Cascaded Self-Supervision to Advance Cardiac MRI Segmentation in Low-Data Regimes
Abstract
1. Introduction
- We propose a novel SSL strategy for cardiac MRI segmentation by performing multi-step pseudo-labeling based on cascaded ST stages.
- We extensively evaluate different individual SSL strategies and their combinations and compare them with our proposed cascaded algorithm on 2D and 3D imaging data.
- We assess the performance of all investigated strategies in a low-data regime by systematically reducing the size of labeled data and comparing with the fully supervised case as well as related work.
2. Related Work
2.1. Consistency Regularization
2.2. Student-Teacher
2.3. Pseudo-Labeling
3. Method
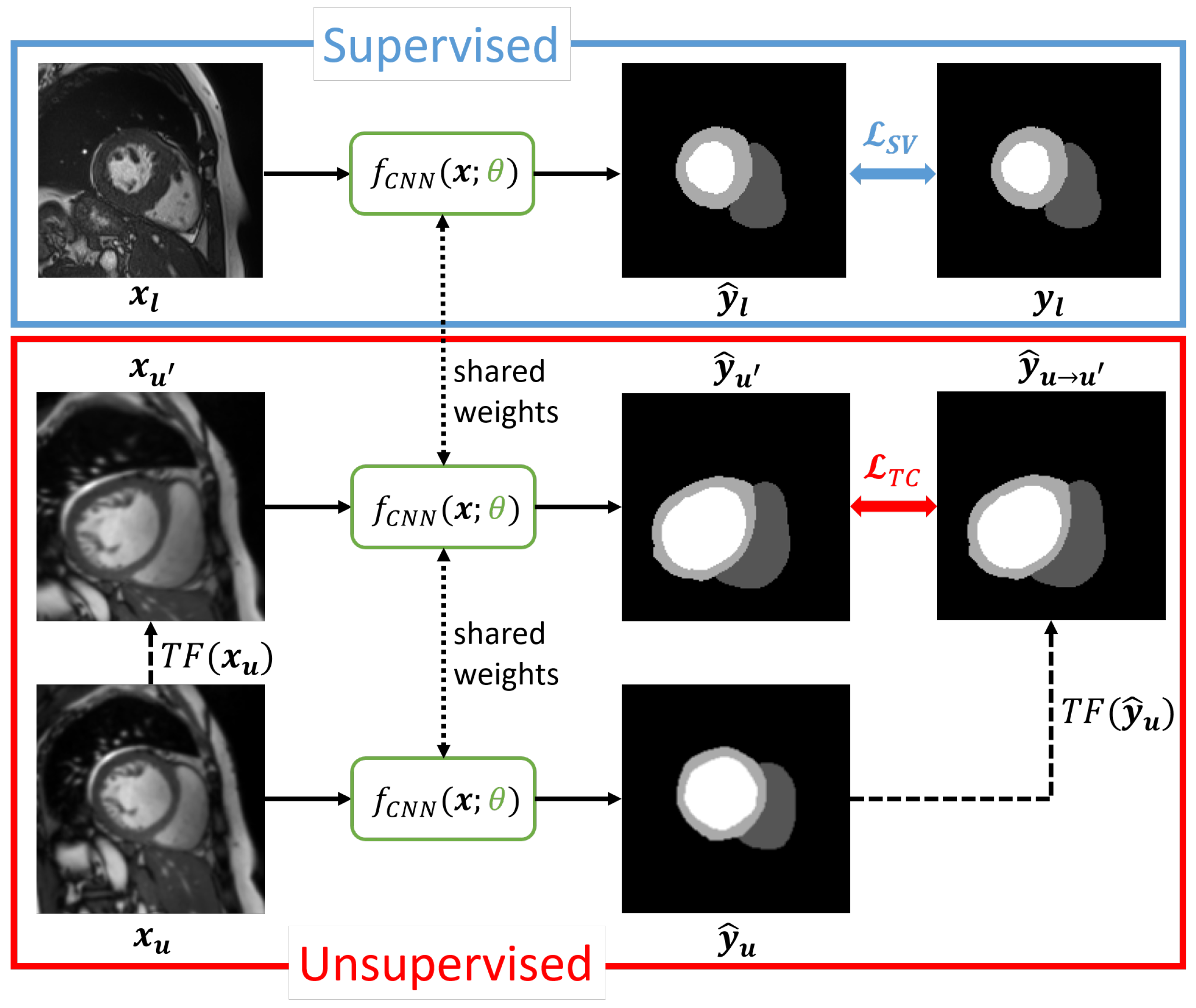
3.1. Transformation Consistency
3.2. Student–Teacher
3.2.1. Student–Teacher Without Transformations
3.2.2. Student–Teacher with Transformations
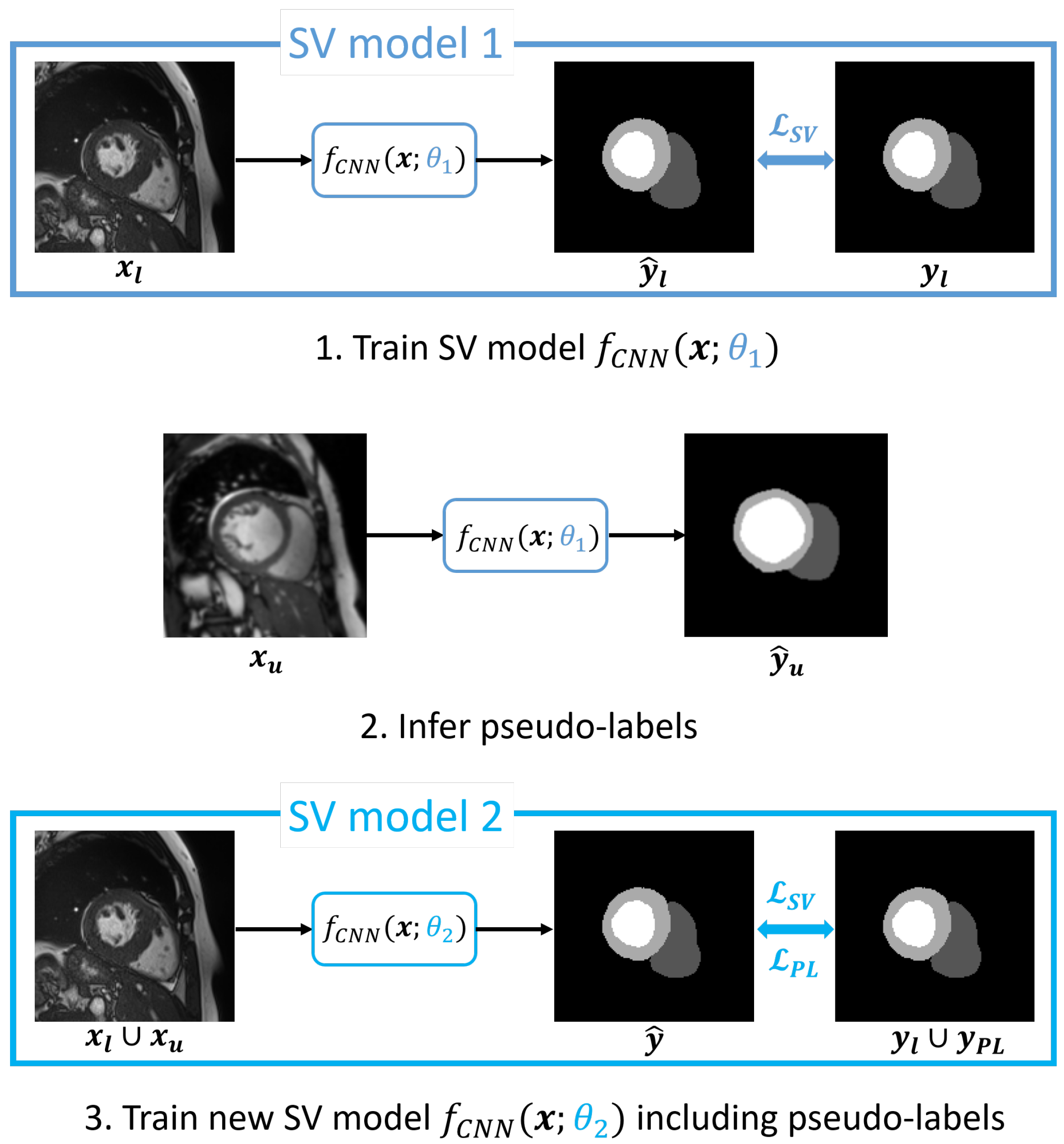
3.3. Self-Training via Pseudo-Labeling
3.4. Cascaded Self-Supervision
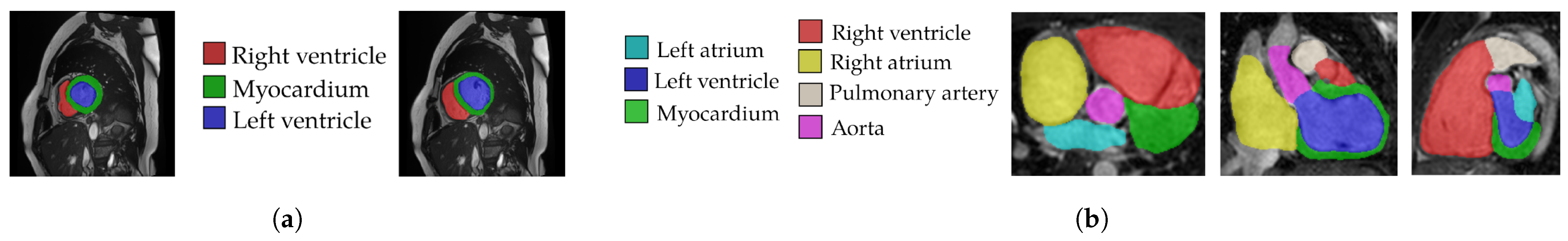
3.5. Datasets
4. Experimental Setup
4.1. Data Preprocessing
4.2. Data Augmentation
4.3. Neural Network Architecture
4.4. Implementation Details
4.5. Evaluation Metrics
4.6. Self-Training Method Variants
4.7. Training Setup
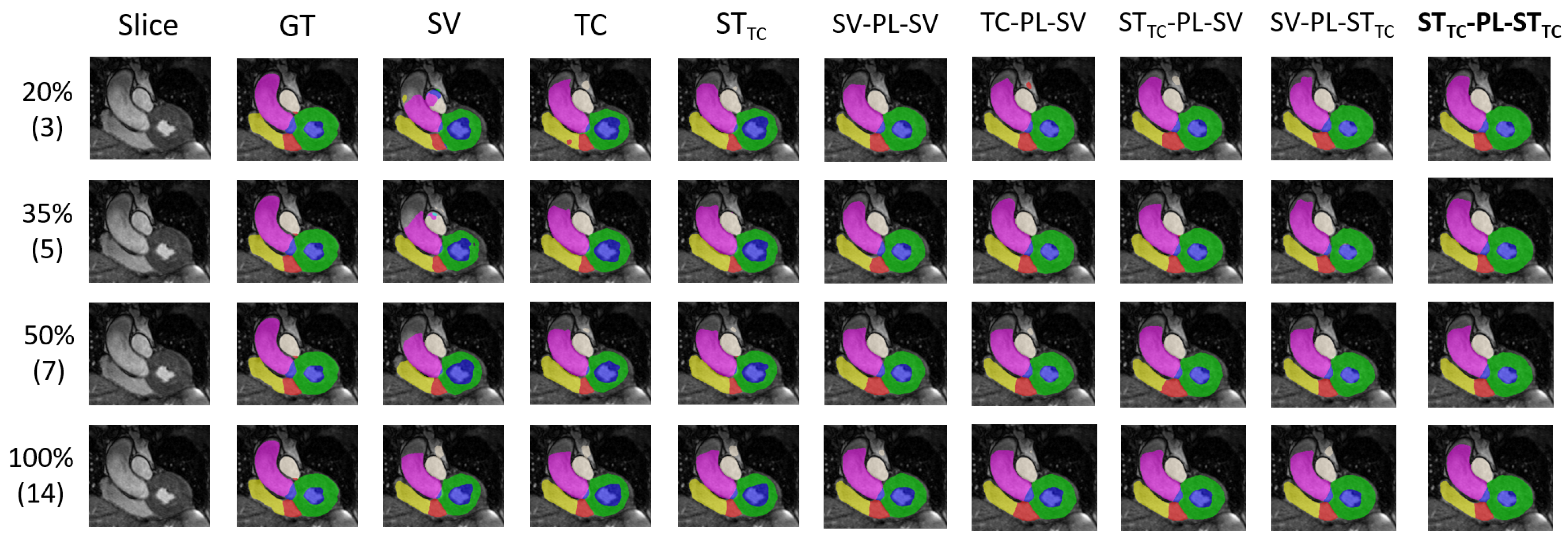
5. Results and Discussion
5.1. Internal Evaluation
5.2. Comparison to Literature
5.3. Limitations
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACDC | Automated Cardiac Diagnosis Challenge |
| AO | Aorta |
| ASSD | Average Symmetric Surface Distance |
| CNN | Convolutional Neural Network |
| CT | Computed Tomography |
| CVD | Cardiovascular Diseases |
| DSC | Dice Similarity Coefficient |
| DTC | Dual Task Consistency |
| EMA | Exponential Moving Average |
| GDL | Generalized Dice Loss |
| LCLPL | Local Contrastive Loss with Pseudo-Labels |
| LA | Left Atrium |
| LV | Left Ventricle |
| MICCAI | Medical Image Computing and Computer Assisted Intervention |
| MMWHS | Multi-Modality Whole Heart Segmentation |
| MRI | Magnetic Resonance Imaging |
| MSE | Mean Squared Error |
| MYO | Myocardium |
| PA | Pulmonary Artery |
| PL | Pseudo-Labeling |
| PLGCL | Pseudo-Label Guided Contrastive Learning |
| RA | Right Atrium |
| RV | Right Ventricle |
| ST | Student–Teacher |
| SV | Supervised |
| TC | Transformation Consistency |
| SSL | Self-Supervised Learning |
References
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of cardiovascular diseases and risk factors, 1990-2019: Update from the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J. Assessing the role of circulating, genetic, and imaging biomarkers in cardiovascular risk prediction. Circulation 2011, 123, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Cawley, P.J.; Maki, J.H.; Otto, C.M. Cardiovascular magnetic resonance imaging for valvular heart disease: Technique and validation. Circulation 2009, 119, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep Learning for Cardiac Image Segmentation: A Review. Front. Cardiovasc. Med. 2020, 7, 25. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Munich, Germany, 5–9 October 2015; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Zhuang, X.; Li, L.; Payer, C.; Stern, D.; Urschler, M.; Heinrich, M.P.; Oster, J.; Wang, C.; Smedby, O.; Bian, C.; et al. Evaluation of algorithms for Multi-Modality Whole Heart Segmentation: An open-access grand challenge. Med. Image Anal. 2019, 58, 101537. [Google Scholar] [CrossRef]
- Zhuang, X.; Shen, J. Multi-scale patch and multi-modality atlases for whole heart segmentation of MRI. Med. Image Anal. 2016, 31, 77–87. [Google Scholar] [CrossRef]
- Chapelle, O.; Schoelkopf, B.; Zien, A. Semi-Supervised Learning; The MIT Press: Cambridge, MA, USA, 2006. [Google Scholar]
- van Engelen, J.E.; Hoos, H.H. A survey on semi-supervised learning. Mach. Learn. 2020, 109, 373–440. [Google Scholar] [CrossRef]
- Yang, X.; Song, Z.; King, I.; Xu, Z. A survey on deep semi-supervised learning. IEEE Trans. Knowl. Data Eng. 2023, 35, 8934–8954. [Google Scholar] [CrossRef]
- Neff, T.; Payer, C.; Štern, D.; Urschler, M. Generative Adversarial Network based Synthesis for Supervised Medical Image Segmentation. In Proceedings of the OAGM & ARW Joint Workshop 2017: Vision, Automation and Robotics, Vienna, Austria, 10–12 May 2017; pp. 140–145. [Google Scholar] [CrossRef]
- Hadzic, A.; Bogensperger, L.; Joham, S.J.; Urschler, M. Synthetic Augmentation for Anatomical Landmark Localization Using DDPMs. In Proceedings of the Simulation and Synthesis in Medical Imaging (SASHIMI 2024), Marrakesh, Morocco, 10 October 2024; Volume 15187, Lecture Notes in Computer Science. pp. 1–12. [Google Scholar] [CrossRef]
- Jiao, R.; Zhang, Y.; Ding, L.; Xue, B.; Zhang, J.; Cai, R.; Jin, C. Learning with limited annotations: A survey on deep semi-supervised learning for medical image segmentation. Comput. Biol. Med. 2024, 169, 107840. [Google Scholar] [CrossRef]
- Bortsova, G.; Dubost, F.; Hogeweg, L.; Katramados, I.; de Bruijne, M. Semi-supervised Medical Image Segmentation via Learning Consistency Under Transformations. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019, Shenzhen, China, 13–17 October 2019; Springer International Publishing: Cham, Switzerland, 2019; pp. 810–818. [Google Scholar] [CrossRef]
- Li, X.; Yu, L.; Chen, H.; Fu, C.W.; Xing, L.; Heng, P.A. Transformation-consistent self-ensembling model for semisupervised medical image segmentation. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 523–534. [Google Scholar] [CrossRef] [PubMed]
- DeVries, T.; Taylor, G.W. Improved Regularization of Convolutional Neural Networks with Cutout. arXiv 2017, arXiv:1708.04552. [Google Scholar]
- Yun, S.; Han, D.; Chun, S.; Oh, S.J.; Yoo, Y.; Choe, J. CutMix: Regularization strategy to train strong classifiers with localizable features. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; IEEE: Piscataway, NJ, USA, 2019. [Google Scholar] [CrossRef]
- French, G.; Laine, S.; Aila, T.; Mackiewicz, M.; Finlayson, G. Semi-supervised semantic segmentation needs strong, varied perturbations. In Proceedings of the British Machine Vision Conference (BMVC), Virtual Event, 7–10 September 2020; pp. 1–14. [Google Scholar] [CrossRef]
- Luo, X.; Chen, J.; Song, T.; Wang, G. Semi-supervised medical image segmentation through dual-task consistency. Proc. Conf. AAAI Artif. Intell. 2021, 35, 8801–8809. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, M.; Ge, Z.; Cai, J.; Zhang, L. Semi-supervised left atrium segmentation with mutual consistency training. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, Strasbourg, France, 27 September–1 October 2021; Lecture Notes in Computer Science. Springer International Publishing: Cham, Switzerland, 2021; pp. 297–306. [Google Scholar] [CrossRef]
- Wu, Y.; Ge, Z.; Zhang, D.; Xu, M.; Zhang, L.; Xia, Y.; Cai, J. Mutual consistency learning for semi-supervised medical image segmentation. Med. Image Anal. 2022, 81, 102530. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chen, Z.; Chen, C.; Lu, M.; Zou, Y. Complementary consistency semi-supervised learning for 3D left atrial image segmentation. Comput. Biol. Med. 2023, 165, 107368. [Google Scholar] [CrossRef]
- Laine, S.; Aila, T. Temporal Ensembling for Semi-Supervised Learning. In Proceedings of the International Conference on Learning Representations (ICLR), Toulon, France, 24–26 April 2017. [Google Scholar]
- Tarvainen, A.; Valpola, H. Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Yu, L.; Wang, S.; Li, X.; Fu, C.W.; Heng, P.A. Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019, Shenzhen, China, 13–17 October 2019; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2019; pp. 605–613. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Tian, J.; Zhong, C.; Shi, Z.; Zhang, Y.; He, Z. Double-uncertainty weighted method for semi-supervised learning. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020, Lima, Peru, 4–8 October 2020; Lecture Notes in Computer Science. Springer International Publishing: Cham, Switzerland, 2020; pp. 542–551. [Google Scholar] [CrossRef]
- Liu, L.; Tan, R.T. Certainty driven consistency loss on multi-teacher networks for semi-supervised learning. Pattern Recognit. 2021, 120, 108140. [Google Scholar] [CrossRef]
- Huang, W.; Chen, C.; Xiong, Z.; Zhang, Y.; Chen, X.; Sun, X.; Wu, F. Semi-supervised neuron segmentation via reinforced consistency learning. IEEE Trans. Med. Imaging 2022, 41, 3016–3028. [Google Scholar] [CrossRef]
- Lei, T.; Zhang, D.; Du, X.; Wang, X.; Wan, Y.; Nandi, A.K. Semi-Supervised Medical Image Segmentation Using Adversarial Consistency Learning and Dynamic Convolution Network. IEEE Trans. Med. Imaging 2023, 42, 1265–1277. [Google Scholar] [CrossRef]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. mixup: Beyond Empirical Risk Minimization. In Proceedings of the International Conference on Learning Representations (ICLR), Toulon, France, 24–26 April 2017. [Google Scholar]
- Basak, H.; Bhattacharya, R.; Hussain, R.; Chatterjee, A. An exceedingly simple consistency regularization method for semi-supervised medical image segmentation. In Proceedings of the 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–4. [Google Scholar] [CrossRef]
- Triguero, I.; García, S.; Herrera, F. Self-labeled techniques for semi-supervised learning: Taxonomy, software and empirical study. Knowl. Inf. Syst. 2015, 42, 245–284. [Google Scholar] [CrossRef]
- Beyer, L.; Zhai, X.; Oliver, A.; Kolesnikov, A. S4L: Self-supervised semi-supervised learning. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; IEEE: Piscataway, NJ, USA, 2019. [Google Scholar] [CrossRef]
- Bai, W.; Oktay, O.; Sinclair, M.; Suzuki, H.; Rajchl, M.; Tarroni, G.; Glocker, B.; King, A.; Matthews, P.M.; Rueckert, D. Semi-supervised learning for network-based cardiac MR image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2017, Quebec City, QC, Canada, 11–13 September 2017; Lecture Notes in Computer Science. Springer International Publishing: Cham, Switzerland, 2017; pp. 253–260. [Google Scholar] [CrossRef]
- Fan, D.P.; Zhou, T.; Ji, G.P.; Zhou, Y.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Inf-Net: Automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imaging 2020, 39, 2626–2637. [Google Scholar] [CrossRef]
- Li, Y.; Chen, J.; Xie, X.; Ma, K.; Zheng, Y. Self-loop uncertainty: A novel pseudo-label for semi-supervised medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020, Lima, Peru, 4–8 October 2020; Lecture Notes in Computer Science. Springer International Publishing: Cham, Switzerland, 2020; pp. 614–623. [Google Scholar] [CrossRef]
- Xie, Q.; Luong, M.T.; Hovy, E.; Le, Q.V. Self-training with noisy student improves ImageNet classification. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 14–19 June 2020; IEEE: Piscataway, NJ, USA, 2020. [Google Scholar] [CrossRef]
- Chaitanya, K.; Erdil, E.; Karani, N.; Konukoglu, E. Local contrastive loss with pseudo-label based self-training for semi-supervised medical image segmentation. Med. Image Anal. 2023, 87, 102792. [Google Scholar] [CrossRef]
- Basak, H.; Yin, Z. Pseudo-label guided contrastive learning for semi-supervised medical image segmentation. In Proceedings of the 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Vancouver, BC, Canada, 17–24 June 2023; IEEE: Piscataway, NJ, USA, 2023. [Google Scholar] [CrossRef]
- Payer, C.; Štern, D.; Bischof, H.; Urschler, M. Multi-label Whole Heart Segmentation Using CNNs and Anatomical Label Configurations. In Proceedings of the Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges, Quebec City, QC, Canada, 10–14 September 2018; Springer International Publishing: Cham, Switzerland, 2018; pp. 190–198. [Google Scholar] [CrossRef]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Jorge Cardoso, M. Generalised Dice overlap as a deep learning loss function for highly unbalanced segmentations. In Proceedings of the Deep Learning in Medical Image Analysis—DLMIA 2017, Quebec City, QC, Canada, 14 September 2017; Springer: Cham, Switzerland, 2017; Volume 10553, Lecture Notes in Computer Science. pp. 240–248. [Google Scholar] [CrossRef]
- Chen, X.; He, K. Exploring simple Siamese representation learning. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; IEEE: Piscataway, NJ, USA, 2021. [Google Scholar] [CrossRef]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Gonzalez Ballester, M.A.; et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Thaler, F.; Gsell, M.A.F.; Plank, G.; Urschler, M. CaRe-CNN: Cascading Refinement CNN for Myocardial Infarct Segmentation with Microvascular Obstructions. In Proceedings of the 19th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2024)—Volume 3: VISAPP, Rome, Italy, 27–29 February 2024; pp. 53–64. [Google Scholar] [CrossRef]
- Thaler, F.; Stern, D.; Plank, G.; Urschler, M. LA-CaRe-CNN: Cascading Refinement CNN for Left Atrial Scar Segmentation. In Proceedings of the MICCAI Challenge on Comprehensive Analysis and Computing of Real-World Medical Images, CARE 2024, Marrakesh, Morocco, 6–10 October 2024; Lecture Notes in Computer Science. Springer: Cham, Switzerland, 2025; Volume 15548, pp. 180–191. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on ImageNet classification. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 13–16 December 2015; pp. 1026–1034. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Payer, C.; Stern, D.; Bischof, H.; Urschler, M. Coarse to Fine Vertebrae Localization and Segmentation with SpatialConfiguration-Net and U-Net. In Proceedings of the 15th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2020)—Volume 5: VISAPP, Valletta, Malta, 27–29 February 2020; pp. 124–133. [Google Scholar] [CrossRef]
- Luo, X.; Wang, G.; Liao, W.; Chen, J.; Song, T.; Chen, Y.; Zhang, S.; Metaxas, D.N.; Zhang, S. Semi-supervised medical image segmentation via uncertainty rectified pyramid consistency. Med. Image Anal. 2022, 80, 102517. [Google Scholar] [CrossRef]


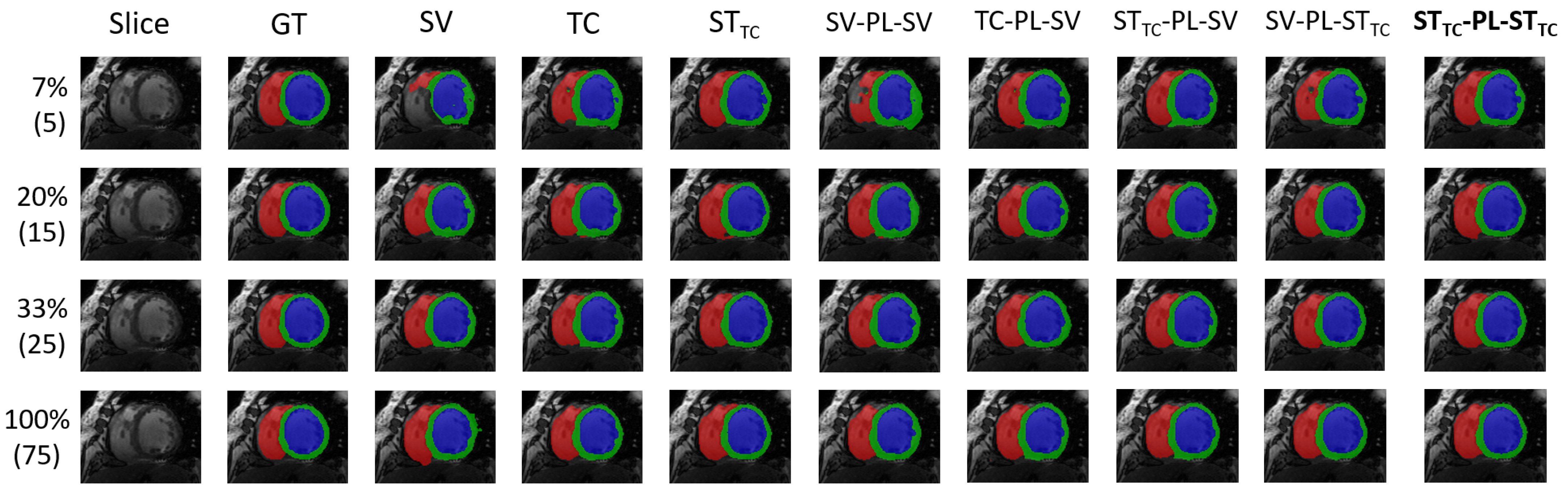
| Method | ACDC: Percentage of Patients in Labeled Set | |||||||
|---|---|---|---|---|---|---|---|---|
| 7% | 20% | 33% | 100% | |||||
| DSC | ASSD | DSC | ASSD | DSC | ASSD | DSC | ASSD | |
| (%)↑ | (mm)↓ | (%)↑ | (mm)↓ | (%)↑ | (mm)↓ | (%)↑ | (mm)↓ | |
| SV | 78.26 ± 19.22 | 2.15 ± 2.35 | 86.57 ± 14.01 | 1.19 ± 1.40 | 87.14 ± 11.22 | 0.99 ± 1.05 | 89.65 ± 7.80 | 0.92 ± 1.06 |
| TC | 84.18 ± 12.67 | 1.54 ± 1.68 | 88.01 ± 10.23 | 1.11 ± 1.33 | 88.81 ± 9.34 | 1.02 ± 1.24 | 89.73 ± 7.43 | 0.92 ± 1.03 |
| 85.19 ± 12.54 | 1.26 ± 1.39 | 88.98 ± 9.64 | 0.91 ± 1.14 | 89.69 ± 8.69 | 0.84 ± 0.96 | 90.62 ± 7.24 | 0.75± 0.86 | |
| 86.90 ± 10.86 | 1.14 ± 1.20 | 89.48± 8.88 | 0.87±0.99 | 89.88± 8.54 | 0.83±0.88 | 90.81±6.88 | 0.75±0.83 | |
| SV−PL−SV | 85.72 ± 11.24 | 1.39± 1.67 | 88.98 ± 8.65 | 1.00 ± 1.19 | 89.44 ± 8.34 | 0.92 ± 1.03 | 89.93 ± 7.29 | 0.86 ± 0.91 |
| TC−PL−SV | 86.55 ± 10.20 | 1.27 ± 1.41 | 88.89 ± 9.08 | 0.99 ± 1.09 | 89.43 ± 8.39 | 0.94 ± 1.16 | 89.90 ± 7.43 | 0.87 ± 0.94 |
| 87.57± 9.77 | 1.15 ± 1.27 | 89.31 ± 8.67 | 0.95 ± 1.12 | 89.66 ± 8.07 | 0.90 ± 1.05 | 90.12 ± 7.30 | 0.86 ± 1.00 | |
| 86.56 ± 10.62 | 1.17 ± 1.36 | 89.37 ± 8.22 | 0.91 ± 1.02 | 89.83 ± 7.38 | 0.86 ± 0.93 | 90.18 ± 6.99 | 0.81 ± 0.86 | |
| 87.10 ± 9.51 | 1.13±1.18 | 89.02 ± 8.74 | 0.95 ± 1.08 | 89.53 ± 7.76 | 0.90 ± 0.99 | 89.89 ± 7.32 | 0.84 ± 0.94 | |
| 88.43±8.90 | 1.02±1.08 | 89.82±7.96 | 0.86±0.96 | 90.04±7.43 | 0.85±0.93 | 90.49±6.82 | 0.79 ± 0.85 | |
| Method | MMWHS: Percentage of Patients in Labeled Set | |||||||
|---|---|---|---|---|---|---|---|---|
| 20% | 35% | 50% | 100% | |||||
| DSC | ASSD | DSC | ASSD | DSC | ASSD | DSC | ASSD | |
| (%)↑ | (mm)↓ | (%)↑ | (mm)↓ | (%)↑ | (mm)↓ | (%)↑ | (mm)↓ | |
| SV | 81.44 ± 6.02 | 3.12 ± 1.78 | 85.04 ± 5.01 | 2.45 ± 1.84 | 85.97 ± 5.13 | 2.06 ± 1.38 | 87.71 ± 3.69 | 1.57 ± 0.90 |
| TC | 85.72 ± 3.20 | 1.65 ± 0.53 | 87.29 ± 2.97 | 1.44 ± 0.50 | 87.63 ± 3.35 | 1.42 ± 0.55 | 88.36 ± 3.07 | 1.30 ± 0.46 |
| 84.08 ± 4.03 | 2.00 ± 0.79 | 87.17 ± 3.24 | 1.51 ± 0.59 | 88.01 ± 3.27 | 1.40 ± 0.58 | 88.84 ± 2.83 | 1.22 ± 0.42 | |
| 86.14 ± 2.92 | 1.55 ± 0.42 | 87.56 ± 2.86 | 1.40 ± 0.48 | 88.21 ± 2.96 | 1.32 ± 0.46 | 88.69 ± 2.91 | 1.24 ± 0.43 | |
| SV−PL−SV | 85.19 ± 4.75 | 2.08 ± 1.04 | 87.74 ± 3.20 | 1.41 ± 0.57 | 88.71 ± 3.45 | 1.27 ± 0.52 | 89.30 ± 2.82 | 1.18 ± 0.43 |
| TC−PL−SV | 87.58 ± 2.61 | 1.36±0.40 | 88.50 ± 2.72 | 1.27±0.45 | 89.06 ± 2.87 | 1.20 ± 0.42 | 89.25 ± 3.04 | 1.19 ± 0.46 |
| 87.68±2.72 | 1.36 ± 0.43 | 88.55±2.66 | 1.29 ± 0.47 | 89.17 ± 2.68 | 1.22 ± 0.42 | 89.42 ± 2.57 | 1.15 ± 0.38 | |
| 85.60 ± 4.73 | 2.02 ± 1.09 | 88.18 ± 3.17 | 1.32 ± 0.53 | 89.30± 2.94 | 1.15± 0.44 | 90.04±2.48 | 1.05±0.31 | |
| 85.67 ± 4.25 | 1.76 ± 0.80 | 87.54 ± 3.21 | 1.35 ± 0.49 | 89.12 ± 2.73 | 1.17 ± 0.41 | 89.76 ± 2.49 | 1.10 ± 0.35 | |
| 88.16± 3.02 | 1.27±0.41 | 89.09±2.53 | 1.18±0.40 | 89.72±2.61 | 1.07±0.36 | 90.00±2.42 | 1.06±0.31 | |
| Method | ACDC: DSC (%) ↑ | MMWHS: DSC (%) ↑ | ||||||
|---|---|---|---|---|---|---|---|---|
| Percentage of Labeled Patients | Percentage of Labeled Patients | |||||||
| 2% | 4% | 16% | 100% | 10% | 20% | 80% | 100% | |
| Supervised from [39] | 61.40 | 70.20 | 84.40 | 91.20 | 45.10 | 63.70 | 78.70 | 88.31 (*) |
| Noisy Student [38] | 63.20 | 73.70 | 83.60 | - | 59.30 | 68.50 | 78.00 | - |
| Mixup [31] | 69.50 | 78.50 | 86.30 | - | 56.10 | 69.00 | 79.60 | - |
| Self-Training [35] | 69.00 | 74.90 | 86.00 | - | 56.30 | 69.10 | 80.10 | - |
| LCLPL (inter) [39] | 75.90 | 83.10 | 88.30 | - | 57.20 | 71.90 | 81.10 | - |
| LCLPL (intra) [39] | 76.10 | 84.50 | 88.10 | - | 59.90 | 72.10 | 80.30 | - |
| (ours) | 84.53 | 87.90 | 89.66 | - | 64.63 | 86.09 | 88.64 | - |
| (ours) | 78.00 | 88.36 | 90.01 | - | 67.54 | 85.81 | 88.56 | - |
| Method | ACDC: DSC (%) ↑ | ||
|---|---|---|---|
| Percentage Labeled | |||
| 10% | 20% | 100% | |
| Supervised (ours) | 86.54 | 88.63 | 91.92 |
| Supervised from [40] | - | - | 92.30 |
| Double-UA [27] | 83.30 | - | - |
| DTC [20] | 82.70 | 86.30 | - |
| MC-Net [21] | 86.30 | 87.80 | - |
| MC-Net+ [22] | 87.10 | 88.50 | - |
| LCLPL [39] | 88.10 | 90.50 | - |
| PLGCL [40] | 89.10 | 91.20 | - |
| (ours) | 91.20 | 91.52 | - |
| (ours) | 91.57 | 91.51 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urschler, M.; Rechberger, E.; Thaler, F.; Štern, D. Cascaded Self-Supervision to Advance Cardiac MRI Segmentation in Low-Data Regimes. Bioengineering 2025, 12, 872. https://doi.org/10.3390/bioengineering12080872
Urschler M, Rechberger E, Thaler F, Štern D. Cascaded Self-Supervision to Advance Cardiac MRI Segmentation in Low-Data Regimes. Bioengineering. 2025; 12(8):872. https://doi.org/10.3390/bioengineering12080872
Chicago/Turabian StyleUrschler, Martin, Elisabeth Rechberger, Franz Thaler, and Darko Štern. 2025. "Cascaded Self-Supervision to Advance Cardiac MRI Segmentation in Low-Data Regimes" Bioengineering 12, no. 8: 872. https://doi.org/10.3390/bioengineering12080872
APA StyleUrschler, M., Rechberger, E., Thaler, F., & Štern, D. (2025). Cascaded Self-Supervision to Advance Cardiac MRI Segmentation in Low-Data Regimes. Bioengineering, 12(8), 872. https://doi.org/10.3390/bioengineering12080872






