Time Series Classification of Autism Spectrum Disorder Using the Light-Adapted Electroretinogram
Abstract
1. Introduction
- To the best of the authors’ knowledge, this is the first application of SHAP explanation to the TSC algorithms’ results for an ERG signal classification task.
- SHAP methods were applied on the TSC models to provide a domain-agnostic explanation, highlighting the important regions of the signals for classification.
2. Materials and Methods
2.1. Dataset
2.2. Time Series Classification Models
- Word extraction for TSC (WEASEL) is a dictionary-based classifier for time-series data. The algorithm is based on the bag-of-patterns representation, which consists of extracting sub-sequences of different lengths from the time series, discretizing each sub-sequence into a coarsely discrete-valued word, then building a histogram from word counts and training a logistic regression classifier on this bag [41]. In the WEASEL case, it was expected that explainability allowed for highlighting which symbolic patterns (e.g., specific wave chunks) or sub-sequences contributed most to class classification.
- Time Series Forest (TSF) is an ensemble method for TSC that builds multiple decision trees. The algorithm selects each tree in the forest through random selection of several intervals with randomized lengths and positional offsets. For each sampled interval, three statistical features are computed: the mean, the standard deviation, and the slope. The features of each interval are then aggregated into a composite feature vector that subsequently serves as the input feature space for the construction of a decision tree. The resulting trees are then integrated into the ensemble model. The random forest-like classifier algorithm is applied to all trees [42]. In the TSF case, it was expected that explainability would allow the identification of the critical intervals that corresponded to a clinically relevant interpretation of ERG signals for classification.
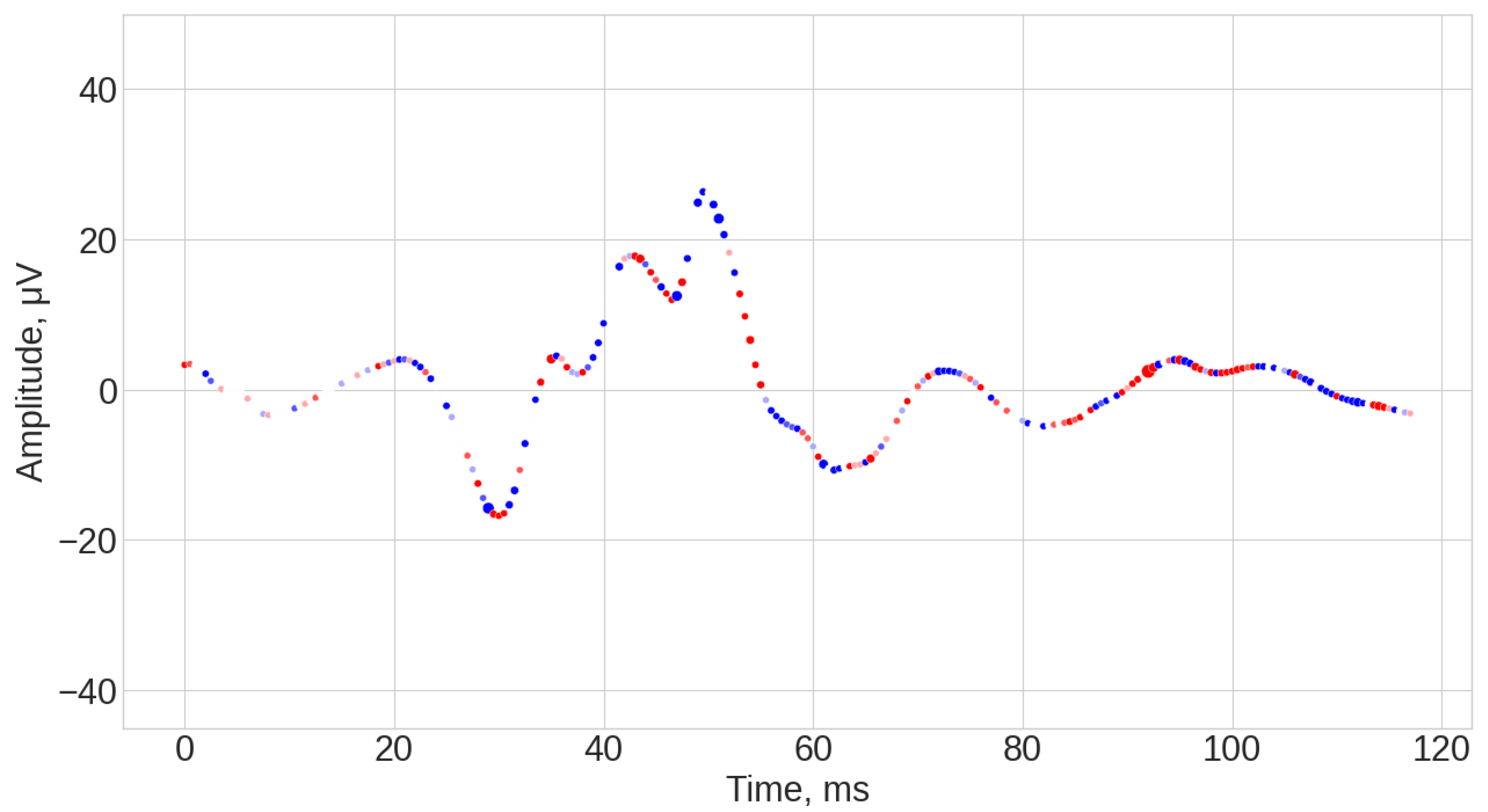
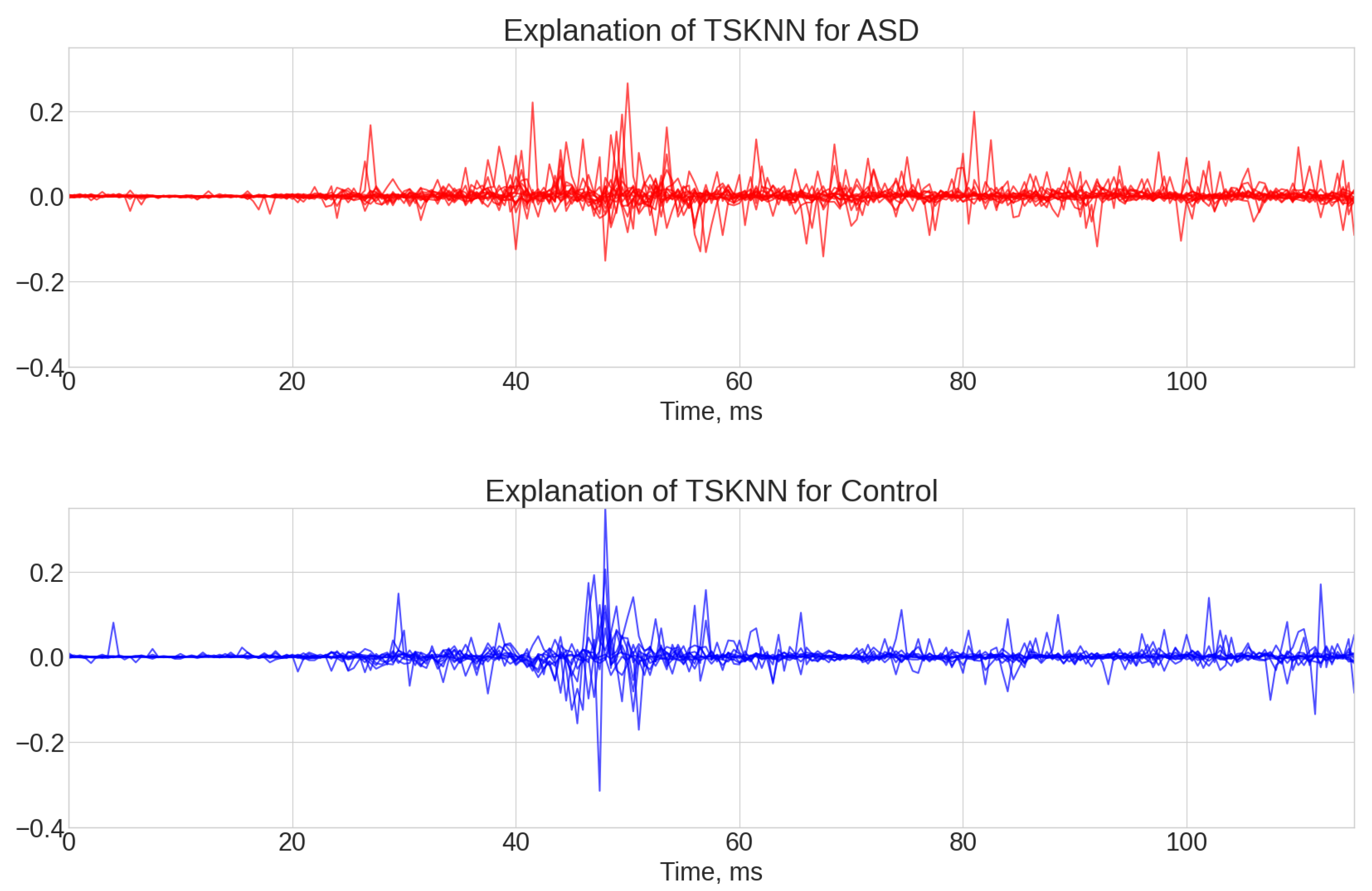
- KNeighborsTimeSeriesClassifier (TS-KNN) is an implementation of the k-nearest neighbors algorithm specifically designed for time series with Dynamic Time Wrapping Distance (DTW). DTW is an elastic distance measure that optimally compares two sequences by warping them non-linearly in time. DTW was applied instead of the traditional Euclidean distance due to the robustness of signal delays (latency) and other distortions in the time domain [43]. In the TS-KNN case, it was expected that explainability would allow for the identification of waveform intervals that corresponded to important patterns of behavior in the ERG signal for classification.
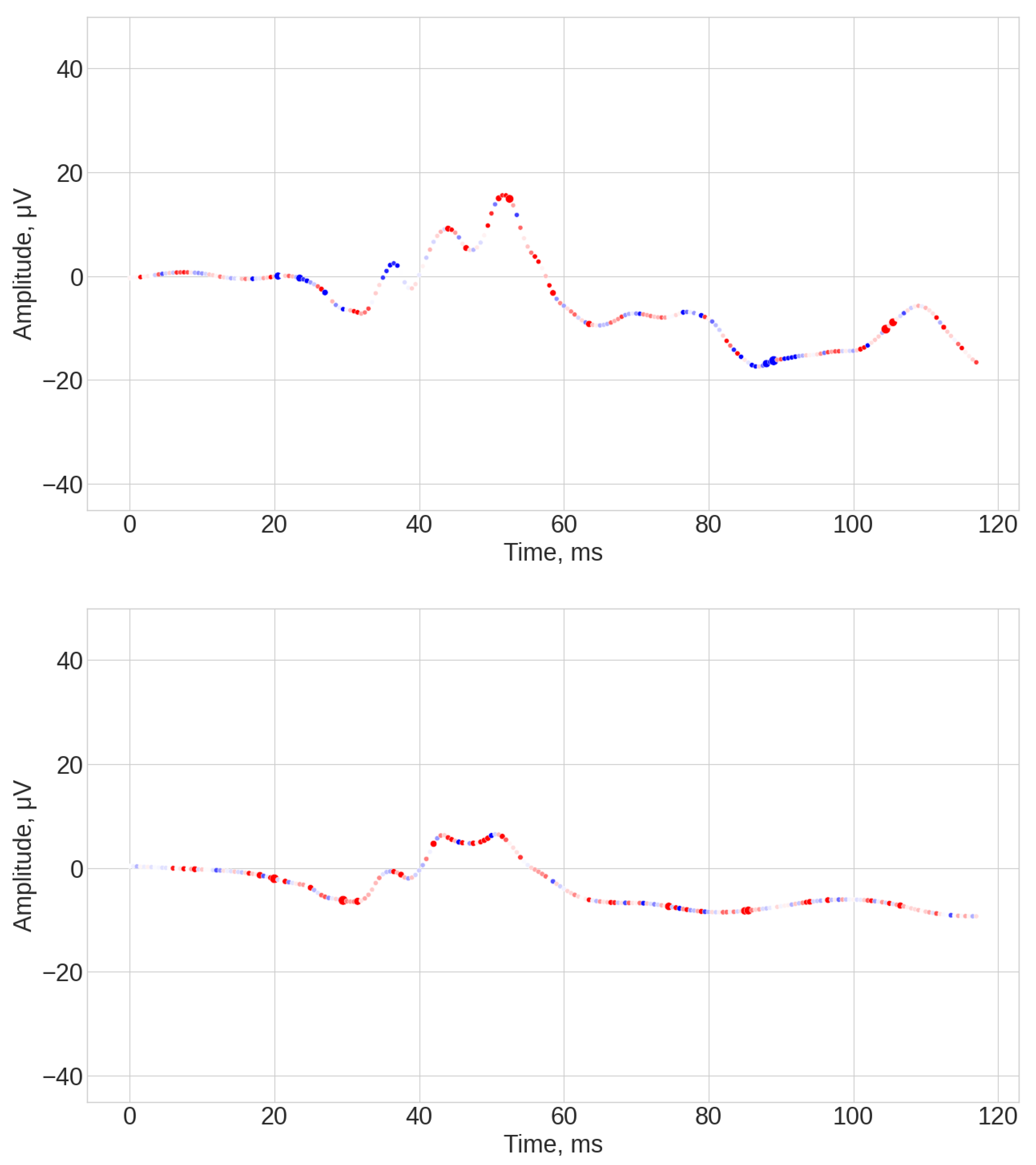
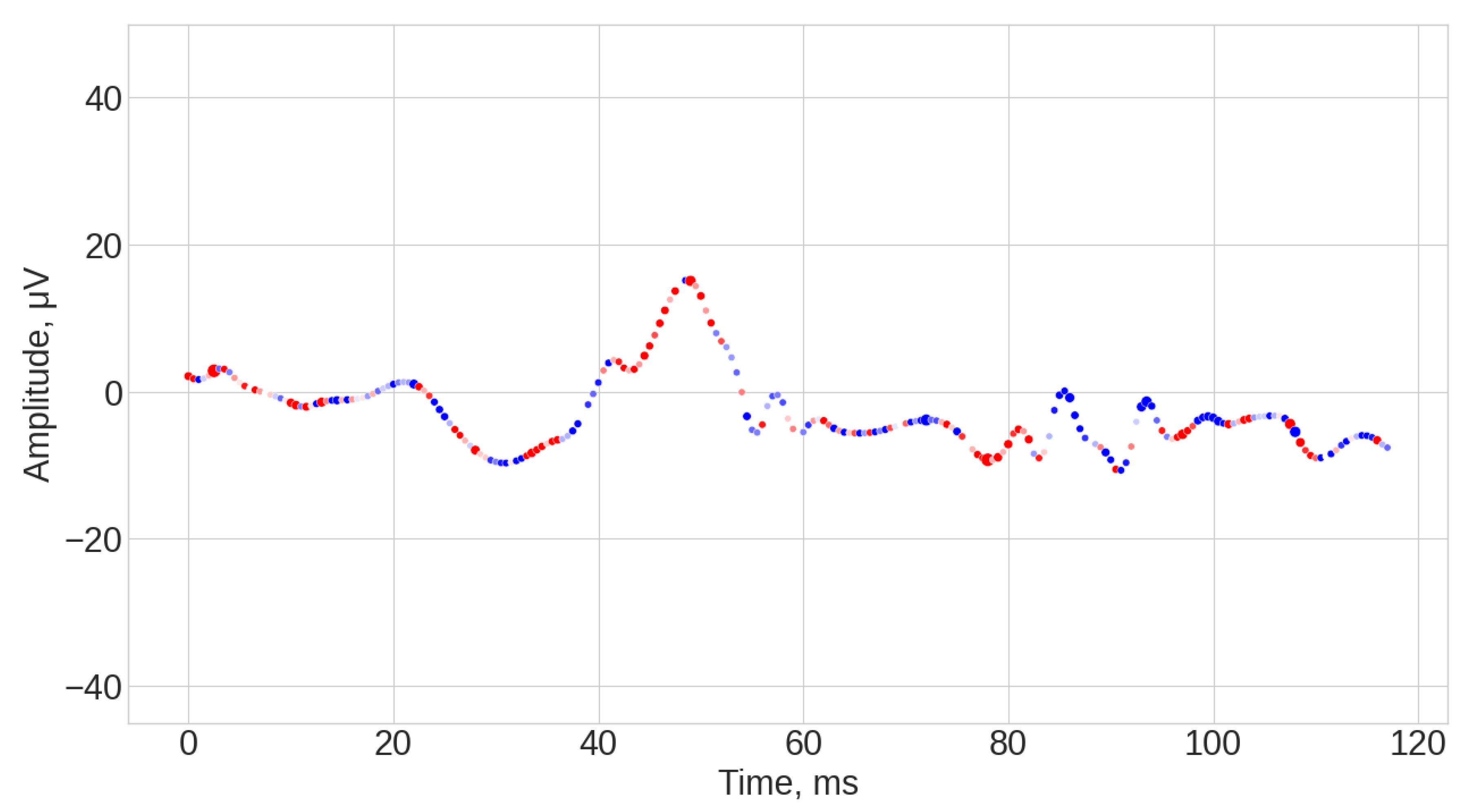
- Random Convolutional Kernel Transform (ROCKET) is a method for TSC based on 1D convolution kernels with random parameter selection. It works by generating a large variety of kernels, each containing different parameters (length, weight, dilation, etc.), and applies these kernels to the data through convolution. Each convolution results in two features, the positive value amount and the maximum value [44]. In this work a random forest was applied to the vector of features obtained for 3000 kernels. In the ROCKET case, it was expected that explainability would identify regions and points (e.g., a-wave amplitude) emphasized by the kernel emphasis, which correspond to differences in retinal signaling between the groups for classification.
2.3. Hyperparameter Selection
- If there was no variation in hyperparameters in the algorithm, then it was run once and the classification metrics values were saved to memory.
- If the algorithm had variations of hyperparameters, then it was run in the amount of these variations and all of the obtained classification metrics were saved to memory.
- —112, 1231, 42, 990, 2500, 467, 777, 89, 258, 24;
- of the TSF algorithm—10, 50, 100, 200, 300, 400, 500;
- of the TS-KNN algorithm—1, 2, 3, 4, 5, 6, 7;
- of the ROCKET algorithm—100, 1000, 10,000, 20,000, 30,000.
- F1-score (separately for control and ASD individuals;
- Balanced accuracy;
- Time for training.
2.4. Explaining TSC Models Using the SHAP Library
- Train a time-series model using sktime.
- Use the trained model to make predictions on the test data.
- Apply SHAP to the model to obtain feature importance values.
3. Results
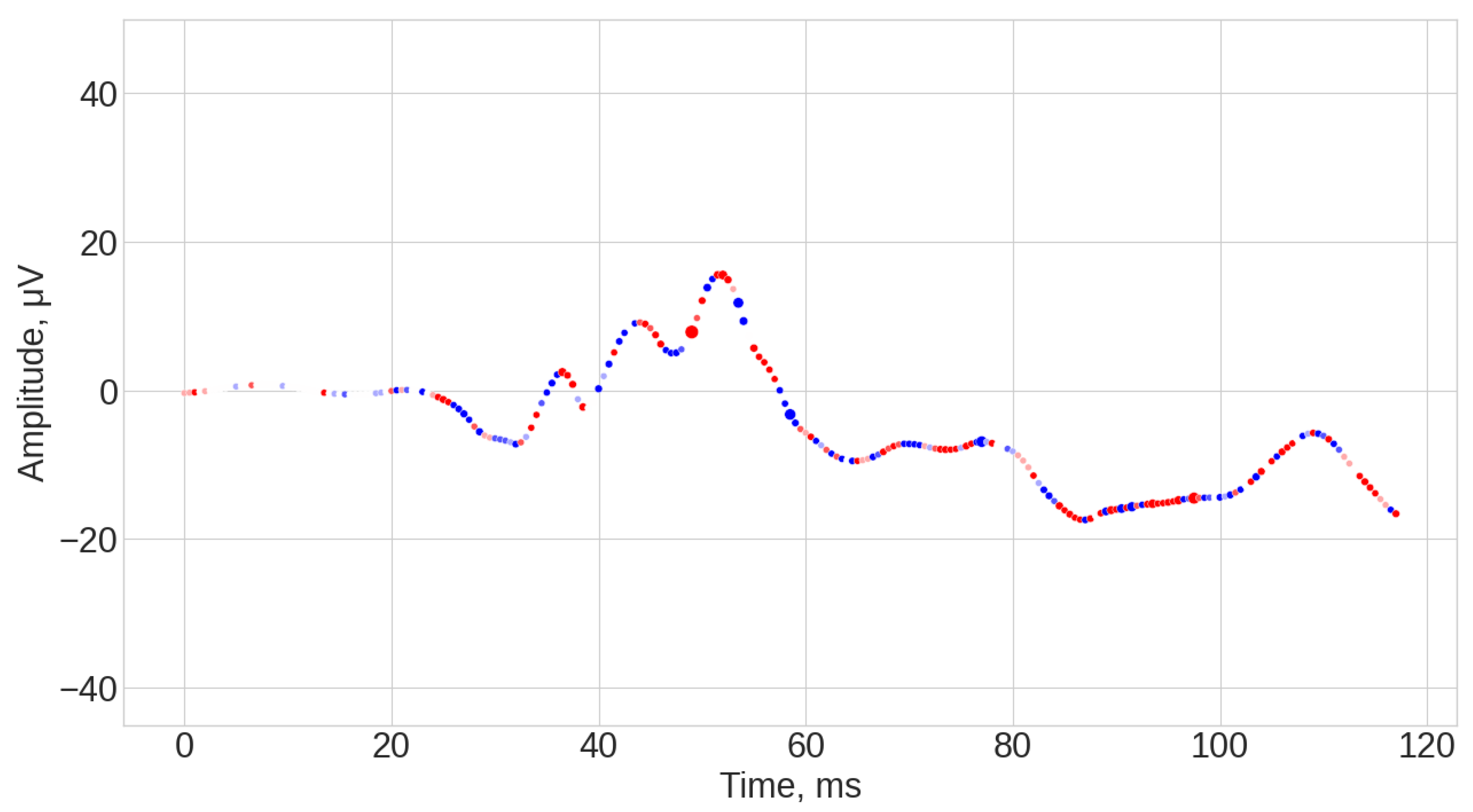
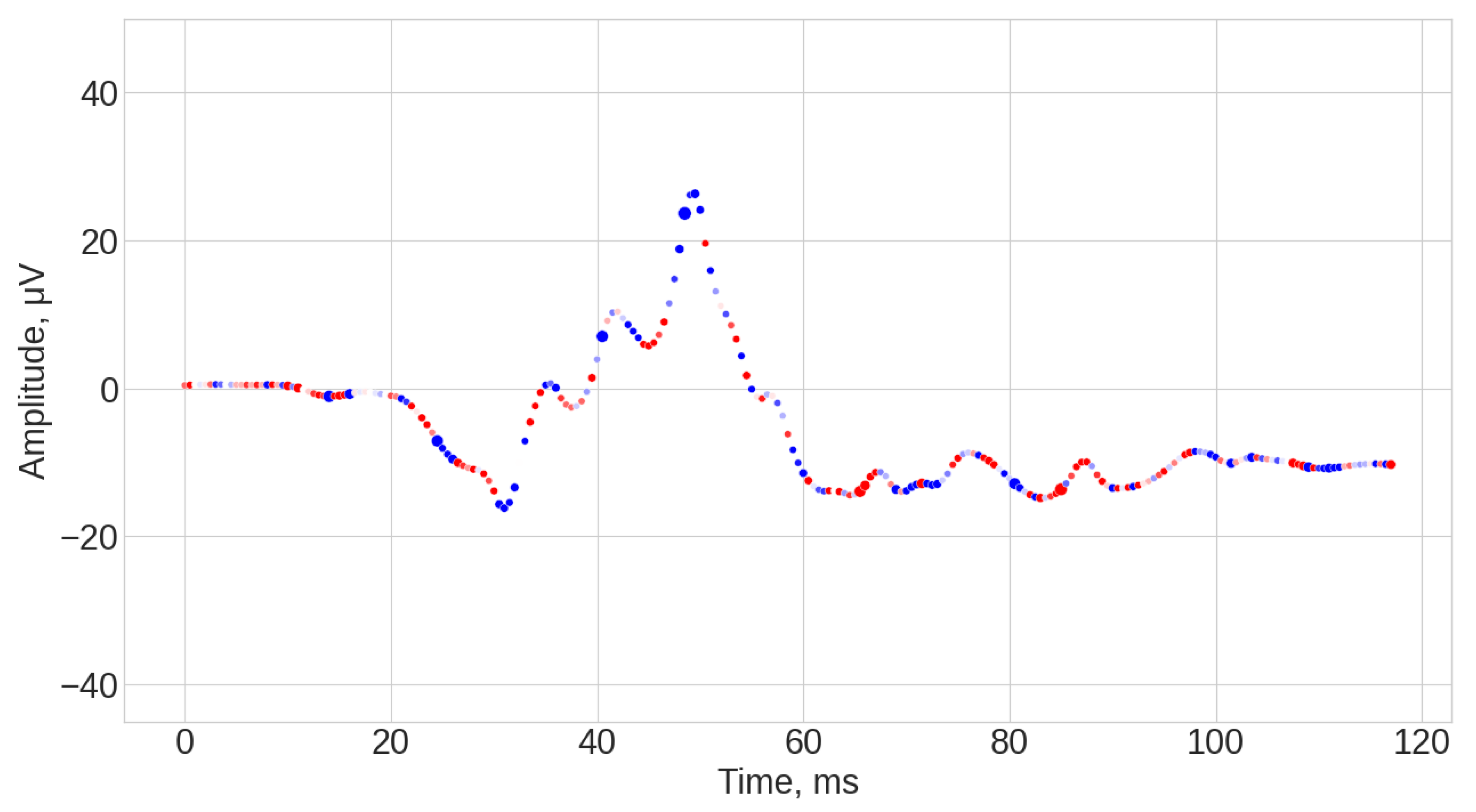
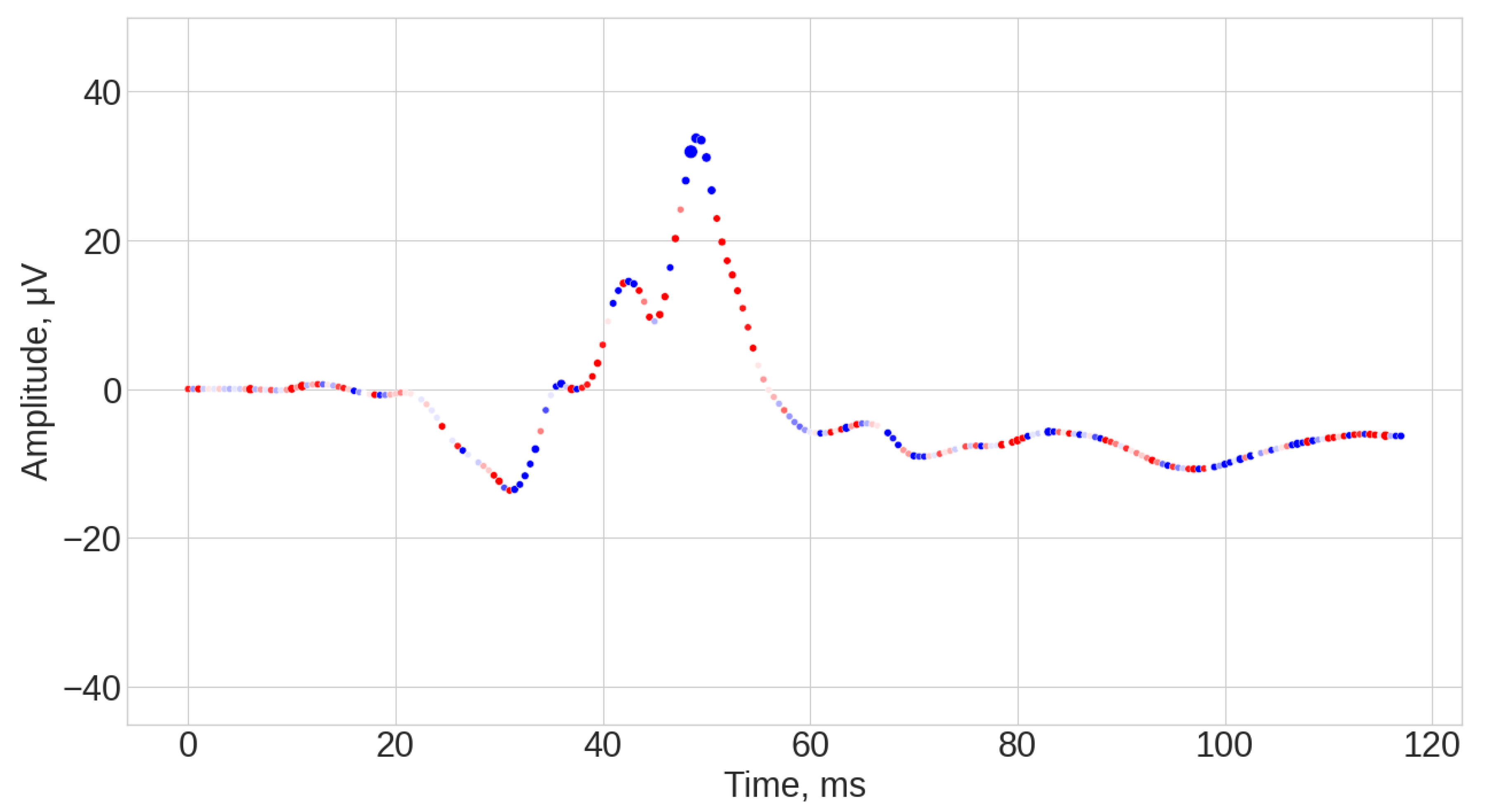
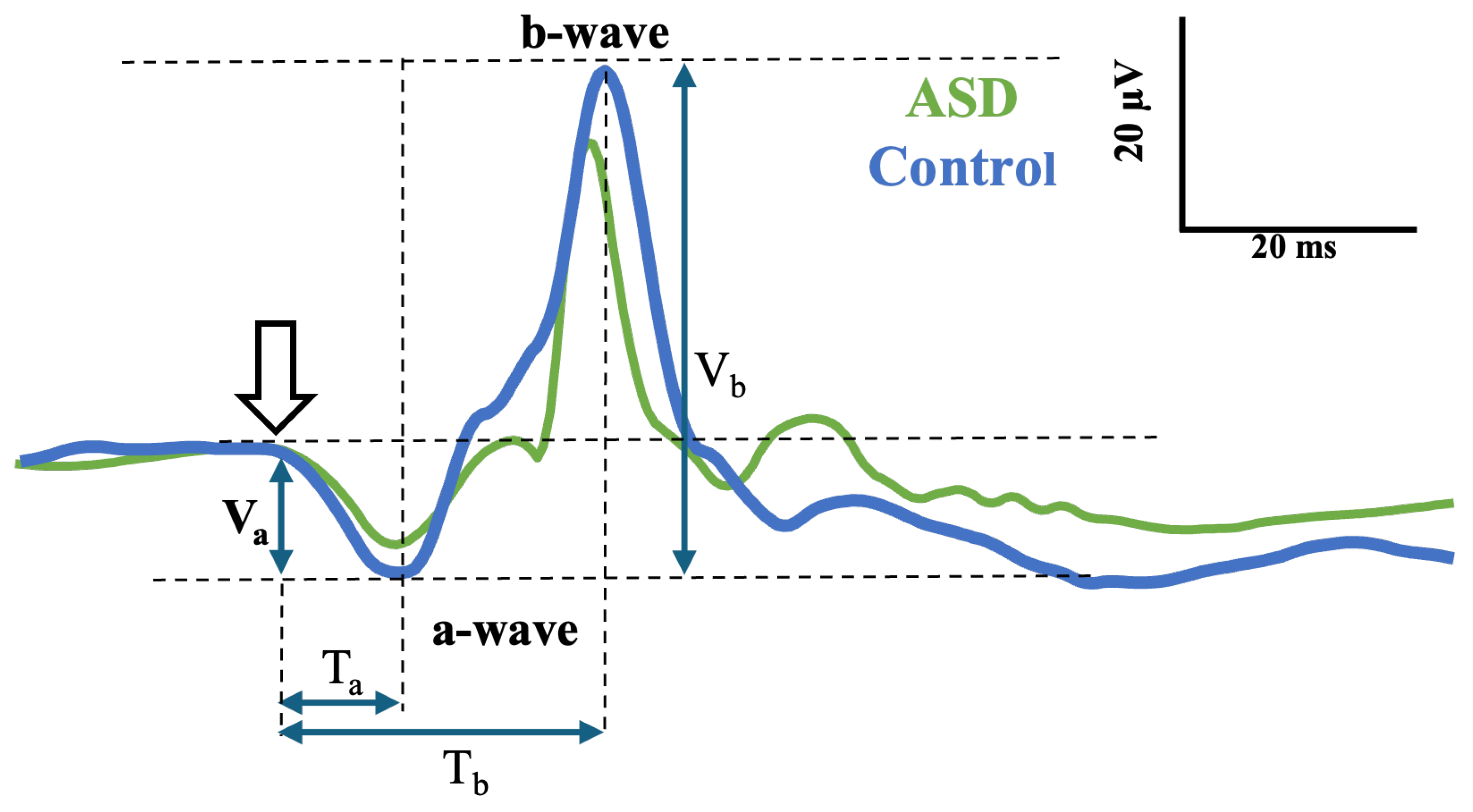
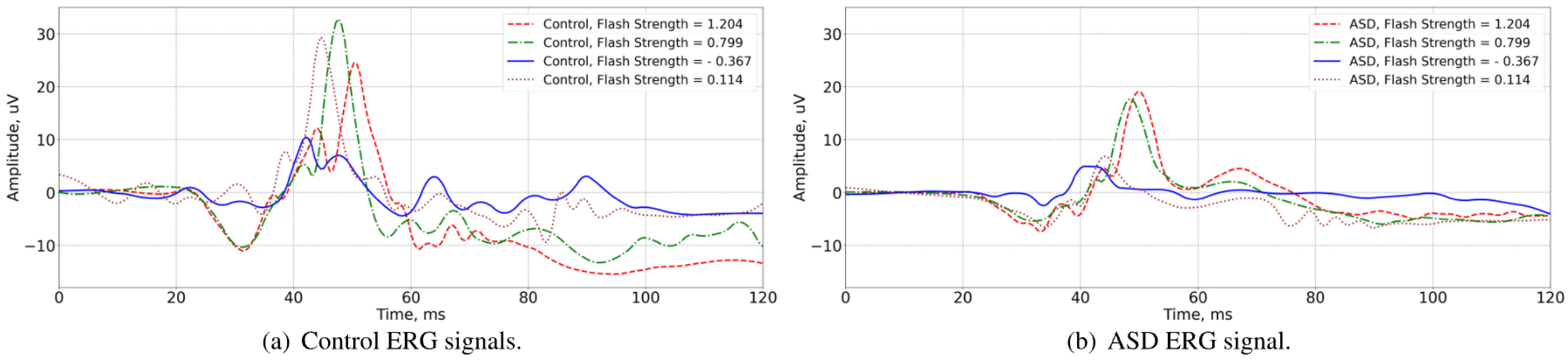
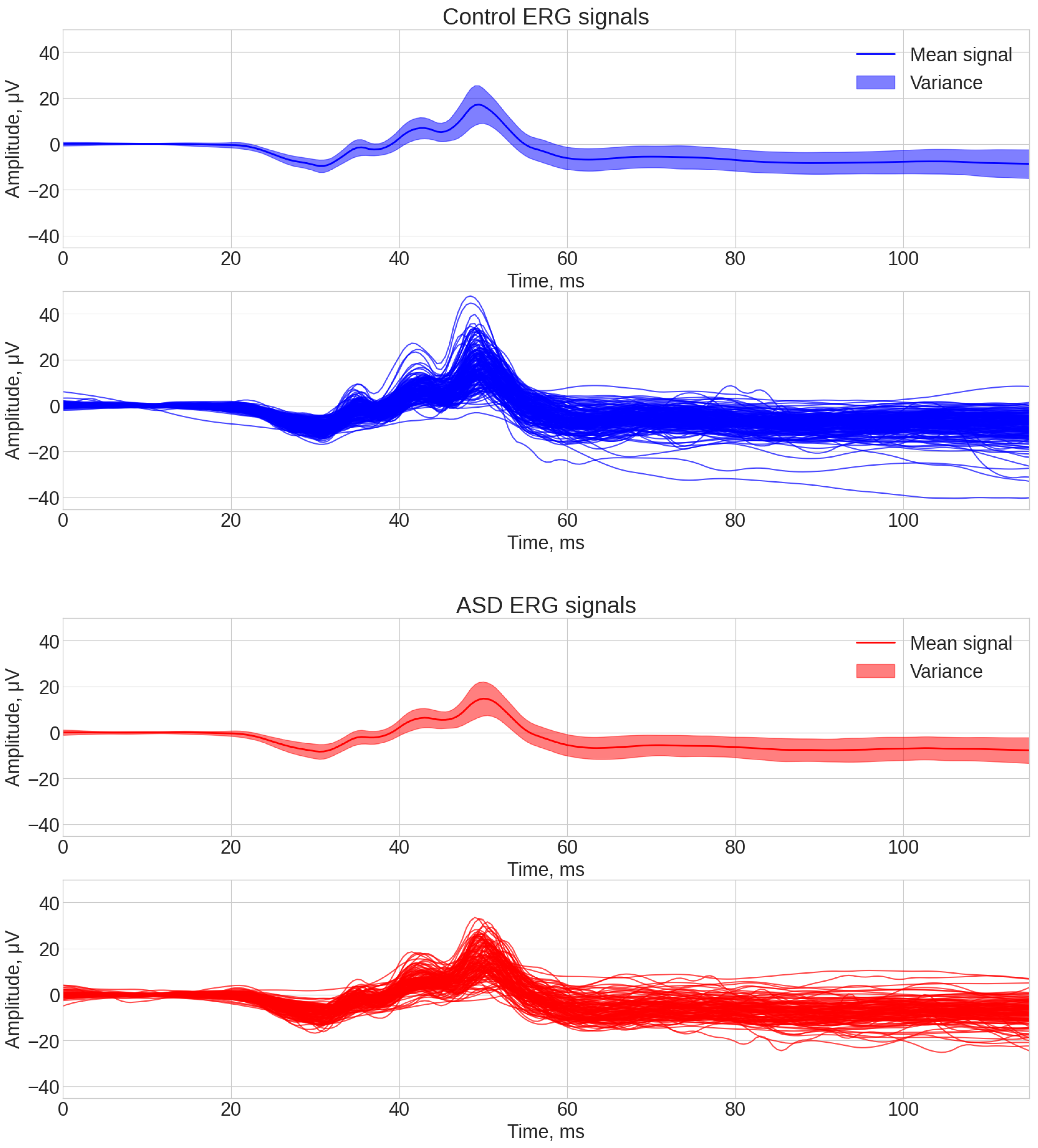
3.1. Visual Inspection of the Signals
3.2. Model Evaluation
- of the TSF algorithm was 10.
- of the TS-KNN algorithm was 3.
- of the ROCKET algorithm was 20,000.
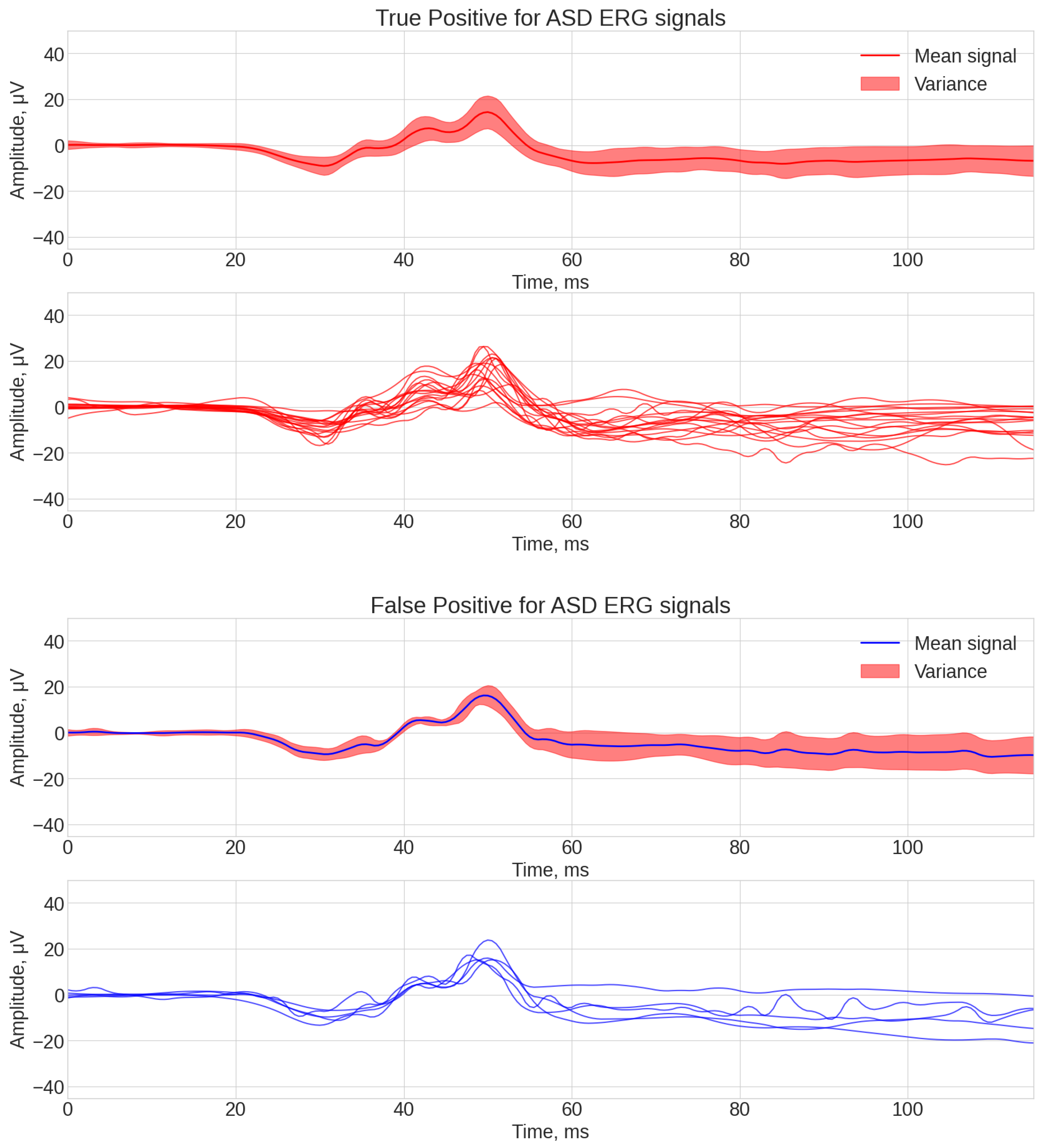
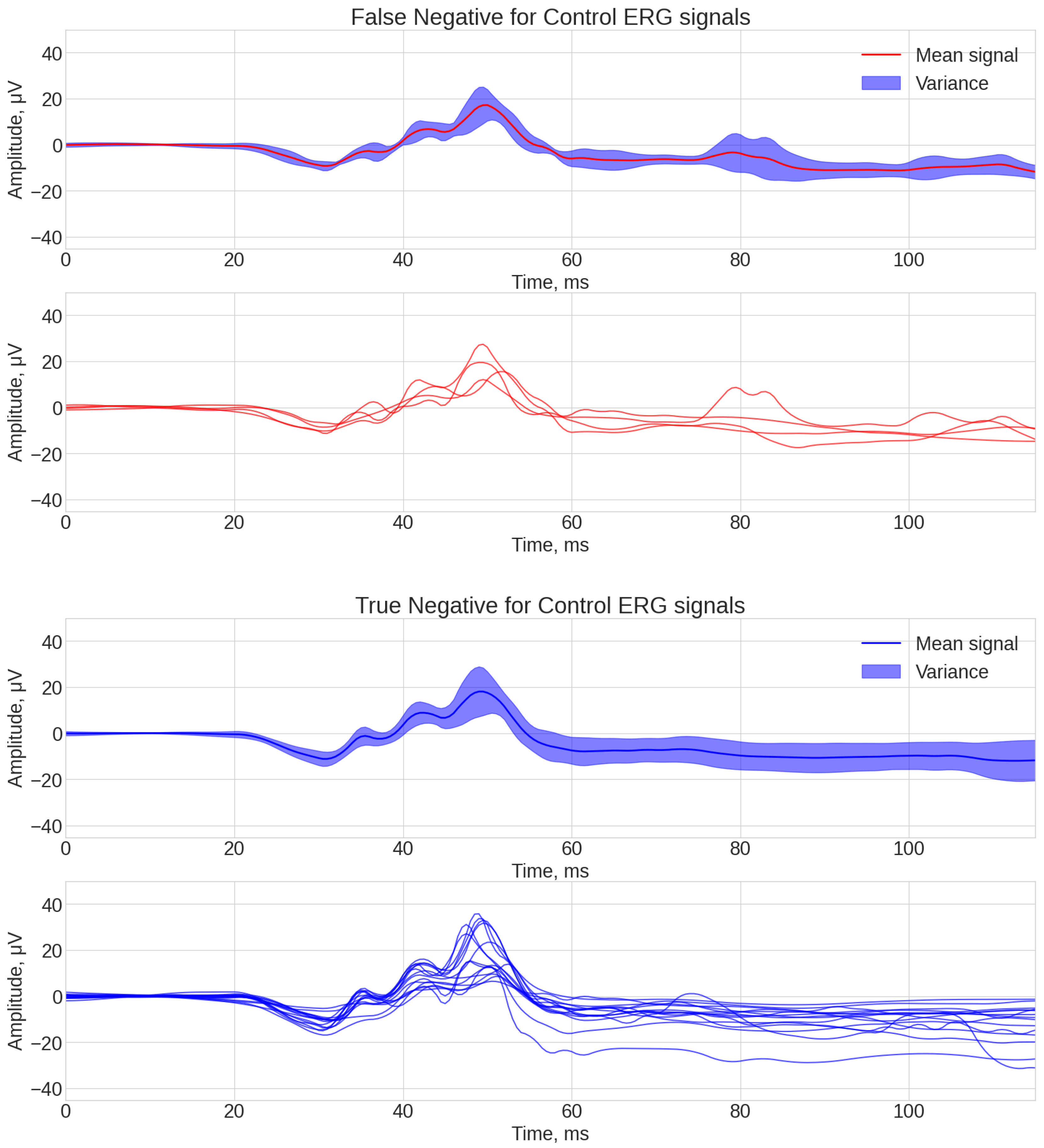
3.3. Analysis of Algorithm Errors
- If was equal to 1 and was equal to 0, then mark the array element as Falsely ASD (or false positive).
- If was 0 and was 1, then mark the array element as Falsely control (or false negative).
- If was 0 and was 0, then mark the array element Correct control (or True Negative).
- If was equal to 1 and was equal to 1, then mark the array element Correct ASD (or true positive).
3.4. Explanation of the Signals
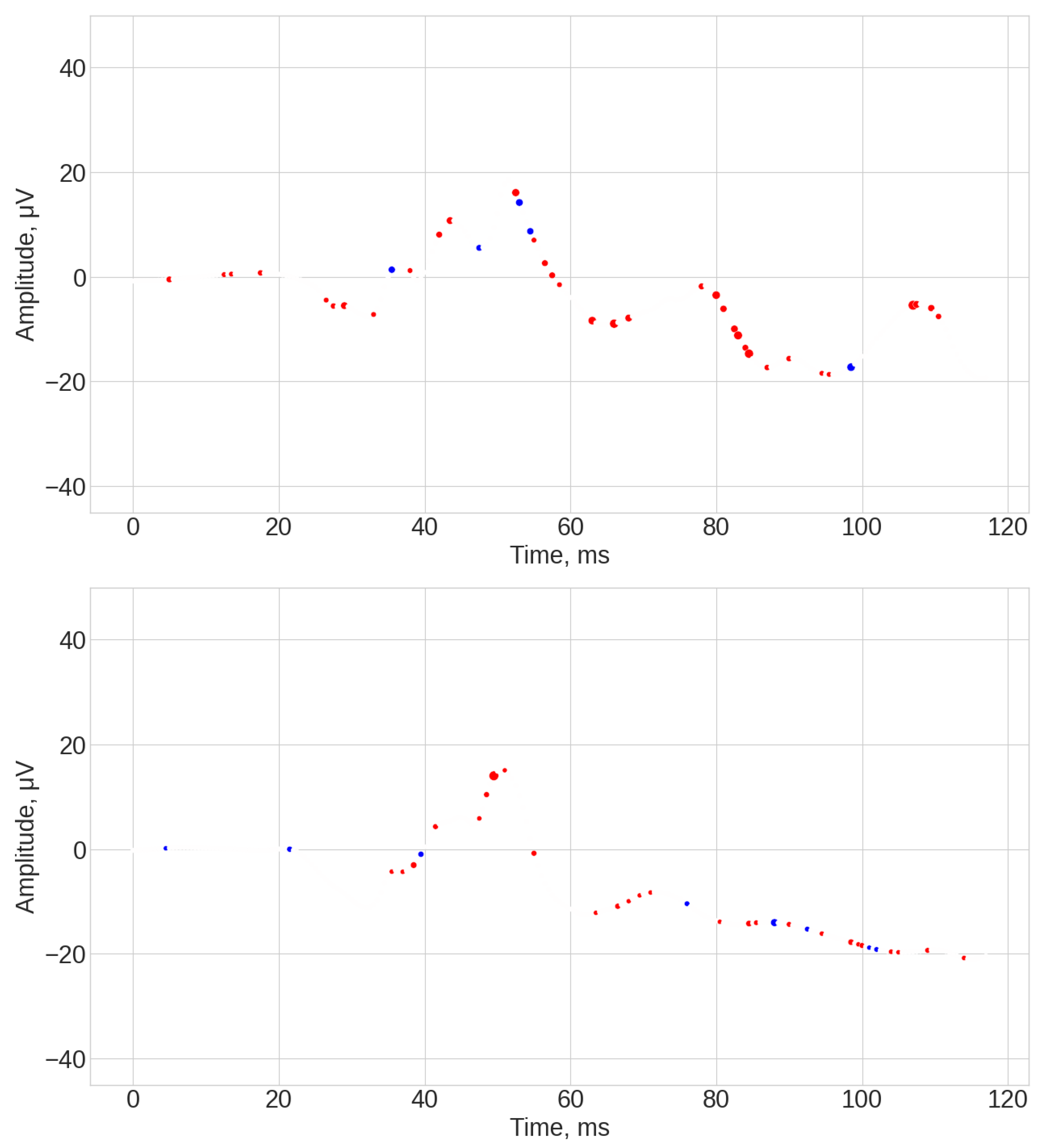
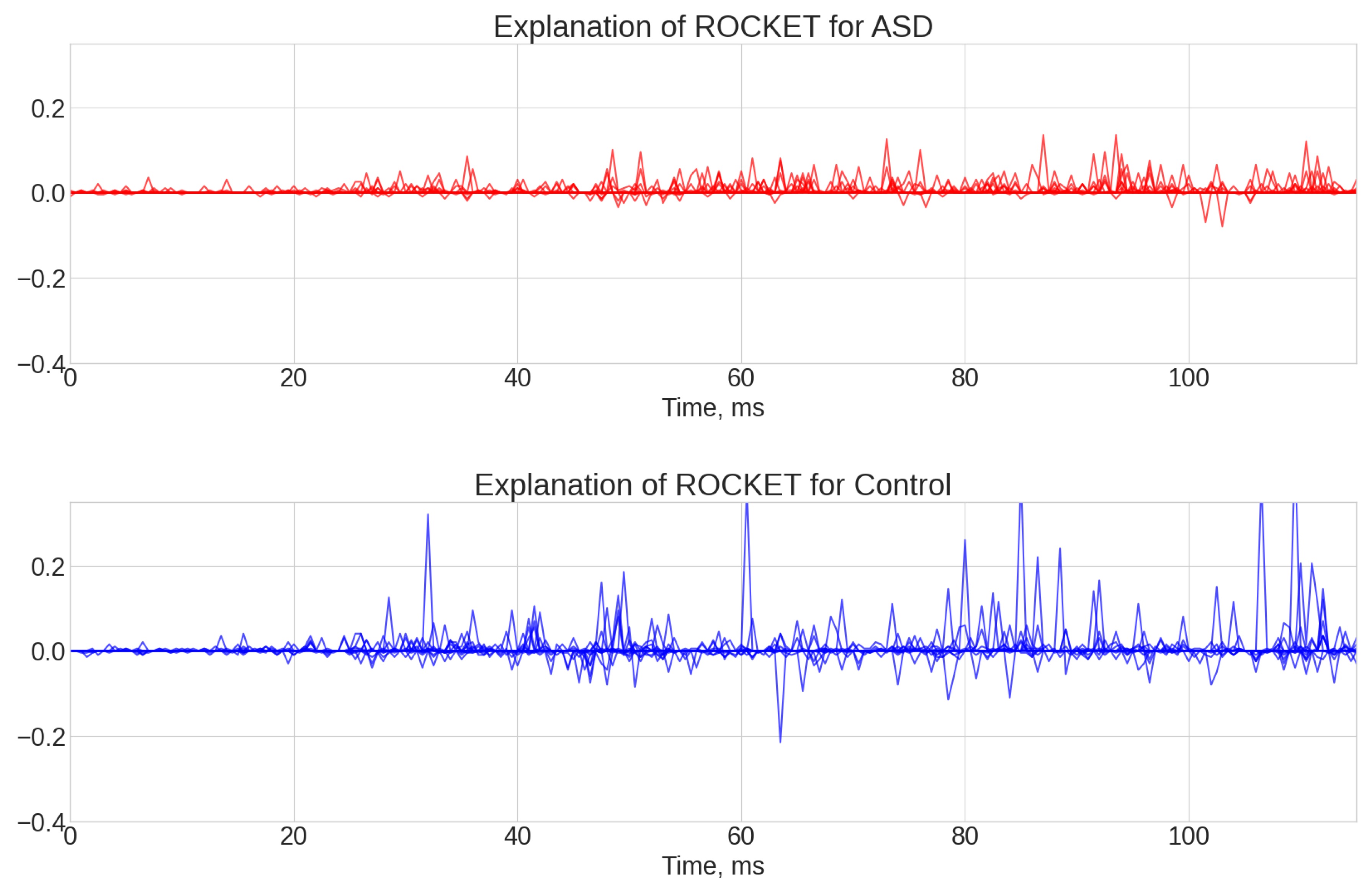
- ROCKET results (see Figure A1, Figure A2 and Figure A3). The explanations of this algorithm were the most sparse and not spread along the whole time series. Similar to the TS-KNN algorithm, the ROCKET algorithm largely ignored the initial baseline part of the signal. The most indicative parts for the predication were associated with the 35 ms and 45 ms time steps, as well as spread throughout the final part of the signal.
- TS-KNN results (see Figure A4, Figure A5 and Figure A6). The first 25 ms of the signal was mostly ignored by the algorithm. The most significant parts of the signals for positive class prediction were associated with the 35–45 ms interval, as well as with the end part of the signal. The significant part for negative class prediction was mostly located in the 35–45 ms region.
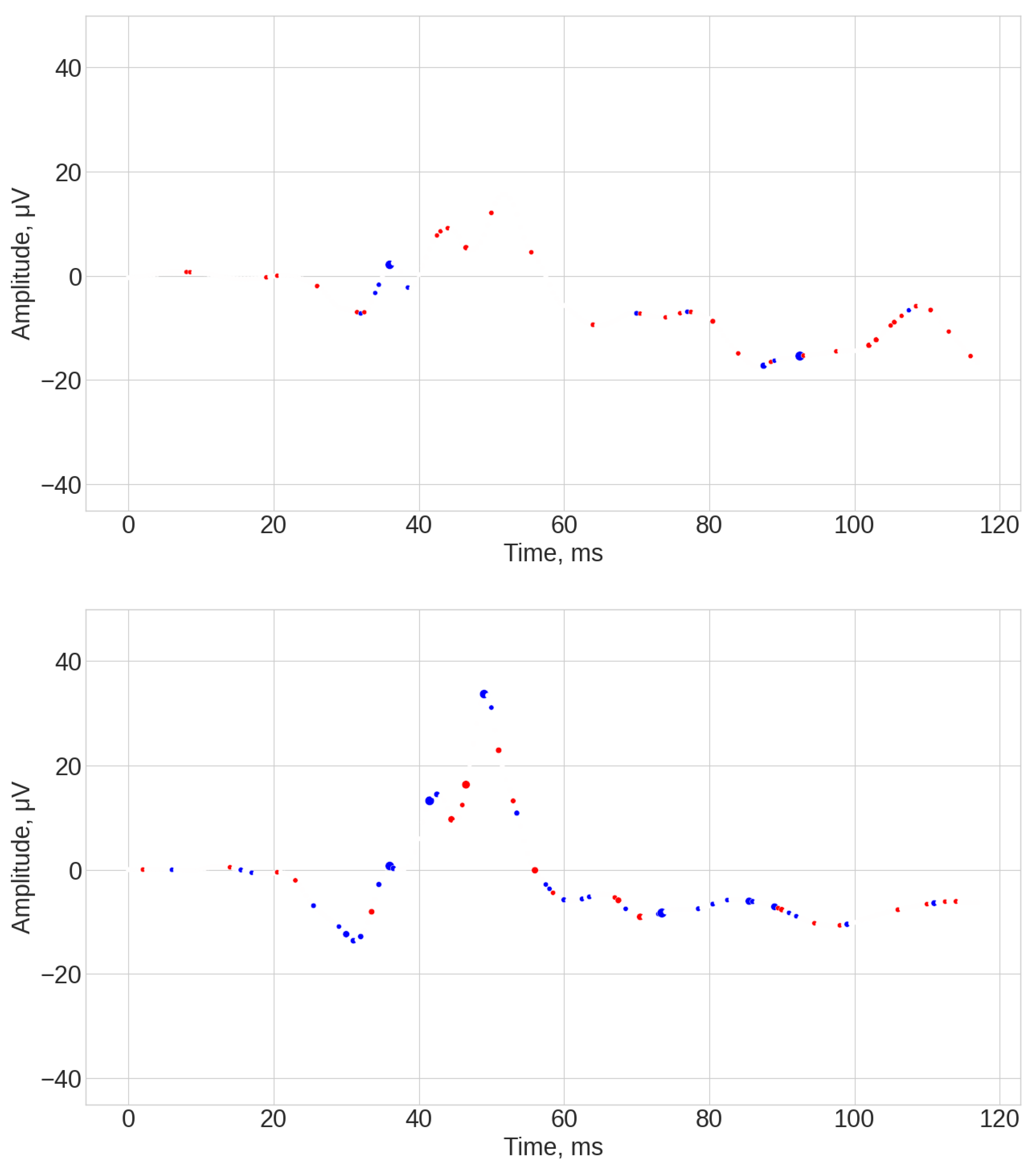
- WEASEL results (see Figure A7, Figure A8 and Figure A9). Throughout the entire signal, there were significant deviations for both control and ASD individuals. The SHAP coefficients were associated with the first part of the signal baseline (from 0 to 25 ms), as well as significant contribution of the middle part (around 50 ms), and the end of the signal (around 85 ms and 100 ms). In general, the WEASEL algorithm failed to highlight the significant parts of the signal associated with the b-wave.
- TSF results (see Figure A10, Figure A11 and Figure A12). Similar to the WEASEL algorithm throughout the entire signal, there were significant deviations for both control and ASD individuals. The most significant signals parts associated with importance for classifications were related to the 50 ms, 60 ms, 75 ms, and 100 ms marks. Overall, the TSF algorithm used the whole signal for prediction but failed to identify specific local significant regions of the signal.
3.5. Discussion
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A






References
- Robson, A.G.; Frishman, L.J.; Grigg, J.; Hamilton, R.; Jeffrey, B.G.; Kondo, M.; Li, S.; McCulloch, D.L. ISCEV Standard for Full-Field Clinical Electroretinography (2022 Update). Doc. Ophthalmol. 2022, 144, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Robson, A.G.; Nilsson, J.; Li, S.; Jalali, S.; Fulton, A.B.; Tormene, A.P.; Holder, G.E.; Brodie, S.E. ISCEV Guide to Visual Electrodiagnostic Procedures. Doc. Ophthalmol. 2018, 136, 1–26. [Google Scholar] [CrossRef]
- Constable, P.A.; Lim, J.K.; Thompson, D.A. Retinal electrophysiology in central nervous system disorders. A review of human and mouse studies. Front. Neurosci. 2023, 17, 1215097. [Google Scholar] [CrossRef]
- Constable, P.A.; Marmolejo-Ramos, F.; Gauthier, M.; Lee, I.O.; Skuse, D.H.; Thompson, D.A. Discrete wavelet transform analysis of the electroretinogram in autism spectrum disorder and attention deficit hyperactivity disorder. Front. Neurosci. 2022, 16, 890461. [Google Scholar] [CrossRef]
- Constable, P.A.; Ritvo, E.R.; Ritvo, A.R.; Lee, I.O.; McNair, M.L.; Stahl, D.; Sowden, J.; Quinn, S.; Skuse, D.H.; Thompson, D.A.; et al. Light-adapted electroretinogram differences in autism spectrum disorder. J. Autism Dev. Disord. 2020, 50, 2874–2885. [Google Scholar] [CrossRef]
- Constable, P.A.; Pinzon-Arenas, J.O.; Mercado Diaz, L.R.; Lee, I.O.; Marmolejo-Ramos, F.; Loh, L.; Zhdanov, A.; Kulyabin, M.; Brabec, M.; Skuse, D.H.; et al. Spectral Analysis of Light-Adapted Electroretinograms in Neurodevelopmental Disorders: Classification with Machine Learning. Bioengineering 2025, 12, 15. [Google Scholar] [CrossRef]
- Robson, J.G.; Saszik, S.M.; Ahmed, J.; Frishman, L.J. Rod and Cone Contributions to the a -wave of the Electroretinogram of the Macaque. J. Physiol. 2003, 547, 509–530. [Google Scholar] [CrossRef]
- Creel, D.J. Chapter 32-Electroretinograms. In Handbook of Clinical Neurology; Levin, K.H., Chauvel, P., Eds.; Clinical Neurophysiology: Basis and Technical Aspects; Elsevier: Amsterdam, The Netherlands, 2019; Volume 160, pp. 481–493. [Google Scholar] [CrossRef]
- Sieving, P.A.; Murayama, K.; Naarendorp, F. Push–Pull Model of the Primate Photopic Electroretinogram: A Role for Hyperpolarizing Neurons in Shaping the b-Wave. Vis. Neurosci. 1994, 11, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.A.; Feather, S.; Stanescu, H.C.; Freudenthal, B.; Zdebik, A.A.; Warth, R.; Ognjanovic, M.; Hulton, S.A.; Wassmer, E.; Van’T Hoff, W.; et al. Altered Electroretinograms in Patients with KCNJ10 Mutations and EAST Syndrome. J. Physiol. 2011, 589, 1681–1689. [Google Scholar] [CrossRef]
- Wachtmeister, L. Oscillatory Potentials in the Retina: What Do They Reveal. Prog. Retin. Eye Res. 1998, 17, 485–521. [Google Scholar] [CrossRef] [PubMed]
- Constable, P.A.; Gaigg, S.B.; Bowler, D.M.; Jägle, H.; Thompson, D.A. Full-Field Electroretinogram in Autism Spectrum Disorder. Doc. Ophthalmol. 2016, 132, 83–99. [Google Scholar] [CrossRef]
- Zhdanov, A.; Dolganov, A.; Zanca, D.; Borisov, V.; Ronkin, M. Advanced Analysis of Electroretinograms Based on Wavelet Scalogram Processing. Appl. Sci. 2022, 12, 12365. [Google Scholar] [CrossRef]
- Gur, M.; Zeevi, Y. Frequency-Domain Analysis of the Human Electroretinogram. J. Opt. Soc. Am. 1980, 70, 53. [Google Scholar] [CrossRef]
- Zhdanov, A.E.; Borisov, V.I.; Dolganov, A.Y.; Lucian, E.; Bao, X.; Kazaijkin, V.N. OculusGraphy: Filtering of Electroretinography Response in Adults. In Proceedings of the 2021 IEEE 22nd International Conference of Young Professionals in Electron Devices and Materials (EDM), Souzga, the Altai Republic, Russia, 30 June–4 July 2021; pp. 395–398. [Google Scholar] [CrossRef]
- Behbahani, S.; Ahmadieh, H.; Rajan, S. Feature Extraction Methods for Electroretinogram Signal Analysis: A Review. IEEE Access 2021, 9, 116879–116897. [Google Scholar] [CrossRef]
- Lindner, M. The ERGtools2 Package: A Toolset for Processing and Analysing Visual Electrophysiology Data. Doc. Ophthalmol. 2025. [Google Scholar] [CrossRef] [PubMed]
- Mahroo, O.A. Visual Electrophysiology and “the Potential of the Potentials”. Eye 2023, 37, 2399–2408. [Google Scholar] [CrossRef]
- Kulyabin, M.; Zhdanov, A.; Dolganov, A.; Ronkin, M.; Borisov, V.; Maier, A. Enhancing electroretinogram classification with multi-wavelet analysis and visual transformer. Sensors 2023, 23, 8727. [Google Scholar] [CrossRef]
- Kulyabin, M.; Constable, P.A.; Zhdanov, A.; Lee, I.O.; Thompson, D.A.; Maier, A. Attention to the electroretinogram: Gated multilayer perceptron for asd classification. IEEE Access 2024. [Google Scholar] [CrossRef]
- Middlehurst, M.; Schäfer, P.; Bagnall, A. Bake off redux: A review and experimental evaluation of recent time series classification algorithms. Data Min. Knowl. Discov. 2024, 38, 1958–2031. [Google Scholar] [CrossRef]
- Dau, H.A.; Bagnall, A.; Kamgar, K.; Yeh, C.C.M.; Zhu, Y.; Gharghabi, S.; Ratanamahatana, C.A.; Keogh, E. The UCR time series archive. IEEE/CAA J. Autom. Sin. 2019, 6, 1293–1305. [Google Scholar] [CrossRef]
- Ruiz, A.P.; Flynn, M.; Large, J.; Middlehurst, M.; Bagnall, A. The great multivariate time series classification bake off: A review and experimental evaluation of recent algorithmic advances. Data Min. Knowl. Discov. 2021, 35, 401–449. [Google Scholar] [CrossRef]
- Wang, W.K.; Chen, I.; Hershkovich, L.; Yang, J.; Shetty, A.; Singh, G.; Jiang, Y.; Kotla, A.; Shang, J.Z.; Yerrabelli, R.; et al. A systematic review of time series classification techniques used in biomedical applications. Sensors 2022, 22, 8016. [Google Scholar] [CrossRef]
- Farahani, M.A.; McCormick, M.; Harik, R.; Wuest, T. Time-series classification in smart manufacturing systems: An experimental evaluation of state-of-the-art machine learning algorithms. Robot. Comput.-Integr. Manuf. 2025, 91, 102839. [Google Scholar] [CrossRef]
- Mbouopda, M.F. Explainable Classification of Uncertain Time Series. Ph.D. Thesis, Université Clermont Auvergne, Clermont-Ferrand, France, 2022. [Google Scholar]
- Theissler, A.; Spinnato, F.; Schlegel, U.; Guidotti, R. Explainable AI for time series classification: A review, taxonomy and research directions. IEEE Access 2022, 10, 100700–100724. [Google Scholar] [CrossRef]
- Cabello, N.; Naghizade, E.; Qi, J.; Kulik, L. Fast, accurate and explainable time series classification through randomization. Data Min. Knowl. Discov. 2024, 38, 748–811. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Le Nguyen, T.; Ifrim, G. Robust explainer recommendation for time series classification. Data Min. Knowl. Discov. 2024, 38, 3372–3413. [Google Scholar] [CrossRef] [PubMed]
- Massoda, S.; Rakkay, H.; Émond, C.; Tellier, V.; Sasseville, A.; Stoica, G.; Chau, A.; Coupland, S.; Hariton, C. High-density retinal signal deciphering in support of diagnosis in psychiatric disorders: A new paradigm. Biomed. Signal Process. Control 2025, 102, 107373. [Google Scholar] [CrossRef]
- Friedel, E.B.N.; Schäfer, M.; Endres, D.; Maier, S.; Runge, K.; Bach, M.; Heinrich, S.P.; Ebert, D.; Domschke, K.; Tebartz Van Elst, L.; et al. Electroretinography in adults with high-functioning autism spectrum disorder. Autism Res. 2022, 15, 2026–2037. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.-A.; Pelletier, C.-A.; Mérette, C.; Jomphe, V.; Turgeon, R.; Bélanger, R.E.; Grondin, S.; Hébert, M. Evaluation of electroretinography (ERG) parameters as a biomarker for ADHD. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2023, 127, 110807. [Google Scholar] [CrossRef]
- Brabec, M.; Marmolejo-Ramos, F.; Loh, L.; Lee, I.O.; Kulyabin, M.; Zhdanov, A.; Posada-Quintero, H.; Thompson, D.A.; Constable, P.A. Remodeling the light-adapted electroretinogram using a bayesian statistical approach. BMC Res. Notes 2025, 18, 33. [Google Scholar] [CrossRef]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Müller, H. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; García, S.; Gil-López, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Bagnall, A.; Lines, J.; Bostrom, A.; Large, J.; Keogh, E. The great time series classification bake off: A review and experimental evaluation of recent algorithmic advances. Data Min. Knowl. Discov. 2017, 31, 606–660. [Google Scholar] [CrossRef] [PubMed]
- Turbé, H.; Bjelogrlic, M.; Lovis, C.; Mengaldo, G. Evaluation of post-hoc interpretability methods in time-series classification. Nat. Mach. Intell. 2023, 5, 250–260. [Google Scholar] [CrossRef]
- Löning, M.; Bagnall, A.; Ganesh, S.; Kazakov, V.; Lines, J.; Király, F.J. sktime: A unified interface for machine learning with time series. arXiv 2019, arXiv:1909.07872. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 30, 4768–4777. [Google Scholar]
- Constable, P.; Marmolejo-Ramos, F.; Thompson, D.; Brabec, M. Electroretinogram Raw Waveforms for Control and Autism. 2022. Available online: https://doi.org/10.25451/flinders.21546210.v1 (accessed on 22 June 2025).
- Schäfer, P.; Leser, U. Fast and accurate time series classification with weasel. In Proceedings of the Proceedings of the 2017 ACM on Conference on Information and Knowledge Management, Singapore, 6–10 November 2017; pp. 637–646. [Google Scholar]
- Deng, H.; Runger, G.; Tuv, E.; Vladimir, M. A time series forest for classification and feature extraction. Inf. Sci. 2013, 239, 142–153. [Google Scholar] [CrossRef]
- Kate, R.J. Using dynamic time warping distances as features for improved time series classification. Data Min. Knowl. Discov. 2016, 30, 283–312. [Google Scholar] [CrossRef]
- Dempster, A.; Petitjean, F.; Webb, G.I. ROCKET: Exceptionally fast and accurate time series classification using random convolutional kernels. Data Min. Knowl. Discov. 2020, 34, 1454–1495. [Google Scholar] [CrossRef]
- Mu, T.; Wang, H.; Zheng, S.; Liang, Z.; Wang, C.; Shao, X.; Liang, Z. TSC-AutoML: Meta-learning for Automatic Time Series Classification Algorithm Selection. In Proceedings of the 2023 IEEE 39th International Conference on Data Engineering (ICDE), Anaheim, CA, USA, 3–7 April 2023; pp. 1032–1044. [Google Scholar]
- Brabec, M.; Constable, P.A.; Thompson, D.A.; Marmolejo-Ramos, F. Group Comparisons of the Individual Electroretinogram Time Trajectories for the Ascending Limb of the B-Wave Using a Raw and Registered Time Series. BMC Res. Notes 2023, 16, 238. [Google Scholar] [CrossRef]
- Durajczyk, M.; Lubiński, W. Congenital Stationary Night Blindness (CSNB)—Case Reports and Review of Current Knowledge. J. Clin. Med. 2025, 14, 1238. [Google Scholar] [CrossRef]
- Zeitz, C.; Robson, A.G.; Audo, I. Congenital Stationary Night Blindness: An Analysis and Update of Genotype–Phenotype Correlations and Pathogenic Mechanisms. Prog. Retin. Eye Res. 2015, 45, 58–110. [Google Scholar] [CrossRef]
- Frishman, L.; Sustar, M.; Kremers, J.; McAnany, J.J.; Sarossy, M.; Tzekov, R.; Viswanathan, S. ISCEV Extended Protocol for the Photopic Negative Response (PhNR) of the Full-Field Electroretinogram. Doc. Ophthalmol. 2018, 136, 207–211. [Google Scholar] [CrossRef]
- Viswanathan, S.; Frishman, L.J.; Robson, J.G.; Walters, J.W. The Photopic Negative Response of the Flash Electroretinogram in Primary Open Angle Glaucoma. Investig. Ophthalmol. Vis. Sci. 2001, 42, 514–522. [Google Scholar]
- Constable, P.A.; Lee, I.O.; Marmolejo-Ramos, F.; Skuse, D.H.; Thompson, D.A. The Photopic Negative Response in Autism Spectrum Disorder. Clin. Exp. Optom. 2021, 104, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Elanwar, R. Retinal Functional and Structural Changes in Patients with Parkinson’s Disease. BMC Neurol. 2023, 23, 330. [Google Scholar] [CrossRef]
- Galetta, K.M.; Morganroth, J.; Moehringer, N.; Mueller, B.; Hasanaj, L.; Webb, N.; Civitano, C.; Cardone, D.A.; Silverio, A.; Galetta, S.L.; et al. Adding Vision to Concussion Testing: A Prospective Study of Sideline Testing in Youth and Collegiate Athletes. J. Neuro-Opthalmology 2015, 35, 235–241. [Google Scholar] [CrossRef]
- Garcia-Martin, E.; Rodriguez-Mena, D.; Satue, M.; Almarcegui, C.; Dolz, I.; Alarcia, R.; Seral, M.; Polo, V.; Larrosa, J.M.; Pablo, L.E. Electrophysiology and Optical Coherence Tomography to Evaluate Parkinson Disease Severity. Investig. Opthalmology Vis. Sci. 2014, 55, 696. [Google Scholar] [CrossRef]
- Ikeda, H.; Head, G.M.; Ellis, C.J.K. Electrophysiological Signs of Retinal Dopamine Deficiency in Recently Diagnosed Parkinson’s Disease and a Follow up Study. Vis. Res. 1994, 34, 2629–2638. [Google Scholar] [CrossRef] [PubMed]
- Firmani, G. Ocular Biomarkers in Alzheimer’s Disease: Insights into Early Detection Through Eye-Based Diagnostics—A Literature Review. Clin. Ter. 2024, 175, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Manjur, S.M.; Diaz, L.R.M.; Lee, I.O.; Skuse, D.H.; Thompson, D.A.; Marmolejos-Ramos, F.; Constable, P.A.; Posada-Quintero, H.F. Detecting Autism Spectrum Disorder and Attention Deficit Hyperactivity Disorder Using Multimodal Time-Frequency Analysis with Machine Learning Using the Electroretinogram from Two Flash Strengths. J. Autism Dev. Disord. 2025, 55, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Posada-Quintero, H.F.; Manjur, S.M.; Hossain, M.B.; Marmolejo-Ramos, F.; Lee, I.O.; Skuse, D.H.; Thompson, D.A.; Constable, P.A. Autism Spectrum Disorder Detection Using Variable Frequency Complex Demodulation of the Electroretinogram. Res. Autism Spectr. Disord. 2023, 109, 102258. [Google Scholar] [CrossRef]
- Donié, C.; Das, N.; Endo, S.; Hirche, S. Estimating Motor Symptom Presence and Severity in Parkinson’s Disease from Wrist Accelerometer Time Series Using ROCKET and InceptionTime. Sci. Rep. 2025, 15, 19140. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.O.; Fritsch, D.M.; Kerz, M.; Sowden, J.C.; Constable, P.A.; Skuse, D.H.; Thompson, D.A. Global motion coherent deficits in individuals with autism spectrum disorder and their family members are associated with retinal function. Sci. Rep. 2025, 15, 28249. [Google Scholar] [CrossRef]
| TSC | F1 Control | F1 ASD | Balanced Accuracy |
|---|---|---|---|
| WEASEL | |||
| TSF | |||
| TS-KNN | |||
| ROCKET |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chistiakov, S.; Dolganov, A.; Constable, P.A.; Zhdanov, A.; Kulyabin, M.; Thompson, D.A.; Lee, I.O.; Albasu, F.; Borisov, V.; Ronkin, M. Time Series Classification of Autism Spectrum Disorder Using the Light-Adapted Electroretinogram. Bioengineering 2025, 12, 951. https://doi.org/10.3390/bioengineering12090951
Chistiakov S, Dolganov A, Constable PA, Zhdanov A, Kulyabin M, Thompson DA, Lee IO, Albasu F, Borisov V, Ronkin M. Time Series Classification of Autism Spectrum Disorder Using the Light-Adapted Electroretinogram. Bioengineering. 2025; 12(9):951. https://doi.org/10.3390/bioengineering12090951
Chicago/Turabian StyleChistiakov, Sergey, Anton Dolganov, Paul A. Constable, Aleksei Zhdanov, Mikhail Kulyabin, Dorothy A. Thompson, Irene O. Lee, Faisal Albasu, Vasilii Borisov, and Mikhail Ronkin. 2025. "Time Series Classification of Autism Spectrum Disorder Using the Light-Adapted Electroretinogram" Bioengineering 12, no. 9: 951. https://doi.org/10.3390/bioengineering12090951
APA StyleChistiakov, S., Dolganov, A., Constable, P. A., Zhdanov, A., Kulyabin, M., Thompson, D. A., Lee, I. O., Albasu, F., Borisov, V., & Ronkin, M. (2025). Time Series Classification of Autism Spectrum Disorder Using the Light-Adapted Electroretinogram. Bioengineering, 12(9), 951. https://doi.org/10.3390/bioengineering12090951








