L. Process development and metabolic engineering for bioethanol production from lignocellulosic biomass. In Biorefinery: From Biomass to Chemicals and Fuels; Aresta, M., Dibenedetto, A., Dumeignil, F., Eds.; Walter de Gruyter: Berlin, Germany, 2012. [Google Scholar]
- Buschke, N.; Schäfer, R.; Becker, J.; Wittmann, C. Metabolic engineering of industrial platform microorganisms for biorefinery applications–optimization of substrate spectrum and process robustness by rational and evolutive strategies. Bioresour. Technol. 2013, 133, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Batche, H.; Valenzuela-Solano, C.; Hernandez-Martinez, R. Production of Conidiospores in Solid-State Fermentation of Trichoderma Harzianum and T. asperellum Isolated from Grapevine in Baja California; Phytopathology: St. Paul, MN, USA, 2014; p. 179. [Google Scholar]
- Barrios-González, J. Solid-state fermentation: Physiology of solid medium, its molecular basis and applications. Process Biochem. 2012, 47, 175–185. [Google Scholar] [CrossRef]
- Hölker, U.; Lenz, J. Solid-state fermentation—Are there any biotechnological advantages? Curr. Opin. Microbiol. 2005, 8, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Kawase, T.; Sammoto, H.; Gomi, K.; Kariyama, M.; Miyake, T. Uniform culture in solid-state fermentation with fungi and its efficient enzyme production. J. Biosci. Bioeng. 2011, 111, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Gomi, K.; Kariyama, M.; Miyake, T. Rapid enzyme production and mycelial growth in solid-state fermentation using the non-airflow box. J. Biosci. Bioeng. 2013, 116, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Singhania, R.R.; Sukumaran, R.K.; Patel, A.K.; Larroche, C.; Pandey, A. Advancement and comparative profiles in the production technologies using solid-state and submerged fermentation for microbial cellulases. Enzyme Microb. Technol. 2010, 46, 541–549. [Google Scholar] [CrossRef]
- Maulini-Duran, C.; Abraham, J.; Rodríguez-Pérez, S.; Cerda, A.; Jiménez-Peñalver, P.; Gea, T. Gaseous emissions during the solid state fermentation of different wastes for enzyme production at pilot scale. Bioresour. Technol. 2015, 179, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Roukas, T. The role of oxidative stress on carotene production by Blakeslea trispora in submerged fermentation. Crit. Rev. Biotechnol. 2016, 36, 424–433. [Google Scholar] [PubMed]
- Gibbs, P.A.; Seviour, R.J.; Schmid, F. Growth of Filamentous Fungi in Submerged Culture: Problems and Possible Solutions. Crit. Rev. Biotechnol. 2000, 20, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Weng, Y.; Xu, H.; Mao, Z. Enzyme immobilization for biodiesel production. Appl. Microbiol. Biotechnol. 2012, 93, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, G.; Krishnan, C. α-Amylase production from catabolite derepressed Bacillus subtilis KCC103 utilizing sugarcane bagasse hydrolysate. Bioresour. Technol. 2008, 99, 3044–3050. [Google Scholar] [CrossRef] [PubMed]
- Cunha, F.M.; Esperanca, M.N.; Zangirolami, T.C.; Badino, A.C.; Farinas, C.S. Sequential solid-state and submerged cultivation of Aspergillus niger on sugarcane bagasse for the production of cellulase. Bioresour. Technol. 2012, 112, 270–274. [Google Scholar] [CrossRef]
- Fakruddin, M.; Mohammad Mazumdar, R.; Bin Mannan, K.S.; Chowdhury, A.; Hossain, M.N. Critical Factors Affecting the Success of Cloning, Expression, and Mass Production of Enzymes by Recombinant E. coli. ISRN Biotechnol. 2013, 2013, 590587. [Google Scholar] [CrossRef] [PubMed]
- Young, C.L.; Britton, Z.T.; Robinson, A.S. Recombinant protein expression and purification: A comprehensive review of affinity tags and microbial applications. Biotechnol. J. 2012, 7, 620–634. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ye, M.; Pan, Y.; Zhang, Y.; Bian, Y.; Sun, Z. Integration of Cell Lysis, Protein Extraction, and Digestion into One Step for Ultrafast Sample Preparation for Phosphoproteome Analysis. Anal. Chem. 2014, 86, 6786–6791. [Google Scholar] [CrossRef] [PubMed]
- Pečová, M.; Šebela, M.; Markova, Z.; Polakova, K.; Čuda, J.; Šafářová, K. Thermostable trypsin conjugates immobilized to biogenic magnetite show a high operational stability and remarkable reusability for protein digestion. Nanotechnology 2013, 24, 125102. [Google Scholar] [CrossRef] [PubMed]
- Malinsky, J.; Opekarová, M.; Grossmann, G.; Tanner, W. Membrane microdomains, rafts, and detergent-resistant membranes in plants and fungi. Annu. Rev. Plant Biol. 2013, 64, 501–529. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, W.; Thomas, G.A. Simultaneously extracting DNA, RNA, and protein using kits: Is sample quantity or quality prejudiced? Anal. Biochem. 2013, 433, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Campden, R.; Pétrin, D.; Robitaille, M.; Audet, N.; Gora, S.; Angers, S.; Hébert, T.E. Tandem Affinity Purification to Identify Cytosolic and Nuclear Gβγ-Interacting Proteins. In Nuclear G-Protein Coupled Receptors; Springer: New York, NY, USA, 2015; pp. 161–184. [Google Scholar]
- Kanada, K.N.; Nakatsuji, T.; Gallo, R.L. Doxycycline indirectly inhibits proteolytic activation of tryptic kallikrein-related peptidases and activation of cathelicidin. J. Investig. Dermatol. 2012, 132, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Hai, F.I.; Nghiem, L.D.; Roddick, F.; Price, W.E. Removal of trace organic contaminants by nitrifying activated sludge and whole-cell and crude enzyme extract of Trametes versicolor. Water Sci. Technol. 2013, 67, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Girgih, A.T.; Malomo, S.A.; Ju, X.; Aluko, R.E. Antioxidant activities of enzymatic rapeseed protein hydrolysates and the membrane ultrafiltration fractions. J. Funct. Foods 2013, 5, 219–227. [Google Scholar] [CrossRef]
- Mirica, K.A.; Lockett, M.R.; Snyder, P.W.; Shapiro, N.D.; Mack, E.T.; Nam, S. Selective precipitation and purification of monovalent proteins using oligovalent ligands and ammonium sulfate. Bioconj. Chem. 2012, 23, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.L.; He, T.; Tscheliessnig, A.; Mueller, M.; Tan, R.B.H.; Jungbauer, A. Protein precipitation by polyethylene glycol: A generalized model based on hydrodynamic radius. J. Biotechnol. 2012, 157, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Hanson, C. Recent Advances in Liquid-Liquid Extraction; Pergamon Press, Elsevier: Oxford, UK, 2013. [Google Scholar]
- Babu, B.R.; Rastogi, N.K.; Raghavarao, K.S.M.S. Liquid–liquid extraction of bromelain and polyphenol oxidase using aqueous two-phase system. Chem. Eng. Process. Process Intensif. 2008, 47, 83–89. [Google Scholar] [CrossRef]
- Scopes, R.K. Protein Purification: Principles and Practice; Springer: Berlin, Germany, 2013. [Google Scholar]
- Kunji, E.R.S.; Harding, M.; Butler, P.J.G.; Akamine, P. Determination of the molecular mass and dimensions of membrane proteins by size exclusion chromatography. Methods 2008, 46, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.M.H.; Kittelmann, J.; Pilgram, F.; Osberghaus, A.; Hubbuch, J. Custom-tailored Adsorbers: A Molecular Dynamics Study on Optimal Design of Ion Exchange Chromatography Material. J. Chromatogr. A 2015, 1413, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Shchukina, O.I.; Zatirakha, A.V.; Smolenkov, A.D.; Nesterenko, P.N.; Shpigun, O.A. Anion exchangers with branched functional ion exchange layers of different hydrophilicity for ion chromatography. J. Chromatogr. A 2015, 1408, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Zhang, H.; Huang, L.; Nishinari, K. Effects of the lyotropic series salts on the gelation of konjac glucomannan in aqueous solutions. Carbohydr. Polym. 2008, 74, 68–78. [Google Scholar] [CrossRef]
- Zhao, K.; Yang, L.; Wang, X.; Bai, Q.; Yang, F.; Wang, F. Preparation of a novel dual-function strong cation exchange/hydrophobic interaction chromatography stationary phase for protein separation. Talanta 2012, 98, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Ayyar, B.V.; Arora, S.; Murphy, C.; O’Kennedy, R. Affinity chromatography as a tool for antibody purification. Methods 2012, 56, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Tropea, J.E.; Cherry, S.; Waugh, D.S. Expression and purification of soluble his 6-tagged TEV protease. In High Throughput Protein Expression and Purification: Methods and ProtocolsI; Humana Press: Totowa, NJ, USA, 2009; pp. 297–307. [Google Scholar]
- Royle, L.; Radcliffe, C.M.; Dwek, R.A.; Rudd, P.M. Detailed structural analysis of n-glycans released from glycoproteins in sds-page gel bands using HPLC combined with exoglycosidase array digestions. In Glycobiology Protocols; Humana Press: Totowa, NJ, USA, 2007; pp. 125–143. [Google Scholar]
- Rothe, G.M. Electrophoresis of Enzymes: Laboratory Methods; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Boeris, V.; Balce, I.; Vennapusa, R.R.; Rodríguez, M.A.; Picó, G.; Lahore, M.F. Production, recovery and purification of a recombinant β-galactosidase by expanded bed anion exchange adsorption. J. Chromatogr. B 2012, 900, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Dong, J.; Ma, Y.; Hovde, S.; Geiger, J.H.; Baker, G.L.; Bruening, M.L. Formation of high-capacity protein-adsorbing membranes through simple adsorption of poly (acrylic acid)-containing films at low pH. Langmuir 2012, 28, 6885–6892. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Hernandez, K.; Fernandez-Lafuente, R. Control of protein immobilization: Coupling immobilization and site-directed mutagenesis to improve biocatalyst or biosensor performance. Enzyme Microb. Technol. 2011, 48, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Fatima, A.; Husain, Q. Polyclonal antibodies mediated immobilization of a peroxidase from ammonium sulphate fractionated bitter gourd (Momordica charantia) proteins. Biomol. Eng. 2007, 24, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bhattacharyya, D.; Bachas, L.G. Orientation specific immobilization of organophosphorus hydrolase on magnetic particles through gene fusion. Biomacromolecules 2001, 2, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Van Tilbeurgh, H.; Egloff, M.-P.; Martinez, C.; Rugani, N.; Verger, R.; Cambillau, C. Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by x-ray crystallography. Nature 1993, 362, 814–820. [Google Scholar] [CrossRef] [PubMed]
- Brzozowski, A.; Derewenda, U.; Derewenda, Z.; Dodson, G.; Lawson, D.; Turkenburg, J.; Bjorkling, F.; Huge-Jensen, B.; Patkar, S.; Thim, L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature 1991, 351, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, Z.S.; Derewenda, U.; Dodson, G.G. The crystal and molecular structure of the rhizomucor miehei triacylglyceride lipase at 1.9 Å resolution. J. Mol. Biol. 1992, 227, 818–839. [Google Scholar] [CrossRef]
- Fernández-Lorente, G.; Palomo, J.M.; Cabrera, Z.; Guisán, J.M.; Fernández-Lafuente, R. Specificity enhancement towards hydrophobic substrates by immobilization of lipases by interfacial activation on hydrophobic supports. Enzyme Microb. Technol. 2007, 41, 565–569. [Google Scholar] [CrossRef]
- Fernandez-Lorente, G.; Cabrera, Z.; Godoy, C.; Fernandez-Lafuente, R.; Palomo, J.M.; Guisan, J.M. Interfacially activated lipases against hydrophobic supports: Effect of the support nature on the biocatalytic properties. Process Biochem. 2008, 43, 1061–1067. [Google Scholar] [CrossRef]
- Manoel, E.A.; dos Santos, J.C.; Freire, D.M.; Rueda, N.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports involves the open form of the enzyme. Enzyme Microb. Technol. 2015, 71, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Palomo, J.M.; Muñoz, G.; Fernández-Lorente, G.; Mateo, C.; Fernández-Lafuente, R.; Guisán, J.M. Interfacial adsorption of lipases on very hydrophobic support (octadecyl–sepabeads): Immobilization, hyperactivation and stabilization of the open form of lipases. J. Mol. Catal. B Enzym. 2002, 19, 279–286. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Rocha-Martin, J.; Godoy, C.; Rodrigues, R.C.; Guisan, J.M. Complete reactivation of immobilized derivatives of a trimeric glutamate dehydrogenase from thermus thermophillus. Process Biochem. 2010, 45, 107–113. [Google Scholar] [CrossRef]
- Barbosa, O.; Torres, R.; Ortiz, C.; Berenguer-Murcia, A.N.; Rodrigues, R.C.; Fernandez-Lafuente, R. Heterofunctional supports in enzyme immobilization: From traditional immobilization protocols to opportunities in tuning enzyme properties. Biomacromolecules 2013, 14, 2433–2462. [Google Scholar] [CrossRef] [PubMed]
- Mateo, C.; Palomo, J.M.; Fuentes, M.; Betancor, L.; Grazu, V.; López-Gallego, F.; Pessela, B.C.; Hidalgo, A.; Fernández-Lorente, G.; Fernández-Lafuente, R. Glyoxyl agarose: A fully inert and hydrophilic support for immobilization and high stabilization of proteins. Enzyme Microb. Technol. 2006, 39, 274–280. [Google Scholar] [CrossRef]
- Mateo, C.; Abian, O.; Bernedo, M.; Cuenca, E.; Fuentes, M.; Fernandez-Lorente, G.; Palomo, J.M.; Grazu, V.; Pessela, B.C.; Giacomini, C. Some special features of glyoxyl supports to immobilize proteins. Enzyme Microb. Technol. 2005, 37, 456–462. [Google Scholar] [CrossRef]
- Pedroche, J.; del Mar Yust, M.; Mateo, C.; Fernández-Lafuente, R.; Girón-Calle, J.; Alaiz, M.; Vioque, J.; Guisán, J.M.; Millán, F. Effect of the support and experimental conditions in the intensity of the multipoint covalent attachment of proteins on glyoxyl-agarose supports: Correlation between enzyme–support linkages and thermal stability. Enzyme Microb. Technol. 2007, 40, 1160–1166. [Google Scholar] [CrossRef]
- Jönsson, L.J.; Alriksson, B.; Nilvebrant, N.-O. Bioconversion of lignocellulose: Inhibitors and detoxification. Biotechnol. Biofuels 2013, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Ranatunga, T.D.; Jervis, J.; Helm, R.F.; McMillan, J.D.; Wooley, R.J. The effect of overliming on the toxicity of dilute acid pretreated lignocellulosics: The role of inorganics, uronic acids and ether-soluble organics. Enzyme Microb. Technol. 2000, 27, 240–247. [Google Scholar] [CrossRef]
- Alriksson, B.; Horvath, I.; Sjöde, A.; Nilvebrant, N.-O.; Jönsson, L. Ammonium Hydroxide Detoxification of Spruce Acid Hydrolysates. In Twenty-Sixth Symposium on Biotechnology for Fuels and Chemicals; Davison, B., Evans, B., Finkelstein, M., McMillan, J., Eds.; Humana Press: Totowa, NJ, USA, 2005; pp. 911–922. [Google Scholar]
- Behera, S.; Arora, R.; Nandhagopal, N.; Kumar, S. Importance of chemical pretreatment for bioconversion of lignocellulosic biomass. Renew. Sustain. Energy Rev. 2014, 36, 91–106. [Google Scholar] [CrossRef]
- Jönsson, L.; Palmqvist, E.; Nilvebrant, N.-O.; Hahn-Hägerdal, B. Detoxification of wood hydrolysates with laccase and peroxidase from the white-rot fungus trametes versicolor. Appl. Microbiol. Biotechnol. 1998, 49, 691–697. [Google Scholar] [CrossRef]
- Cao, G.; Ximenes, E.; Nichols, N.N.; Zhang, L.; Ladisch, M. Biological abatement of cellulase inhibitors. Bioresource Technol. 2013, 146, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Ximenes, E.; Mosier, N.S.; Ladisch, M.R. Soluble inhibitors/deactivators of cellulase enzymes from lignocellulosic biomass. Enzyme Microb. Technol. 2011, 48, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, D.; Guerra, L.; Camassola, M.; Dillon, A.J. Secretion of endoglucanases and β-glucosidases by penicillium echinulatum 9A02S1 in presence of different carbon sources. Ind. Crops Prod. 2013, 50, 882–886. [Google Scholar] [CrossRef]
- Yang, S.; Xiong, H.; Yang, H.; Yan, Q.; Jiang, Z. High-level production of β-1, 3-1, 4-glucanase by rhizomucor miehei under solid-state fermentation and its potential application in the brewing industry. J. Appl. Microbiol. 2015, 118, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Irshad, M.; Anwar, Z.; Afroz, A. Characterization of exo 1, 4-[beta] glucanase produced from tricoderma viridi through solid-state bio-processing of orange peel waste. Adv. Biosci. Biotechnol. 2012, 3, 580–584. [Google Scholar] [CrossRef]
- Dhillon, G.S.; Kaur, S.; Brar, S.K.; Verma, M. Potential of apple pomace as a solid substrate for fungal cellulase and hemicellulase bioproduction through solid-state fermentation. Ind. Crops. Prod. 2012, 38, 6–13. [Google Scholar] [CrossRef]
- Sun, H.-Y.; Li, J.; Zhao, P.; Peng, M. Banana peel: A novel substrate for cellulase production under solid-state fermentation. Afr. J. Biotechnol. 2011, 10, 17887–17890. [Google Scholar]
- Dilipkumar, M.; Rajasimman, M.; Rajamohan, N. Utilization of copra waste for the solid state fermentatative production of inulinase in batch and packed bed reactors. Carbohydr. Polym. 2014, 102, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Onilude, A.A.; Fadaunsi, I.F.; Garuba, E.O. Inulinase production by saccharomyces sp. in solid state fermentation using wheat bran as substrate. Ann. Microbiol. 2012, 62, 843–848. [Google Scholar] [CrossRef]
- Paixão, S.M.; Teixeira, P.D.; Silva, T.P.; Teixeira, A.V.; Alves, L. Screening of novel yeast inulinases and further application to bioprocesses. New Biotechnol. 2013, 30, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Rashad, M.M.; Nooman, M.U. Production, purification and characterization of extracellular invertase from saccharomyses cerevisiae NRRL Y-12632 by solid state fermentation of red carrot residue. Aust. J. Basic Appl. Sci. 2009, 3, 1910–1919. [Google Scholar]
- Veana, F.; Martínez-Hernández, J.; Aguilar, C.; Rodríguez-Herrera, R.; Michelena, G. Utilization of molasses and sugar cane bagasse for production of fungal invertase in solid state fermentation using aspergillus niger GH1. Braz. J. Microbiol. 2014, 45, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Uma, C.; Gomathi, D.; Ravikumar, G.; Kalaiselvi, M.; Palaniswamy, M. Production and properties of invertase from a cladosporium cladosporioides in SmF using pomegranate peel waste as substrate. Asian Pac. J. Trop. Biomed. 2012, 2, S605–S611. [Google Scholar] [CrossRef]
- Akolkar, S.; Sajgure, A.; Lele, S. Lactase production from lactobacillus acidophilus. World J. Microb. Biot. 2005, 21, 1119–1122. [Google Scholar] [CrossRef]
- Yin, J.-S.; Liang, Q.-L.; Li, D.-M.; Sun, Z.-T. Optimization of production conditions for β-mannanase using apple pomace as raw material in solid-state fermentation. Ann. Microbiol. 2013, 63, 101–108. [Google Scholar] [CrossRef]
- Rashid, J.I.A.; Samat, N.; Yusoff, W.M.W. Studies on extraction of mannanase enzyme by aspergillus terreus SUK-1 from fermented palm kernel cake. Pak. J. Biol. Sci. 2013, 16, 933. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.; Lima, V.; Giloni-Lima, P.; Knob, A. Passion fruit peel as novel substrate for enhanced β-glucosidases production by penicillium verruculosum: Potential of the crude extract for biomass hydrolysis. Biomass Bioenergy 2015, 72, 216–226. [Google Scholar] [CrossRef]
- Li, P.-J.; Xia, J.-L.; Shan, Y.; Nie, Z.-Y.; Su, D.-L.; Gao, Q.-R.; Zhang, C.; Ma, Y.-L. Optimizing production of pectinase from orange peel by penicillium oxalicum PJ02 using response surface methodology. Waste Biomass Valoriz. 2015, 6, 13–22. [Google Scholar] [CrossRef]
- Patil, S.R.; Dayanand, A. Production of pectinase from deseeded sunflower head by aspergillus niger in submerged and solid-state conditions. Bioresour. Technol. 2006, 97, 2054–2058. [Google Scholar] [CrossRef] [PubMed]
- Murthy, P.S.; Naidu, M.M. Production and application of xylanase from penicillium sp. Utilizing coffee by-products. Food Bioprocess Technol. 2012, 5, 657–664. [Google Scholar] [CrossRef]
- Nascimento, R.P.D.; Alves Junior, N.; Coelho, R.R.R. Brewer’s spent grain and corn steep liquor as alternative culture medium substrates for proteinase production by Streptomyces malaysiensis amt-3. Braz. J. Microbiol. 2011, 42, 1384–1389. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Misiewicz, A. Microbial transglutaminase and its application in the food industry. A review. Folia Microbiol. 2014, 59, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Songulashvili, G.; Spindler, D.; Jimenez-Tobon, G.A.; Jaspers, C.; Kerns, G.; Penninckx, M.J. Production of a high level of laccase by submerged fermentation at 120-L scale of Cerrena unicolor C-139 grown on wheat bran. C. R. Biol. 2015, 338, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Gerber, C.B.; Kaufmann, F.; Nicoletti, G.; Dalla Costa, M.; Kempka, A.P. Production of lipase using cassava peel and sunflower oil in solid-state fermentation: Preliminary study. J. Agr. Sci. Technol. A 2013, 3, 948. [Google Scholar]
- Jadhav, M.S.; Chougule, D.; Rampure, S. Lipase production from banana peel extract and potato peel extract. Int. J. Res. Pure Appl. Micrbiol. 2013, 3, 11–13. [Google Scholar]
- Mittal, A.; Singh, G.; Goyal, V.; Yadav, A.; Aggarwal, N.K. Production of phytase by acido-thermophilic strain of Klebsiella sp. DB-3FJ711774. 1 using orange peel flour under submerged fermentation. Innov. Rom. Food Biotechnol. 2012, 10, 18. [Google Scholar]
- Demir, H.; Göğüş, N.; Tari, C.; Heerd, D.; Lahore, M.F. Optimization of the process parameters for the utilization of orange peel to produce polygalacturonase by solid-state fermentation from an Aspergillus sojae mutant strain. Turk. J. Biol. 2012, 36, 394–404. [Google Scholar]
- Demir, H.; Tarı, C. Valorization of wheat bran for the production of polygalacturonase in SSF of Aspergillus sojae. Ind. Crops Prod. 2014, 54, 302–309. [Google Scholar] [CrossRef]
- Sharma, R.; Rawat, R.; Bhogal, R.S.; Oberoi, H.S. Multi-component thermostable cellulolytic enzyme production by Aspergillus niger HN-1 using pea pod waste: Appraisal of hydrolytic potential with lignocellulosic biomass. Process Biochem. 2015, 50, 696–704. [Google Scholar] [CrossRef]
- Inoue, H.; Kitao, C.; Yano, S.; Sawayama, S. Production of β-xylosidase from Trichoderma asperellum KIF125 and its application in efficient hydrolysis of pretreated rice straw with fungal cellulase. World J. Microb. Biotechnol. 2016, 32, 186. [Google Scholar] [CrossRef] [PubMed]
- Dapper, T.B.; Arfelli, V.C.; Henn, C.; Simões, M.R.; dos Santos, M.R.; Torre, C.L.D.; Silva, J.L.C.; Simão, R.C.G.; Kadowaki, M.K. Fructofuranosidase production by Aspergillus versicolor isolated from Atlantic forest and grown on apple pomace. Afr. J. Microbiol. Res. 2016, 25, 938–948. [Google Scholar]

| Enzyme | Feedstock | Bioprocessing Conditions | Microbial Strain | Application | Reference |
|---|---|---|---|---|---|
| Amylase | Brewers’ spent grain hydrolysate | Submerged fermentation mode, hydrolysate used as additive in amylase production media, fermentation time: 30 h | Catabolite-repressed Bacillus subtilis KCC103 | Baking, brewing, animal nutrition, aquaculture, biofuel, dishwashing and laundry detergents | [58] |
| β-glucanase | Oatmeal Orange peel | Solid state fermentation mode, 50% w/w moisture, temperature: 30 °C, pH: 5.5, fermentation time: 4 days Solid state fermentation mode, orange peel with basal media, incubated at 30 °C, pH: 5.5, fermentation time: 4 days | Rhizomucor miehei CAU432 Tricoderma viride MBL | Brewing, bioethanol | [109,110,111] |
| Cellulase | Apple pomace Banana peel | Solid state fermentation mode, 75% initial moisture content, initial temperature: 30 °C, fermentation time: 48–72 h Solid state fermentation mode, 50% moisture content, temperature: 30 °C, fermentation time: 192 h | Aspergillus niger NRRL-567 Trichoderma viride GIM 3.0010 | Detergents, bleaching, deinking, refining, starch modification, drainage improvement, decolourization of dyes in effluent, cellulosic and starch based ethanol, biodiesel | [112,113] |
| Inulinase | Yacon juice Banana peel, wheat bran, rice bran, orange peel, bagasse Soy bean cake | Submerged batch fermentation mode, temperature: 30 °C, pH: 5, fermentation time: 7 days Solid State fermentation mode, 65% moisture content, temperature: 35 °C, fermentation time: 72 h Solid state fermentation mode, 60% moisture content, K2HPO4, ZnSO4·7H2O, temperature: 30 °C, fermentation time: 48 h | Aspergillus kawachii Saccharomyces sp. Pencillium rugulosum (MTCC-3487) | Production of high-fructose corn syrup | [32,114,115,116] |
| Invertase | Red carrot jam processing residue Sugarcane bagasse Orange peel, pineapple peel waste | Solid state fermentation mode, temperature: 30 °C, fermentation time: 72 h Solid state fermentation mode, temperature: 30 °C, fermentation time: 72 h Solid state fermentation mode, temperature: 30 °C, fermentation time: 32 h, initial pH: 5.5 | S. cerevisiae NRRL Y-12632 Aspergillus niger GH1, Cladosporium cladosporioides | Sucrose hydrolysis | [117,118,119] |
| Lactase | Fermented ragi | Submerged fermentation mode, media supplemented by 0.75% lactose and 1% ragi, fermentation time 12 h, pH 5.5 | Lactobacillus acidophilus | Dairy, preparation of lactose-free food products | [120] |
| Mannanase | Apple pomace and cotton seed powder mixture | Solid state fermentation mode, 50% initial moisture content, pH 5.5, temperature 30 °C, fermentation time 48 h | Aspergillus niger SN-09 | Paper and pulp, textile, pharmaceuticals | [121,122,123] |
| Palm kernel cake | Solid state fermentation mode in stainless steel horizontal bioreactor, initial moisture content 1:0.75 (w/v), temperature 30 °C, fermentation time 4 days. | Aspergillus terreus SUK-1 | |||
| Passion fruit peel | Submerged fermentation mode, pH 6.5, 8.6 days | Pencillium verruculosum | |||
| Pectinase | Orange peel | Submerged fermentation mode, temperature 35 °C, pH 5.2, fermentation time: 3 days | Pencillium oxalicum PJ02 Aspergillus niger | Processing of starch and wine, juice processing | [124,125] |
| Deseeded sunflower head | Solid state fermentation mode, temperature: 34 °C, initial moisture content: 65%, fermentation time: 120 h Submerged fermentation mode, temperature: 34 °C, pH: 5.0, fermentation time: 120 h | ||||
| Xylanase | Coffee by-products | Solid state fermentation mode, initial moisture content: 50%, temperature: 30 °C, fermentation time: 5 days. | Pencillium sp. CFR 303 | Bleaching and deinking of paper, baking, animal nutrition | [126] |
| Protease | Brewer’s spent grain, corn steep liqour | Submerged fermentation mode, temperature: 28 °C, fermentation time: 6 days | Streptomyces malaysiensis AMT-3, | Food, pharmaceutical, animal feed, leather, diagnostics, waste management | [127] |
| Transglutaminase | Industrial fibrous soy residue | Solid state fermentation mode, temperature: 33 °C, fermentation time: 48 h | Bacillus circulans BL32 | Meat processing, dairy products, baking, edible film, leather finishing, cosmetics | [128] |
| Laccase | Wheat bran | Stirred bioreactor working volume: 120 L, temperature: 30 °C, pH: 6.0, fermentation time: 4 days | Cerrena unicolor C-139 | Bleaching, deinking of paper, polishing and preparation of textiles | [129] |
| Lipase | Banana peel, potato peel, cassava peel | Solid state fermentation mode, initial moisture content: 55%, temperature: 30.5 °C, fermentation time: 60 h | Aspergillus niger | Meat processing, detergents, degreasing, dehairing of leather | [130,131] |
| Phytase | Orange and citrus peel | Solid state fermentation mode, temperature: 50 °C, fermentation time: 72 h | Klebsiella sp. DB-3FJ711774.1 | Animal nutrition | [132] |
| Polygalacturonase | Orange peel, wheat bran | Solid state fermentation mode, temperature: 22.4 °C to 27.5 °C, incubation period: 3.8 to 5.5 days | Aspergillus sojae M3 | Catalyzes the hydrolysis of α-1,4-glycosidic linkages in pectic acid. Used in food industry. | [133,134] |
| Cellulase | Pea pod waste | Solid state fermentation mode, moisture content of 70% made up by Mendel Weber medium, temperature: 30 °C, fermentation time: 96 h | Aspergillus niger HN-1 | Detergents, bleaching, deinking, refining, starch modification, drainage improvement, decolourization of dyes in effluent, cellulosic and starch based ethanol, biodiesel | [135] |
| β-Xylosidase | Wet disc milling rice straw (WDMRS) | Submerged fermentation mode, working volume: 1 L in 2 L bioreactor, pH: 4.8, media composition: 20 g WDMRS, 5 g polypeptone, 4 g urea, 2 g (NH4)2SO4, 5 g (NH4)2HPO4, 5 g KNO3, 15 g KH2PO4, 1 g Tween 80, fermentation time: 96 h | Trichoderma asperellum KIF125 | Baking, improving digestibility of animal feed, production of d-xylose for xylitol manufacture, deinking of recycled paper | [136] |
| β-fructofuranosidase (invertase) | Apple pomace | Submerged fermentation mode, working volume: 25 mL in 125 mL Erlenmeyer flask, initial pH: 7.5, fermentation time: 12 days, room temperature | Aspergillus versicolor | Food additive | [137] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravindran, R.; Jaiswal, A.K. Microbial Enzyme Production Using Lignocellulosic Food Industry Wastes as Feedstock: A Review. Bioengineering 2016, 3, 30. https://doi.org/10.3390/bioengineering3040030
Ravindran R, Jaiswal AK. Microbial Enzyme Production Using Lignocellulosic Food Industry Wastes as Feedstock: A Review. Bioengineering. 2016; 3(4):30. https://doi.org/10.3390/bioengineering3040030
Chicago/Turabian StyleRavindran, Rajeev, and Amit K. Jaiswal. 2016. "Microbial Enzyme Production Using Lignocellulosic Food Industry Wastes as Feedstock: A Review" Bioengineering 3, no. 4: 30. https://doi.org/10.3390/bioengineering3040030
APA StyleRavindran, R., & Jaiswal, A. K. (2016). Microbial Enzyme Production Using Lignocellulosic Food Industry Wastes as Feedstock: A Review. Bioengineering, 3(4), 30. https://doi.org/10.3390/bioengineering3040030