Focused Screening of ECM-Selective Adhesion Peptides on Cellulose-Bound Peptide Microarrays
Abstract
:1. Introduction
2. Materials and Methods
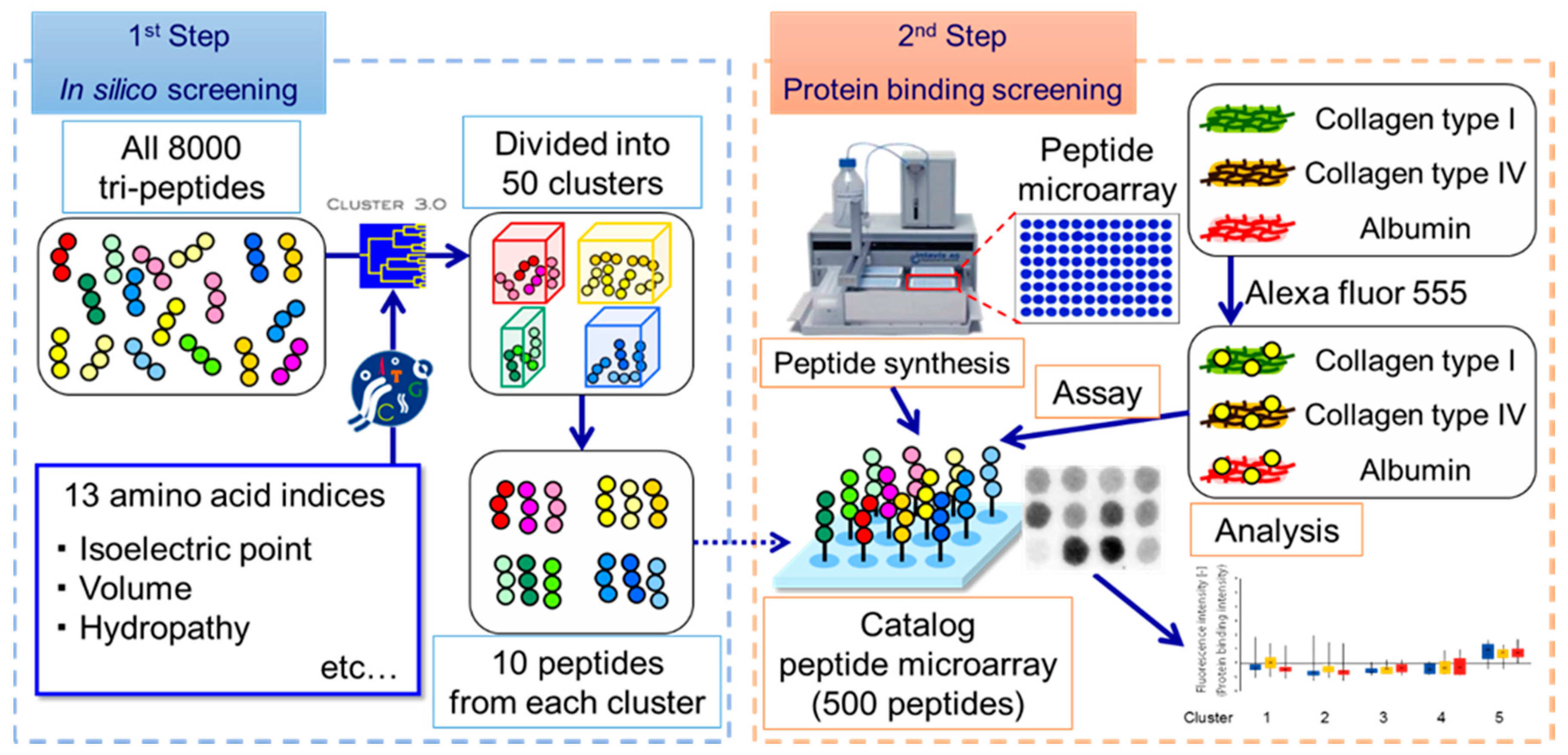
2.1. Design of Custom SPOT Peptide Microarray
2.2. Protein Accumulation Assay on Peptide Microarray
2.3. Cell Culture
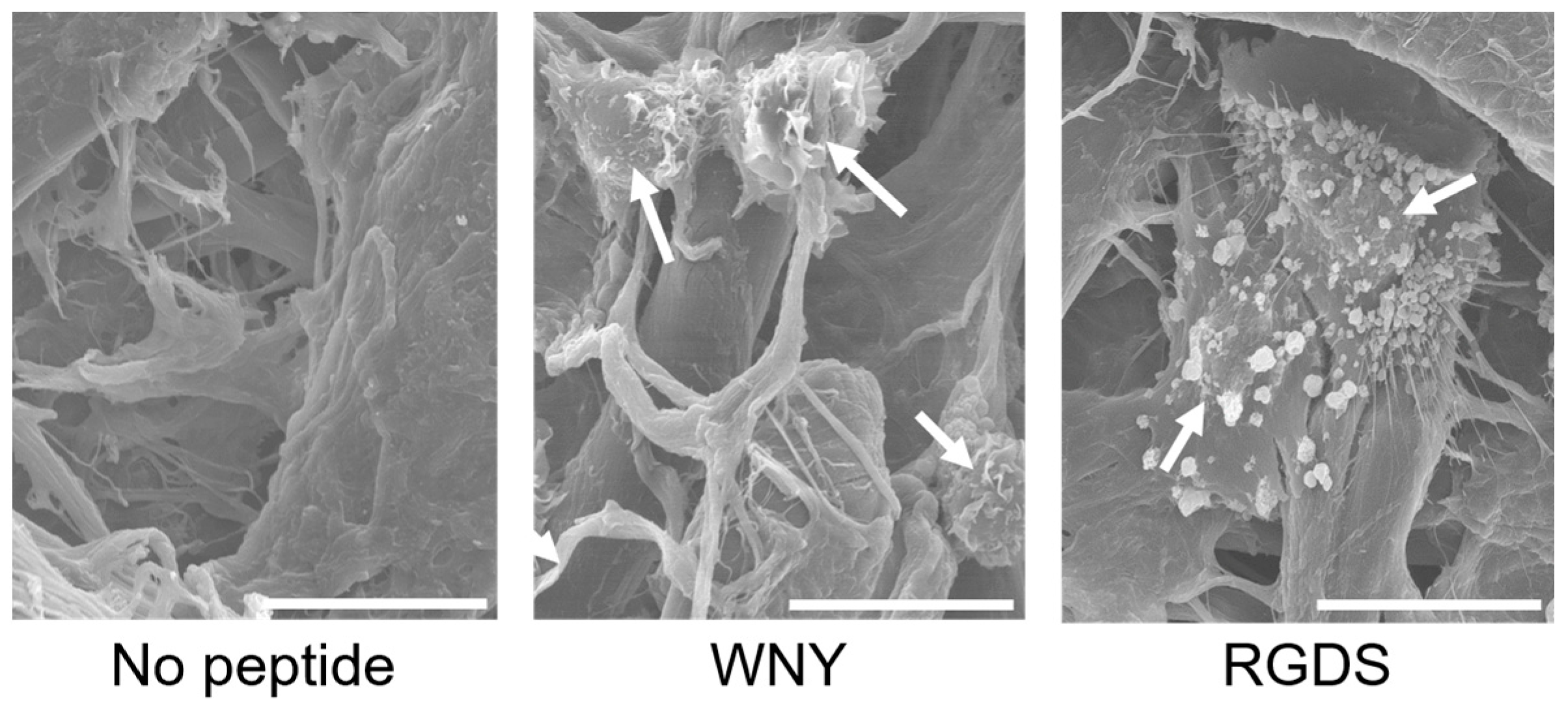
2.4. Scanning Electron Microscopy
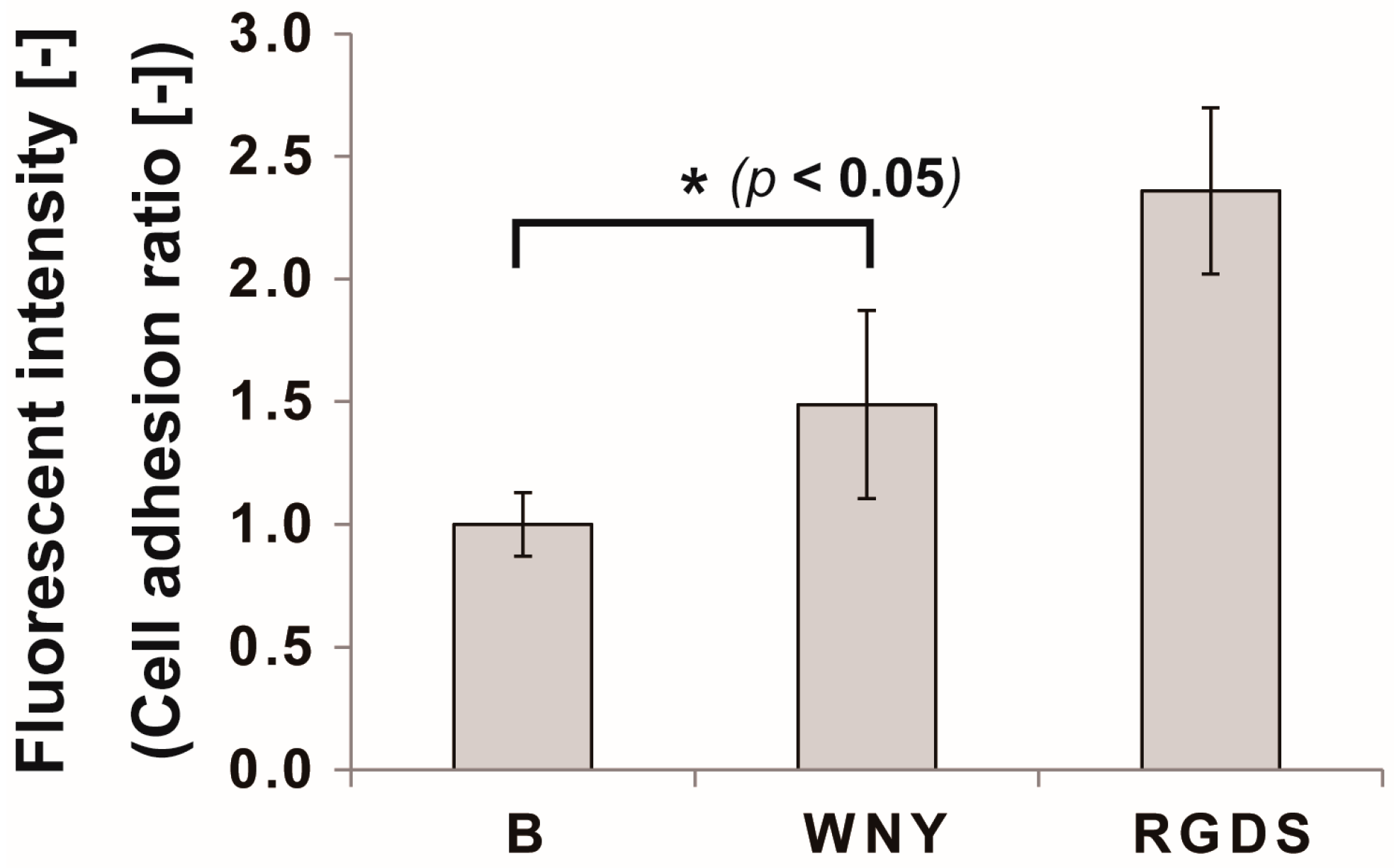
2.5. Cell Adhesion Assay
3. Results and Discussion
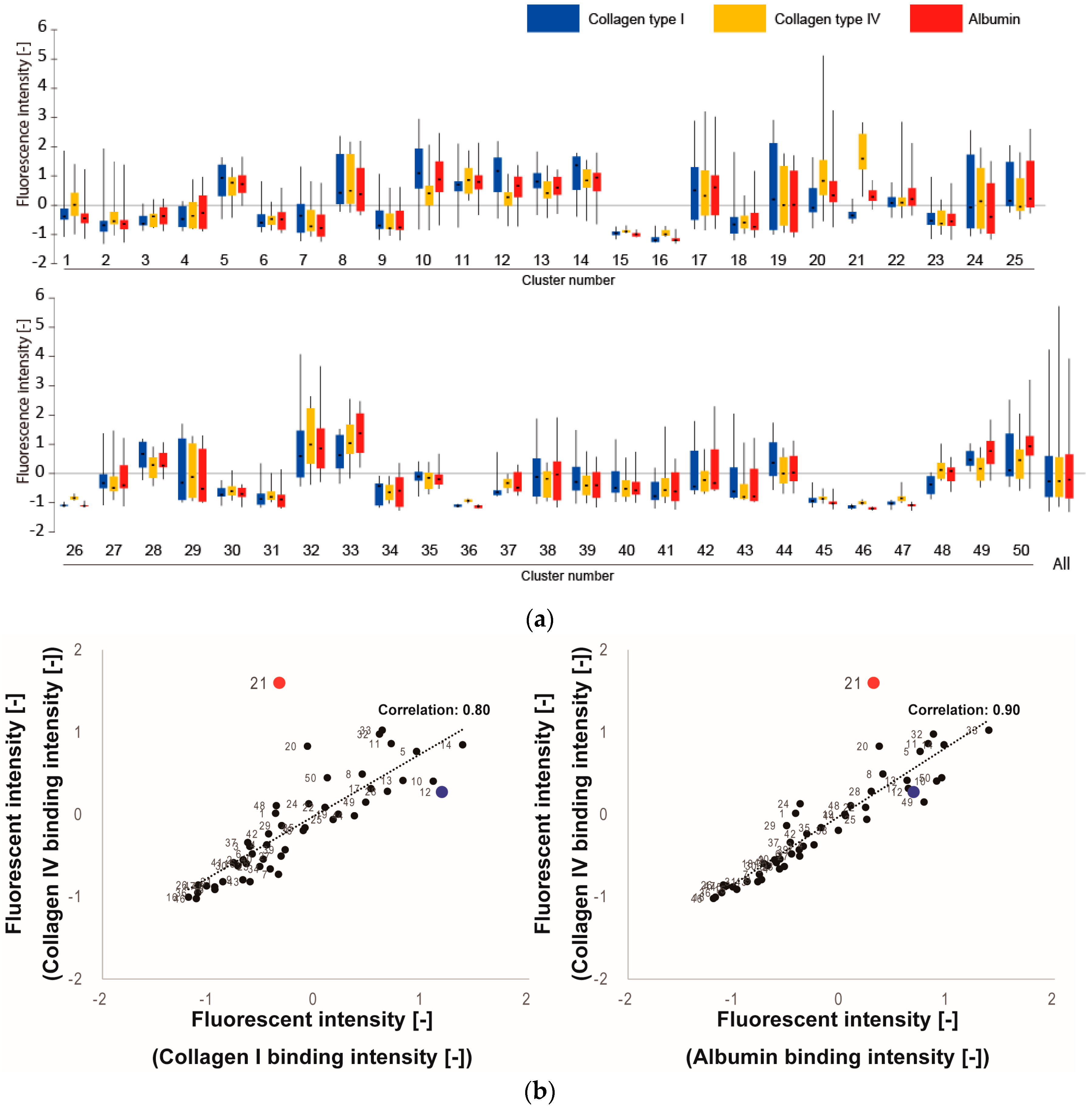
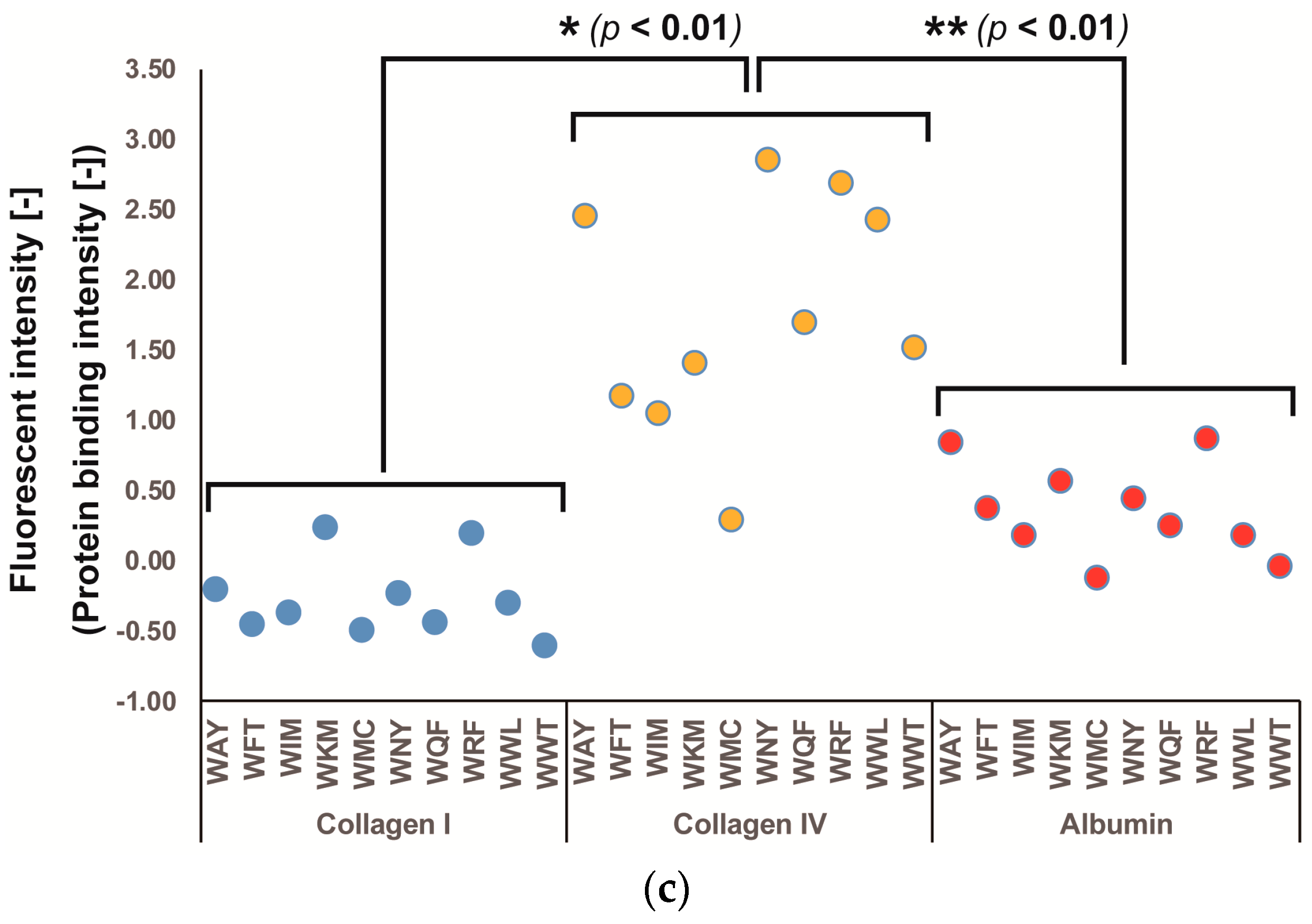
3.1. Screening of Col IV-Selective Adhesion Peptides
3.2. Physicochemical Properties of Col IV-Selective Adhesion Peptides
3.3. Bio-Safety of Col IV-Selective Adhesion Peptides with ECs
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bourassa, M.G. Fate of venous grafts: The past, the present and the future. J. Am. Coll. Cardiol. 1991, 17, 1081–1083. [Google Scholar] [CrossRef]
- Rafat, M.; Matsuura, T.; Li, F.; Griffith, M. Surface modification of collagen-based artificial cornea for reduced endothelialization. J. Biomed. Mater. Res. A 2009, 88, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Petrie, T.A.; Reyes, C.D.; Burns, K.L.; Garcia, A.J. Simple application of fibronectin-mimetic coating enhances osseointegration of titanium implants. J. Cell. Mol. Med. 2009, 13, 2602–2612. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.K.; Kleinert, L.B.; Patula-Steinbrenner, V. Accelerated neovascularization and endothelialization of vascular grafts promoted by covalently bound laminin type 1. J. Biomed. Mater. Res. A 2011, 99A, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Stellos, K.; Langer, H.; Daub, K.; Schoenberger, T.; Gauss, A.; Geisler, T.; Bigalke, B.; Mueller, I.; Schumm, M.; Schaefer, I.; et al. Platelet-derived stromal cell-derived factor-1 regulates adhesion and promotes differentiation of human CD34+ cells to endothelial progenitor cells. Circulation 2008, 117, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Rodenberg, E.J.; Pavalko, F.M. Peptides derived from fibronectin type III connecting segments promote endothelial cell adhesion but not platelet adhesion: Implications in tissue-engineered vascular grafts. Tissue Eng. 2007, 13, 2653–2666. [Google Scholar] [CrossRef] [PubMed]
- Larsen, C.C.; Kligman, F.; Tang, C.; Kottke-Marchant, K.; Marchant, R.E. A biomimetic peptide fluorosurfactant polymer for endothelialization of ePTFE with limited platelet adhesion. Biomaterials 2007, 28, 3537–3548. [Google Scholar] [CrossRef] [PubMed]
- Blindt, R.; Vogt, F.; Astafieva, I.; Fach, C.; Hristov, M.; Krott, N.; Seitz, B.; Kapurniotu, A.; Kwok, C.; Dewor, M.; et al. A novel drug-eluting stent coated with an integrin-binding cyclic Arg-Gly-Asp peptide inhibits neointimal hyperplasia by recruiting endothelial progenitor cells. J. Am. Coll. Cardiol. 2006, 47, 1786–1795. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Yuan, Y.; Liu, C.; Wang, J. Development of mussel adhesive polypeptide mimics coating for in-situ inducing re-endothelialization of intravascular stent devices. Biomaterials 2009, 30, 2764–2773. [Google Scholar] [CrossRef] [PubMed]
- Kanie, K.; Narita, Y.; Zhao, Y.; Kuwabara, F.; Satake, M.; Honda, S.; Kaneko, H.; Yoshioka, T.; Okochi, M.; Honda, H.; et al. Collagen type IV-specific tripeptides for selective adhesion of endothelial and smooth muscle cells. Biotechnol. Bioeng. 2012, 109, 1808–1816. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, F.; Narita, Y.; Yamawaki-Ogata, A.; Kanie, K.; Kato, R.; Satake, M.; Kaneko, H.; Oshima, H.; Usui, A.; Ueda, Y. Novel small-caliber vascular grafts with trimeric Peptide for acceleration of endothelialization. Ann. Thorac. Surg. 2012, 93, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Kato, R.; Kaga, C.; Kunimatsu, M.; Kobayashi, T.; Honda, H. Peptide array-based interaction assay of solid-bound peptides and anchorage-dependant cells and its effectiveness in cell-adhesive peptide design. J. Biosci. Bioeng. 2006, 101, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Kato, R.; Kaga, C.; Kanie, K.; Kunimatsu, M.; Okochi, M.; Honda, H. Peptide array-based peptide-cell interaction analysis. Mini Rev. Org. Chem. 2011, 8, 171–177. [Google Scholar] [CrossRef]
- Hubbell, J.A.; Massia, S.P.; Desai, N.P.; Drumheller, P.D. Endothelial cell-selective materials for tissue engineering in the vascular graft via a new receptor. Biotechnology (N.Y.) 1991, 9, 568–572. [Google Scholar] [CrossRef]
- Gobin, A.S.; West, J.L. Val-ala-pro-gly, an elastin-derived non-integrin ligand: Smooth muscle cell adhesion and specificity. J. Biomed. Mater. Res. A 2003, 67, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Li, J.A.; Zhang, K.; Wu, F.; He, Z.K.; Yang, P.; Huang, N. Constructing bio-functional layers of hyaluronan and type IV collagen on titanium surface for improving endothelialization. J. Mater. Sci. 2015, 50, 3226–3236. [Google Scholar] [CrossRef]
- Frank, R. Spot-synthesis—An easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 1992, 48, 9217–9232. [Google Scholar] [CrossRef]
- AAindex. Available online: http://www.genome.ad.jp/dbget/aaindex.html (accessed on 18 November 2016).
- Kawashima, S.; Pokarowski, P.; Pokarowska, M.; Kolinski, A.; Katayama, T.; Kanehisa, M. Aaindex: Amino acid index database, progress report 2008. Nucleic Acids Res. 2008, 36, D202–D205. [Google Scholar] [CrossRef] [PubMed]
- Software. Available online: http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm#ctv (accessed on 18 November 2016).
- Maple Tree. Available online: http://mapletree.sourceforge.net (accessed on 18 November 2016).
- Zimmerman, J.M.; Eliezer, N.; Simha, R. The characterization of amino acid sequences in proteins by statistical methods. J. Theor. Boil. 1968, 21, 170–201. [Google Scholar] [CrossRef]
- Fauchere, J.L.; Charton, M.; Kier, L.B.; Verloop, A.; Pliska, V. Amino acid side chain parameters for correlation studies in biology and pharmacology. Int. J. Pept. Protein Res. 1988, 32, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Geisow, M.J.; Roberts, R.D.B. Amino-acid preferences for secondary structure vary with protein class. Int. J. Biol. Macromol. 1980, 2, 387–389. [Google Scholar] [CrossRef]
- Takano, K.; Yutani, K. A new scale for side-chain contribution to protein stability based on the empirical stability analysis of mutant proteins. Protein Eng. 2001, 14, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.Y.; Zhou, Y.Q. Quantifying the effect of burial of amino acid residues on protein stability. Proteins 2004, 54, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Crawford, J.L.; Lipscomb, W.N.; Schellma, C.G. Reverse turn as a polypeptide conformation in globular proteins. Proc. Natl. Acad. Sci. USA 1973, 70, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Munoz, V.; Serrano, L. Intrinsic secondary structure propensities of the amino-acids, using statistical phi-psi matrices: Comparison with experimental scales. Proteins 1994, 20, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Grantham, R. Amino-acid difference formula to help explain protein evolution. Science 1974, 185, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Krigbaum, W.R.; Komoriya, A. Local interactions as a structure determinant for protein molecules: II. Biochim. Biophys. Acta. 1979, 576, 204–228. [Google Scholar] [CrossRef]
- Jukes, T.H.; Holmquist, R.; Moise, H. Amino-acid composition of proteins—Selection against genetic code. Science 1975, 189, 50–51. [Google Scholar] [CrossRef] [PubMed]
- Kanie, K.; Kato, R.; Zhao, Y.; Narita, Y.; Okochi, M.; Honda, H. Amino acid sequence preferences to control cell-specific organization of endothelial cells, smooth muscle cells, and fibroblasts. J. Pept. Sci. 2011, 17, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Arrabal, P.M.; Visser, R.; Santos-Ruiz, L.; Becerra, J.; Cifuentes, M. Osteogenic molecules for clinical applications: Improving the bmp-collagen system. Biol. Res. 2013, 46, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Reissmann, S.; Gillen, C.M.; Fulde, M.; Bergmann, R.; Nerlich, A.; Rajkumari, R.; Brahmadathan, K.N.; Chhatwal, G.S.; Nitsche-Schmitz, D.P. Region specific and worldwide distribution of collagen-binding m proteins with parf motifs among human pathogenic streptococcal isolates. PLoS ONE 2012, 7, e30122. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Kim, S.D.; Lee, H.Y.; Lee, S.Y.; Shim, J.W.; Yun, J.; Kim, J.M.; Min, D.S.; Yoo, Y.H.; Bae, Y.S. Collagen-binding motif peptide, a cleavage product of osteopontin, stimulates human neutrophil chemotaxis via pertussis toxin-sensitive G protein-mediated signaling. Febs. Lett. 2008, 582, 3379–3384. [Google Scholar] [CrossRef] [PubMed]
- Takagi, J.; Asai, H.; Saito, Y. A collagen gelatin-binding decapeptide derived from bovine propolypeptide of vonwillebrand-factor. Biochemistry 1992, 31, 8530–8534. [Google Scholar] [CrossRef] [PubMed]
- Dinkla, K.; Nitsche-Schmitz, D.P.; Barroso, V.; Reissmann, S.; Johansson, H.M.; Frick, I.M.; Rohde, M.; Chhatwal, G.S. Identification of a streptococcal octapeptide motif involved in acute rheumatic fever. J. Biol. Chem. 2007, 282, 18686–18693. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Choo, J.E.; Choi, Y.S.; Park, J.B.; Min, D.S.; Lee, S.J.; Rhyu, H.K.; Jo, I.H.; Chung, C.P.; Park, Y.J. Assembly of collagen-binding peptide with collagen as a bioactive scaffold for osteogenesis in vitro and in vivo. Biomaterials 2007, 28, 4257–4267. [Google Scholar] [CrossRef] [PubMed]
- Helms, B.A.; Reulen, S.W.A.; Nijhuis, S.; De Graaf-Heuvelmans, P.T.H.M.; Merkx, M.; Meijer, E.W. High-affinity peptide-based collagen targeting using synthetic phage mimics: From phage display to dendrimer display. J. Am. Chem Soc. 2009, 131, 11683–11685. [Google Scholar] [CrossRef] [PubMed]

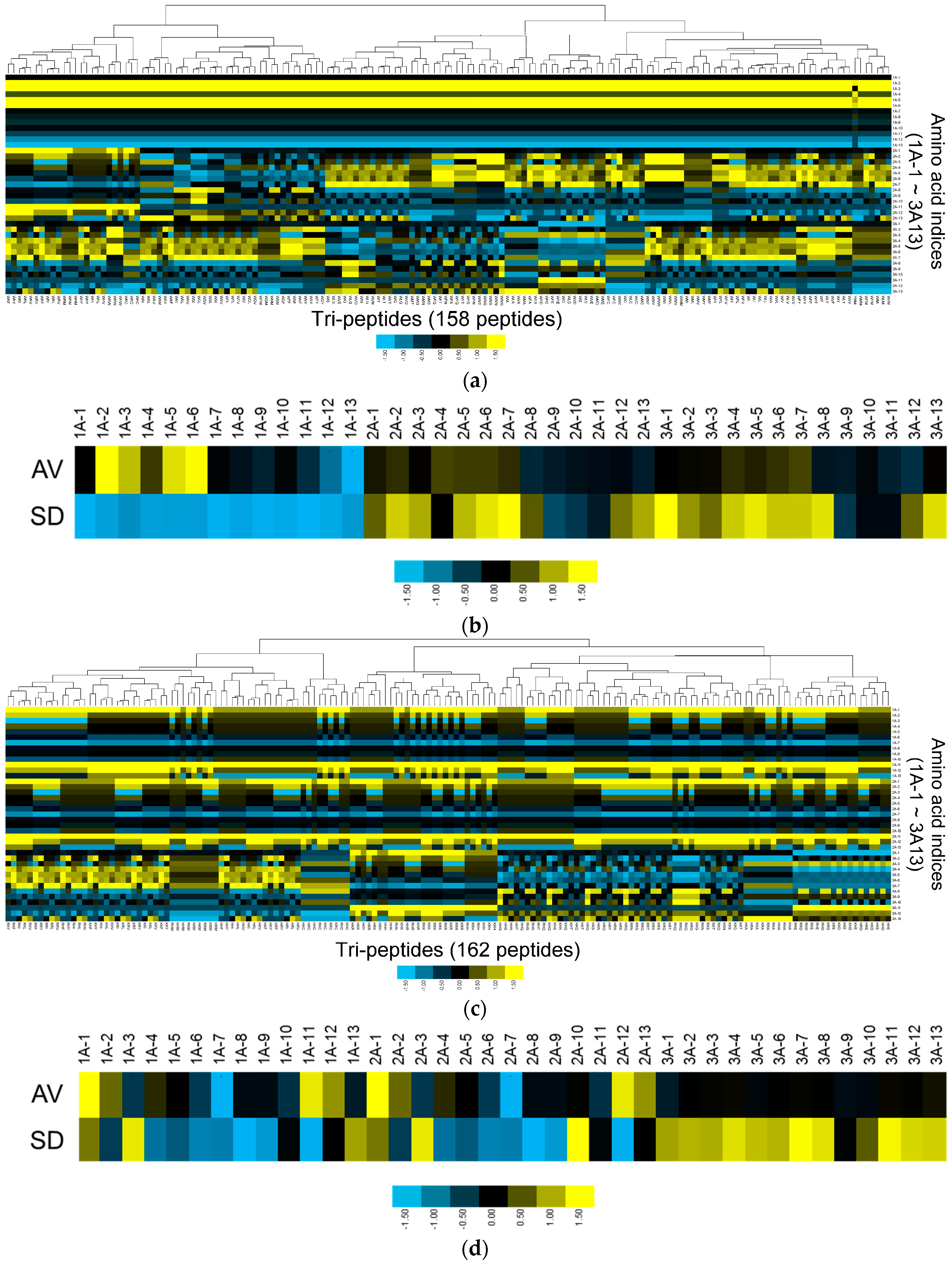
| Index Number | Description | Reference |
|---|---|---|
| 1 | Isoelectric Point | [22] |
| 2 | Normalized van der Waals Volume | [23] |
| 3 | Alpha-Helix Indices for Beta-Proteins | [24] |
| 4 | Beta-Strand Indices for Beta-Proteins | [24] |
| 5 | Side-Chain Contribution To Protein Stability | [25] |
| 6 | The Stability Scale from the Knowledge-Based Atom–Atom Potential | [26] |
| 7 | Hydropathy Index | [27] |
| 8 | Normalized Frequency of Turn | [28] |
| 9 | Free Energy in Beta-Strand Region | [29] |
| 10 | Free Energy in Alpha-Helical Region | [29] |
| 11 | Polarity | [30] |
| 12 | Side Chain Interaction Parameter | [31] |
| 13 | Amino Acid Distribution | [32] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanie, K.; Kondo, Y.; Owaki, J.; Ikeda, Y.; Narita, Y.; Kato, R.; Honda, H. Focused Screening of ECM-Selective Adhesion Peptides on Cellulose-Bound Peptide Microarrays. Bioengineering 2016, 3, 31. https://doi.org/10.3390/bioengineering3040031
Kanie K, Kondo Y, Owaki J, Ikeda Y, Narita Y, Kato R, Honda H. Focused Screening of ECM-Selective Adhesion Peptides on Cellulose-Bound Peptide Microarrays. Bioengineering. 2016; 3(4):31. https://doi.org/10.3390/bioengineering3040031
Chicago/Turabian StyleKanie, Kei, Yuto Kondo, Junki Owaki, Yurika Ikeda, Yuji Narita, Ryuji Kato, and Hiroyuki Honda. 2016. "Focused Screening of ECM-Selective Adhesion Peptides on Cellulose-Bound Peptide Microarrays" Bioengineering 3, no. 4: 31. https://doi.org/10.3390/bioengineering3040031
APA StyleKanie, K., Kondo, Y., Owaki, J., Ikeda, Y., Narita, Y., Kato, R., & Honda, H. (2016). Focused Screening of ECM-Selective Adhesion Peptides on Cellulose-Bound Peptide Microarrays. Bioengineering, 3(4), 31. https://doi.org/10.3390/bioengineering3040031






