PHSP-Net: Personalized Habitat-Aware Deep Learning for Multi-Center Glioblastoma Survival Prediction Using Multiparametric MRI
Abstract
1. Introduction
2. Materials and Methods
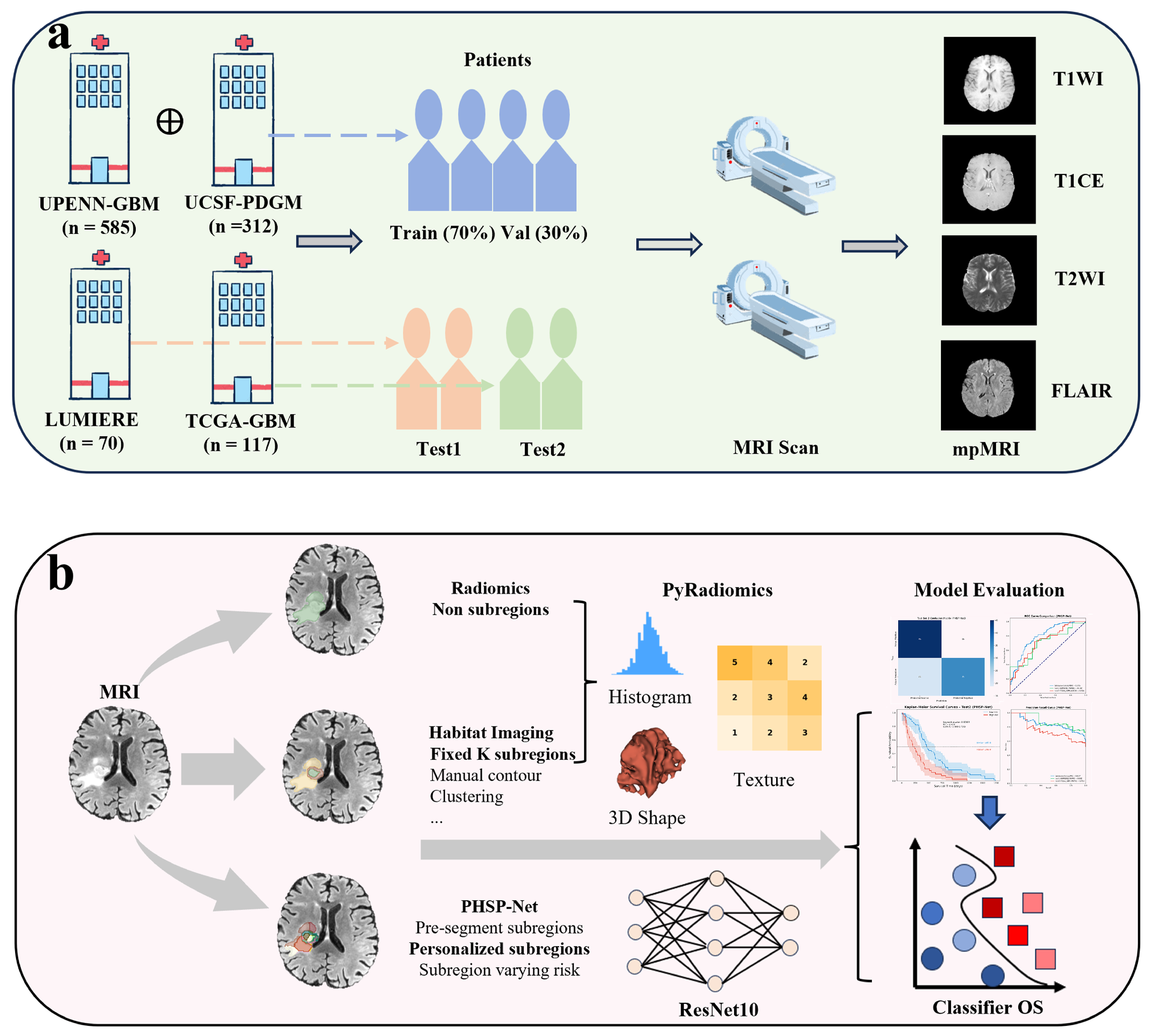
2.1. Data Sources and Description
2.2. Radiomics and Habitat Imaging Feature Extraction
2.3. Feature Selection
2.4. Conventional Machine Learning Models
2.5. Personalized Habitat-Aware Survival Prediction Network
2.6. Model Evaluation
3. Results
3.1. Characteristics of GBM Patients
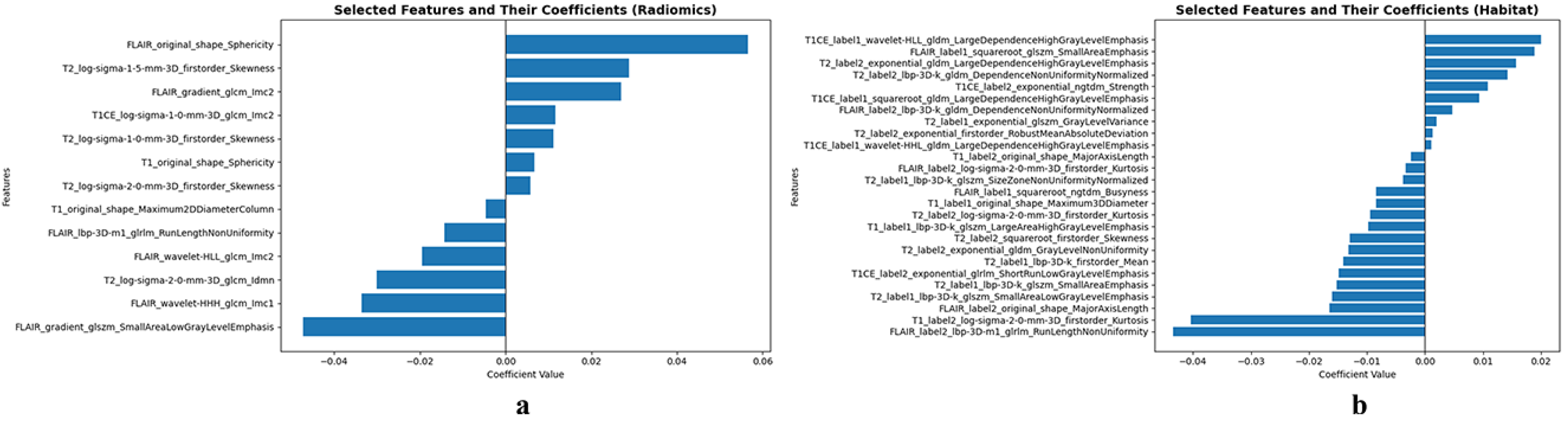
3.2. Feature Selection Analysis
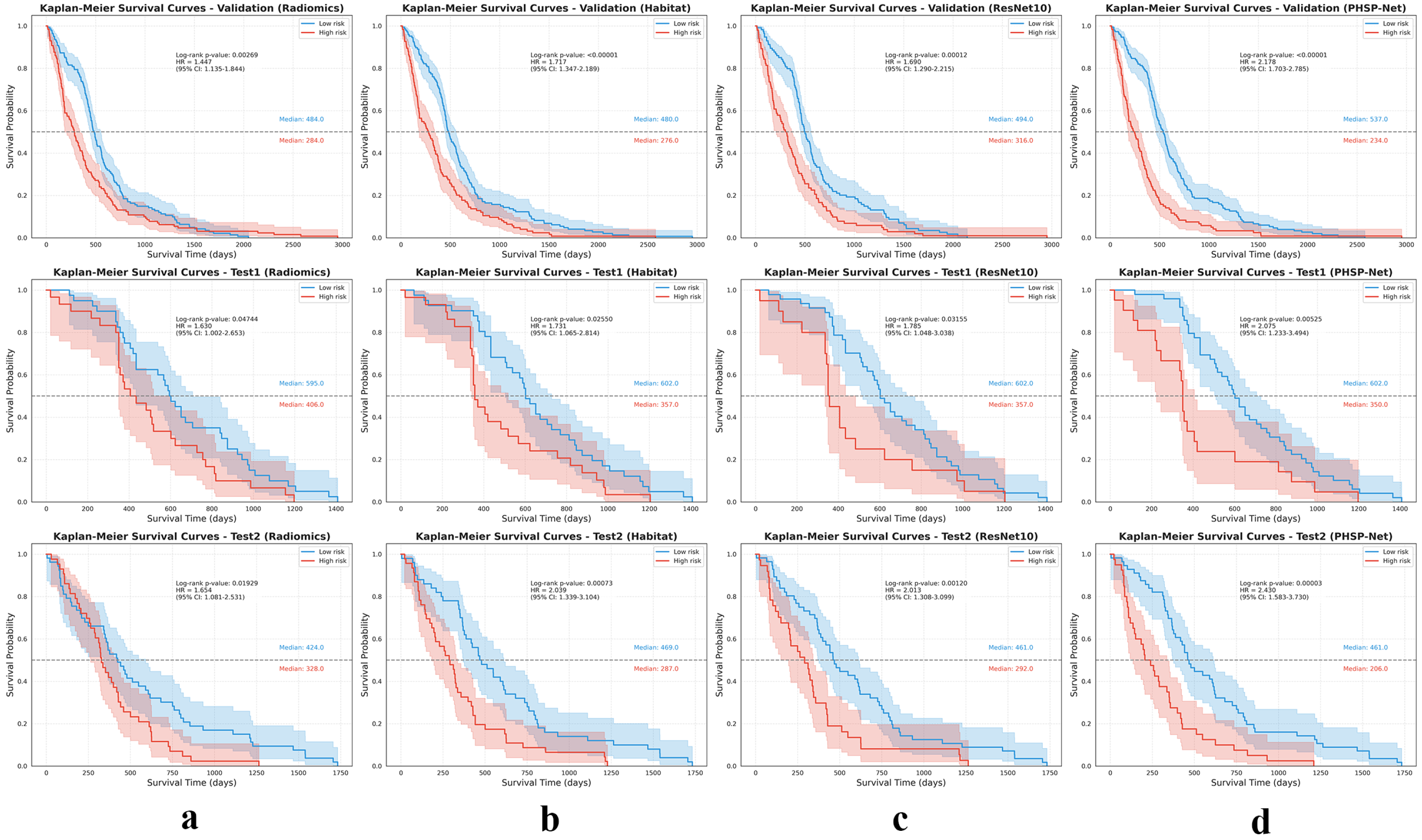
3.3. Model Analysis
3.4. Model Visualization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GBM | Glioblastoma |
| OS | Overall Survival |
| PHSP-Net | Personalized Habitat-Aware Survival Prediction Network |
| MRI | Magnetic Resonance Imaging |
| CNNs | Convolutional Neural Networks |
| AUROC | Area Under the Receiver Operating Characteristic curve |
| AUPRC | Area Under the Precision–Recall Curve |
References
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter-and intra-tumoral heterogeneity. Cell 2020, 180, 188–204. [Google Scholar] [CrossRef]
- Van den Bent, M.J.; Geurts, M.; French, P.J.; Smits, M.; Capper, D.; Bromberg, J.E.; Chang, S.M. Primary brain tumours in adults. Lancet 2023, 402, 1564–1579. [Google Scholar] [CrossRef]
- Verdugo, E.; Puerto, I.; Medina, M.Á. An update on the molecular biology of glioblastoma, with clinical implications and progress in its treatment. Cancer Commun. 2022, 42, 1083–1111. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and molecular epidemiology of adult diffuse glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Vigneswaran, K.; Neill, S.; Hadjipanayis, C.G. Beyond the World Health Organization grading of infiltrating gliomas: Advances in the molecular genetics of glioma classification. Ann. Transl. Med. 2015, 3, 95. [Google Scholar] [PubMed]
- Haller, S.; Haacke, E.M.; Thurnher, M.M.; Barkhof, F. Susceptibility-weighted imaging: Technical essentials and clinical neurologic applications. Radiology 2021, 299, 3–26. [Google Scholar] [CrossRef]
- Keall, P.J.; Brighi, C.; Glide-Hurst, C.; Liney, G.; Liu, P.Z.; Lydiard, S.; Paganelli, C.; Pham, T.; Shan, S.; Tree, A.C.; et al. Integrated MRI-guided radiotherapy—Opportunities and challenges. Nat. Rev. Clin. Oncol. 2022, 19, 458–470. [Google Scholar] [CrossRef]
- Le, V.H.; Minh, T.N.T.; Kha, Q.H.; Le, N.Q.K. A transfer learning approach on MRI-based radiomics signature for overall survival prediction of low-grade and high-grade gliomas. Med. Biol. Eng. Comput. 2023, 61, 2699–2712. [Google Scholar] [CrossRef]
- Duman, A.; Sun, X.; Thomas, S.; Powell, J.R.; Spezi, E. Reproducible and interpretable machine learning-based radiomic analysis for overall survival prediction in glioblastoma multiforme. Cancers 2024, 16, 3351. [Google Scholar] [CrossRef]
- Wang, H. Multimodal MRI radiomics based on habitat subregions of the tumor microenvironment for predicting risk stratification in glioblastoma. PLoS ONE 2025, 20, e0326361. [Google Scholar] [CrossRef]
- Macyszyn, L.; Akbari, H.; Pisapia, J.M.; Da, X.; Attiah, M.; Pigrish, V.; Bi, Y.; Pal, S.; Davuluri, R.V.; Roccograndi, L.; et al. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro-Oncology 2015, 18, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, Y.; Pan, X.; Yang, Q.; Qiang, Y.; Qi, X.S. A Subregion-based survival prediction framework for GBM via multi-sequence MRI space optimization and clustering-based feature bundling and construction. Phys. Med. Biol. 2023, 68, 125005. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zamarud, A.; Marianayagam, N.J.; Park, D.J.; Yener, U.; Soltys, S.G.; Chang, S.D.; Meola, A.; Jiang, H.; Lu, W.; et al. Deep learning-based overall survival prediction in patients with glioblastoma: An automatic end-to-end workflow using pre-resection basic structural multiparametric MRIs. Comput. Biol. Med. 2025, 185, 109436. [Google Scholar] [CrossRef]
- Ben Ahmed, K.; Hall, L.O.; Goldgof, D.B.; Gatenby, R. Ensembles of convolutional neural networks for survival time estimation of high-grade glioma patients from multimodal MRI. Diagnostics 2022, 12, 345. [Google Scholar] [CrossRef]
- Tang, Z.; Xu, Y.; Jin, L.; Aibaidula, A.; Lu, J.; Jiao, Z.; Wu, J.; Zhang, H.; Shen, D. Deep learning of imaging phenotype and genotype for predicting overall survival time of glioblastoma patients. IEEE Trans. Med. Imaging 2020, 39, 2100–2109. [Google Scholar] [CrossRef]
- Bakas, S.; Sako, C.; Akbari, H.; Bilello, M.; Sotiras, A.; Shukla, G.; Rudie, J.D.; Santamaría, N.F.; Kazerooni, A.F.; Pati, S.; et al. The University of Pennsylvania glioblastoma (UPenn-GBM) cohort: Advanced MRI, clinical, genomics, & radiomics. Sci. Data 2022, 9, 453. [Google Scholar] [CrossRef]
- Calabrese, E.; Villanueva-Meyer, J.E.; Rudie, J.D.; Rauschecker, A.M.; Baid, U.; Bakas, S.; Cha, S.; Mongan, J.T.; Hess, C.P. The University of California San Francisco preoperative diffuse glioma MRI dataset. Radiol. Artif. Intell. 2022, 4, e220058. [Google Scholar] [CrossRef] [PubMed]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar] [CrossRef]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Suter, Y.; Knecht, U.; Valenzuela, W.; Notter, M.; Hewer, E.; Schucht, P.; Wiest, R.; Reyes, M. The LUMIERE dataset: Longitudinal Glioblastoma MRI with expert RANO evaluation. Sci. Data 2022, 9, 768. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Stat. Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Hajianfar, G.; Haddadi Avval, A.; Hosseini, S.A.; Nazari, M.; Oveisi, M.; Shiri, I.; Zaidi, H. Time-to-event overall survival prediction in glioblastoma multiforme patients using magnetic resonance imaging radiomics. La Radiol. Medica 2023, 128, 1521–1534. [Google Scholar] [CrossRef]
- Jia, X.; Zhai, Y.; Song, D.; Wang, Y.; Wei, S.; Yang, F.; Wei, X. A multiparametric MRI-based radiomics nomogram for preoperative prediction of survival stratification in glioblastoma patients with standard treatment. Front. Oncol. 2022, 12, 758622. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; He, X.; Li, Y.; Pang, P.; Shu, Z.; Gong, X. The nomogram of MRI-based radiomics with complementary visual features by machine learning improves stratification of glioblastoma patients: A multicenter study. J. Magn. Reson. Imaging 2021, 54, 571–583. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Grove, O.; Gillies, R.J. Quantitative imaging in cancer evolution and ecology. Radiology 2013, 269, 8–14. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, D.; Gao, P.; Tian, Q.; Lu, H.; Xu, X.; He, X.; Liu, Y. Survival-relevant high-risk subregion identification for glioblastoma patients: The MRI-based multiple instance learning approach. Eur. Radiol. 2020, 30, 5602–5610. [Google Scholar] [CrossRef]
- Patrício, C.; Neves, J.C.; Teixeira, L.F. Explainable deep learning methods in medical image classification: A survey. ACM Comput. Surv. 2023, 56, 1–41. [Google Scholar] [CrossRef]
- Cai, L.; Fang, H.; Xu, N.; Ren, B. Counterfactual causal-effect intervention for interpretable medical visual question answering. IEEE Trans. Med. Imaging 2024, 43, 4430–4441. [Google Scholar]
- Wu, X.; Zhang, Y.T.; Lai, K.W.; Yang, M.Z.; Yang, G.L.; Wang, H.H. A novel centralized federated deep fuzzy neural network with multi-objectives neural architecture search for epistatic detection. IEEE Trans. Fuzzy Syst. 2024, 33, 94–107. [Google Scholar] [CrossRef]
- Luckett, P.H.; Olufawo, M.; Lamichhane, B.; Park, K.Y.; Dierker, D.; Verastegui, G.T.; Yang, P.; Kim, A.H.; Chheda, M.G.; Snyder, A.Z.; et al. Predicting survival in glioblastoma with multimodal neuroimaging and machine learning. J. Neuro-Oncol. 2023, 164, 309–320. [Google Scholar] [CrossRef]
- Kaur, G.; Rana, P.S.; Arora, V. Deep learning and machine learning-based early survival predictions of glioblastoma patients using pre-operative three-dimensional brain magnetic resonance imaging modalities. Int. J. Imaging Syst. Technol. 2023, 33, 340–361. [Google Scholar]
- Zheng, Y.; Carrillo-Perez, F.; Pizurica, M.; Heiland, D.H.; Gevaert, O. Spatial cellular architecture predicts prognosis in glioblastoma. Nat. Commun. 2023, 14, 4122. [Google Scholar] [CrossRef]
- Martin, S.A.; Townend, F.J.; Barkhof, F.; Cole, J.H. Interpretable machine learning for dementia: A systematic review. Alzheimer’s Dement. 2023, 19, 2135–2149. [Google Scholar] [CrossRef]
- Salahuddin, Z.; Woodruff, H.C.; Chatterjee, A.; Lambin, P. Transparency of deep neural networks for medical image analysis: A review of interpretability methods. Comput. Biol. Med. 2022, 140, 105111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.P.; Zhang, X.Y.; Cheng, Y.T.; Li, B.; Teng, X.Z.; Zhang, J.; Lam, S.; Zhou, T.; Ma, Z.R.; Sheng, J.B.; et al. Artificial intelligence-driven radiomics study in cancer: The role of feature engineering and modeling. Mil. Med. Res. 2023, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, K.; Qayyum, A.; Ghaly, M.; Al-Fuqaha, A.; Razi, A.; Qadir, J. Explainable, trustworthy, and ethical machine learning for healthcare: A survey. Comput. Biol. Med. 2022, 149, 106043. [Google Scholar] [CrossRef]
- Camalan, S.; Mahmood, H.; Binol, H.; Araujo, A.L.D.; Santos-Silva, A.R.; Vargas, P.A.; Lopes, M.A.; Khurram, S.A.; Gurcan, M.N. Convolutional neural network-based clinical predictors of oral dysplasia: Class activation map analysis of deep learning results. Cancers 2021, 13, 1291. [Google Scholar] [CrossRef]
- Ghorbani, A.; Ouyang, D.; Abid, A.; He, B.; Chen, J.H.; Harrington, R.A.; Liang, D.H.; Ashley, E.A.; Zou, J.Y. Deep learning interpretation of echocardiograms. NPJ Digit. Med. 2020, 3, 10. [Google Scholar] [CrossRef]
- Izadyyazdanabadi, M.; Belykh, E.; Cavallo, C.; Zhao, X.; Gandhi, S.; Moreira, L.B.; Eschbacher, J.; Nakaji, P.; Preul, M.C.; Yang, Y. Weakly-supervised learning-based feature localization for confocal laser endomicroscopy glioma images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 300–308. [Google Scholar]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef]
- Pereira, S.; Meier, R.; Alves, V.; Reyes, M.; Silva, C.A. Automatic brain tumor grading from MRI data using convolutional neural networks and quality assessment. In Proceedings of the International Workshop on Machine Learning in Clinical Neuroimaging, Granada, Spain, 16–20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 106–114. [Google Scholar]
- Sayres, R.; Taly, A.; Rahimy, E.; Blumer, K.; Coz, D.; Hammel, N.; Krause, J.; Narayanaswamy, A.; Rastegar, Z.; Wu, D.; et al. Using a deep learning algorithm and integrated gradients explanation to assist grading for diabetic retinopathy. Ophthalmology 2019, 126, 552–564. [Google Scholar] [CrossRef]
- Alexander, B.M.; Cloughesy, T.F. Adult glioblastoma. J. Clin. Oncol. 2017, 35, 2402–2409. [Google Scholar] [CrossRef]
- Van Tellingen, O.; Yetkin-Arik, B.; De Gooijer, M.; Wesseling, P.; Wurdinger, T.; De Vries, H. Overcoming the blood–Brain tumor barrier for effective glioblastoma treatment. Drug Resist. Updat. 2015, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Parillo, M.; Quattrocchi, C.C. Brain tumor reporting and data system (BT-RADS) for the surveillance of adult-type diffuse gliomas after surgery. Surgeries 2024, 5, 764–773. [Google Scholar] [CrossRef]
- Li, X.; Morgan, P.S.; Ashburner, J.; Smith, J.; Rorden, C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 2016, 264, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.; Woolrich, M.W.; Smith, S.M. FSL. NeuroImage 2012, 62, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Rohlfing, T.; Zahr, N.M.; Sullivan, E.V.; Pfefferbaum, A. The SRI24 multichannel atlas of normal adult human brain structure. Hum. Brain Mapp. 2010, 31, 798–819. [Google Scholar] [CrossRef] [PubMed]
- Avants, B.B.; Epstein, C.L.; Grossman, M.; Gee, J.C. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008, 12, 26–41. [Google Scholar] [CrossRef]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]


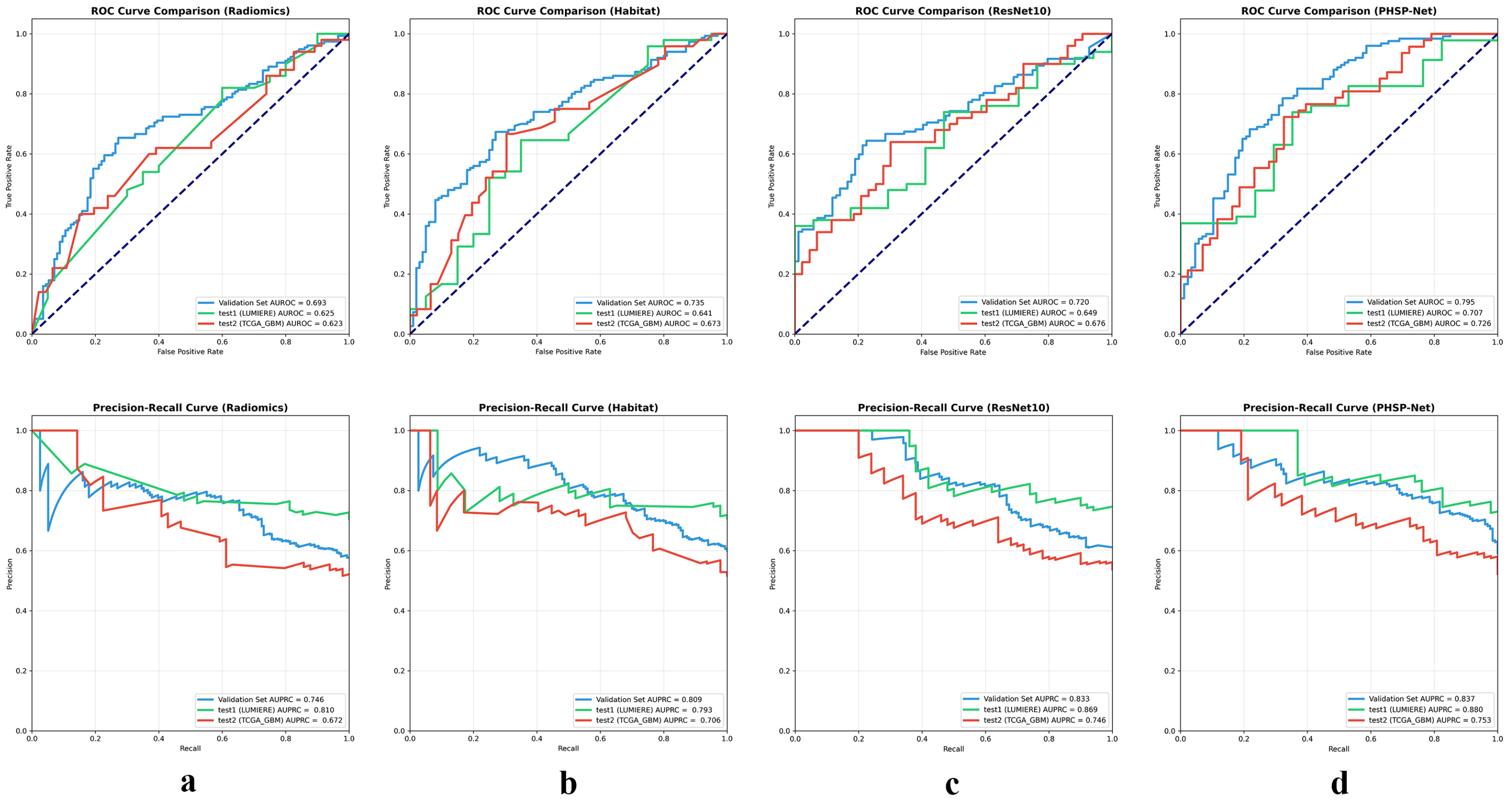
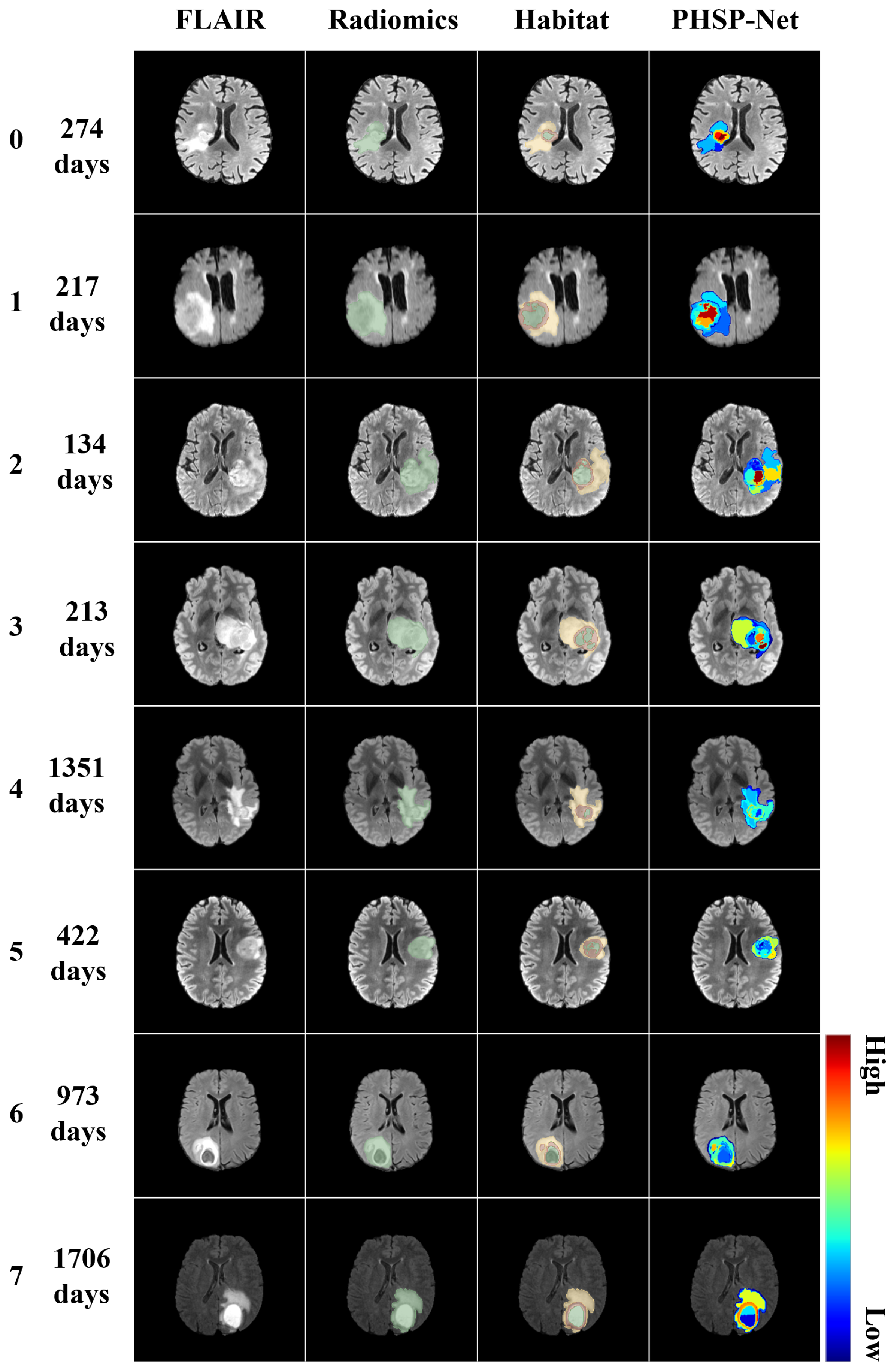
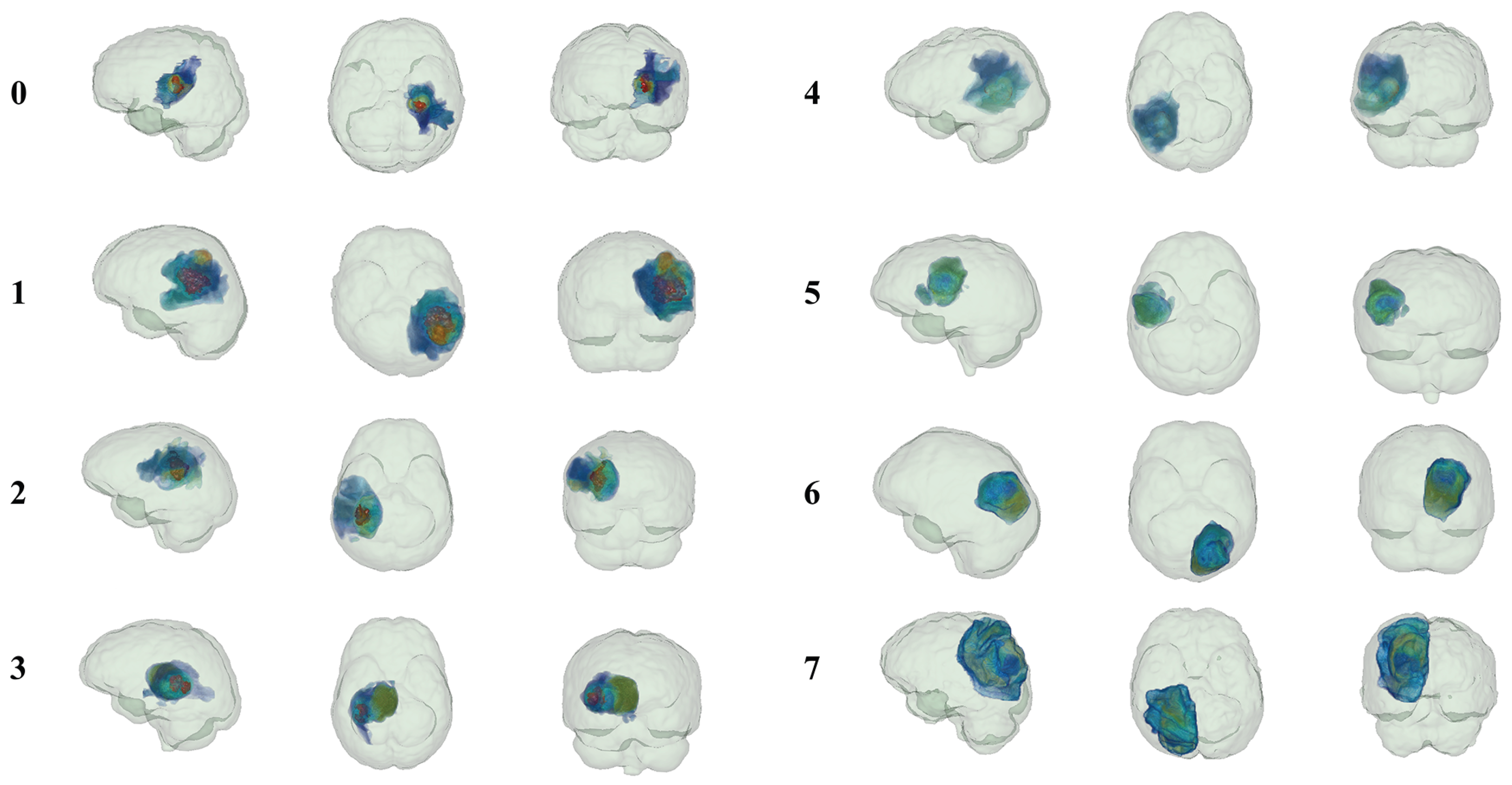
| Model | Cohort | Performance Metrics (95% CI) | ||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Specificity | F1-Score | AUROC | AUPRC | |||
| Validation | 0.670 (0.618–0.731) | 0.738 (0.664–0.811) | 0.670 (0.618–0.731) | 0.675 (0.585–0.761) | 0.700 (0.641–0.760) | 0.693 (0.629–0.756) | 0.746 (0.667–0.825) | |
| Radiomics | Test set 1 (LUMIERE) | 0.571 (0.457–0.692) | 0.778 (0.687–0.859) | 0.571 (0.457–0.692) | 0.600 (0.480–0.720) | 0.651 (0.542–0.757) | 0.625 (0.483–0.770) | 0.810 (0.719–0.905) |
| Test set 2 (TCGA-GBM) | 0.573 (0.469–0.667) | 0.585 (0.475–0.702) | 0.573 (0.469–0.667) | 0.518 (0.383–0.667) | 0.603 (0.519–0.698) | 0.623 (0.506–0.730) | 0.672 (0.583–0.767) | |
| Validation | 0.681 (0.624–0.740) | 0.759 (0.688–0.831) | 0.681 (0.624–0.740) | 0.667 (0.574–0.758) | 0.715 (0.664–0.779) | 0.735 (0.672–0.792) | 0.809 (0.737–0.872) | |
| Habitat Imaging | Test set 1 (LUMIERE) | 0.643 (0.529–0.765) | 0.816 (0.702–0.909) | 0.643 (0.529–0.765) | 0.650 (0.565–0.742) | 0.719 (0.595–0.824) | 0.641 (0.530–0.758) | 0.793 (0.695–0.887) |
| Test set 2 (TCGA-GBM) | 0.675 (0.575–0.766) | 0.681 (0.566–0.808) | 0.675 (0.575–0.766) | 0.679 (0.574–0.759) | 0.683 (0.558–0.774) | 0.673 (0.558–0.784) | 0.706 (0.625–0.783) | |
| Validation | 0.685 (0.625–0.745) | 0.759 (0.702–0.815) | 0.667 (0.582–0.745) | 0.711 (0.613–0.802) | 0.710 (0.655–0.780) | 0.720 (0.652–0.786) | 0.833 (0.784–0.882) | |
| ResNet10 | Test set 1 (LUMIERE) | 0.646 (0.522–0.689) | 0.791 (0.684–0.896) | 0.680 (0.551–0.804) | 0.550 (0.412–0.689) | 0.731 (0.622–0.828) | 0.649 (0.513–0.778) | 0.869 (0.802–0.937) |
| Test set 2 (TCGA-GBM) | 0.607 (0.516–0.697) | 0.615 (0.509–0.720) | 0.656 (0.526–0.782) | 0.554 (0.424–0.685) | 0.635 (0.543–0.726) | 0.676 (0.566–0.787) | 0.746 (0.653–0.837) | |
| Validation | 0.719 (0.662–0.776) | 0.780 (0.705–0.853) | 0.719 (0.662–0.776) | 0.702 (0.607–0.797) | 0.750 (0.694–0.806) | 0.795 (0.731–0.852) | 0.837 (0.785–0.883) | |
| PHSP–Net | Test set 1 (LUMIERE) | 0.714 (0.603–0.825) | 0.833 (0.760–0.903) | 0.714 (0.603–0.825) | 0.600 (0.483–0.727) | 0.792 (0.698–0.886) | 0.707 (0.632–0.779) | 0.880 (0.812–0.945) |
| Test set 2 (TCGA-GBM) | 0.684 (0.600–0.768) | 0.686 (0.563–0.804) | 0.684 (0.600–0.768) | 0.625 (0.530–0.720) | 0.709 (0.632–0.786) | 0.726 (0.649–0.797) | 0.753 (0.673–0.828) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Zheng, Y.; Chen, C.; Wei, J.; Huang, D.; Feng, Y.; Liu, Y. PHSP-Net: Personalized Habitat-Aware Deep Learning for Multi-Center Glioblastoma Survival Prediction Using Multiparametric MRI. Bioengineering 2025, 12, 978. https://doi.org/10.3390/bioengineering12090978
Liu T, Zheng Y, Chen C, Wei J, Huang D, Feng Y, Liu Y. PHSP-Net: Personalized Habitat-Aware Deep Learning for Multi-Center Glioblastoma Survival Prediction Using Multiparametric MRI. Bioengineering. 2025; 12(9):978. https://doi.org/10.3390/bioengineering12090978
Chicago/Turabian StyleLiu, Tianci, Yao Zheng, Chengwei Chen, Jie Wei, Dong Huang, Yuefei Feng, and Yang Liu. 2025. "PHSP-Net: Personalized Habitat-Aware Deep Learning for Multi-Center Glioblastoma Survival Prediction Using Multiparametric MRI" Bioengineering 12, no. 9: 978. https://doi.org/10.3390/bioengineering12090978
APA StyleLiu, T., Zheng, Y., Chen, C., Wei, J., Huang, D., Feng, Y., & Liu, Y. (2025). PHSP-Net: Personalized Habitat-Aware Deep Learning for Multi-Center Glioblastoma Survival Prediction Using Multiparametric MRI. Bioengineering, 12(9), 978. https://doi.org/10.3390/bioengineering12090978








