Plug-and-Play Self-Supervised Denoising for Pulmonary Perfusion MRI
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sample and Imaging Protocol
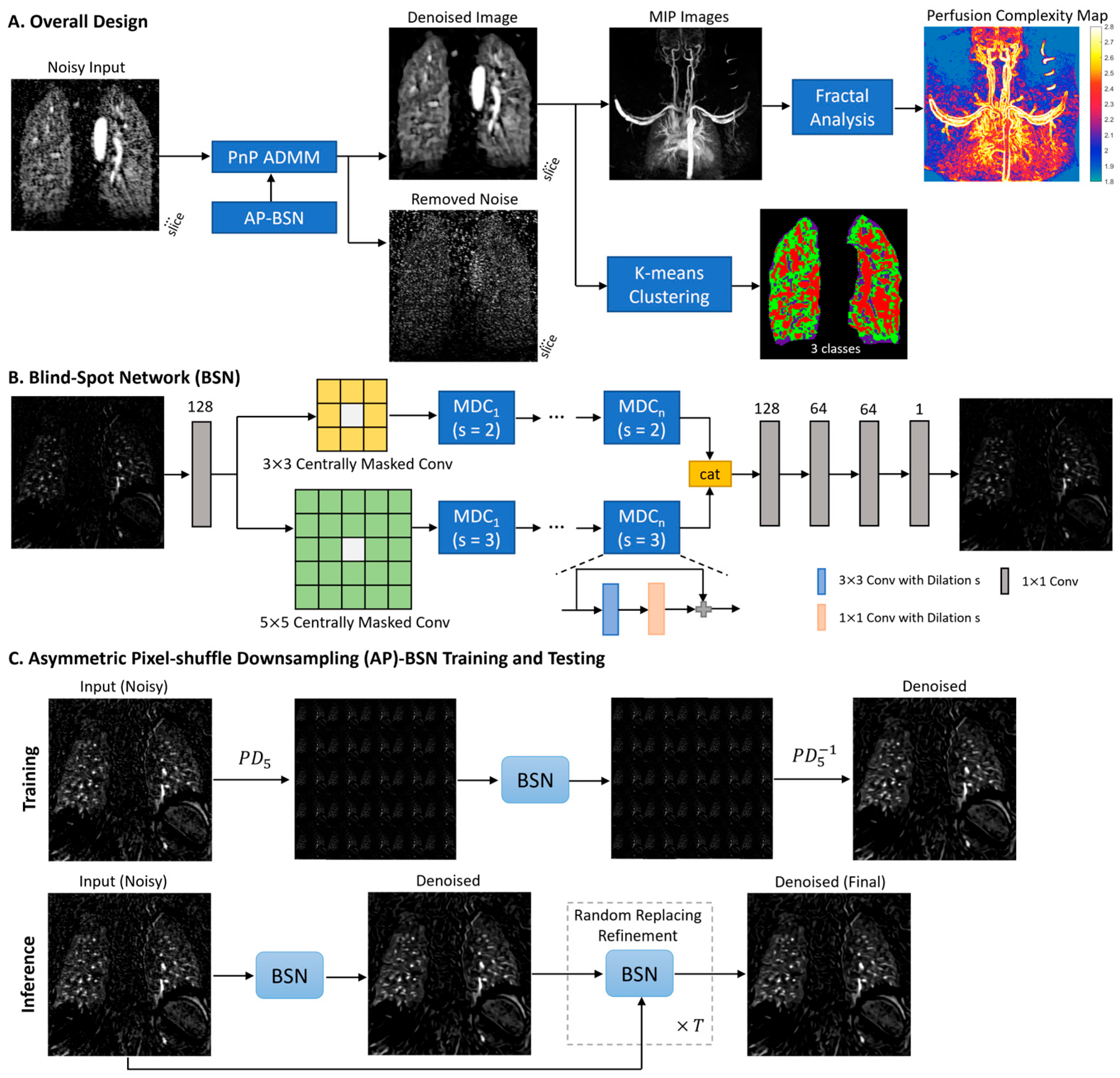
2.2. Model Design
2.3. Perfusion Complexity Analysis Using Fractal Analysis
2.4. Data Processing and Evaluation
3. Results
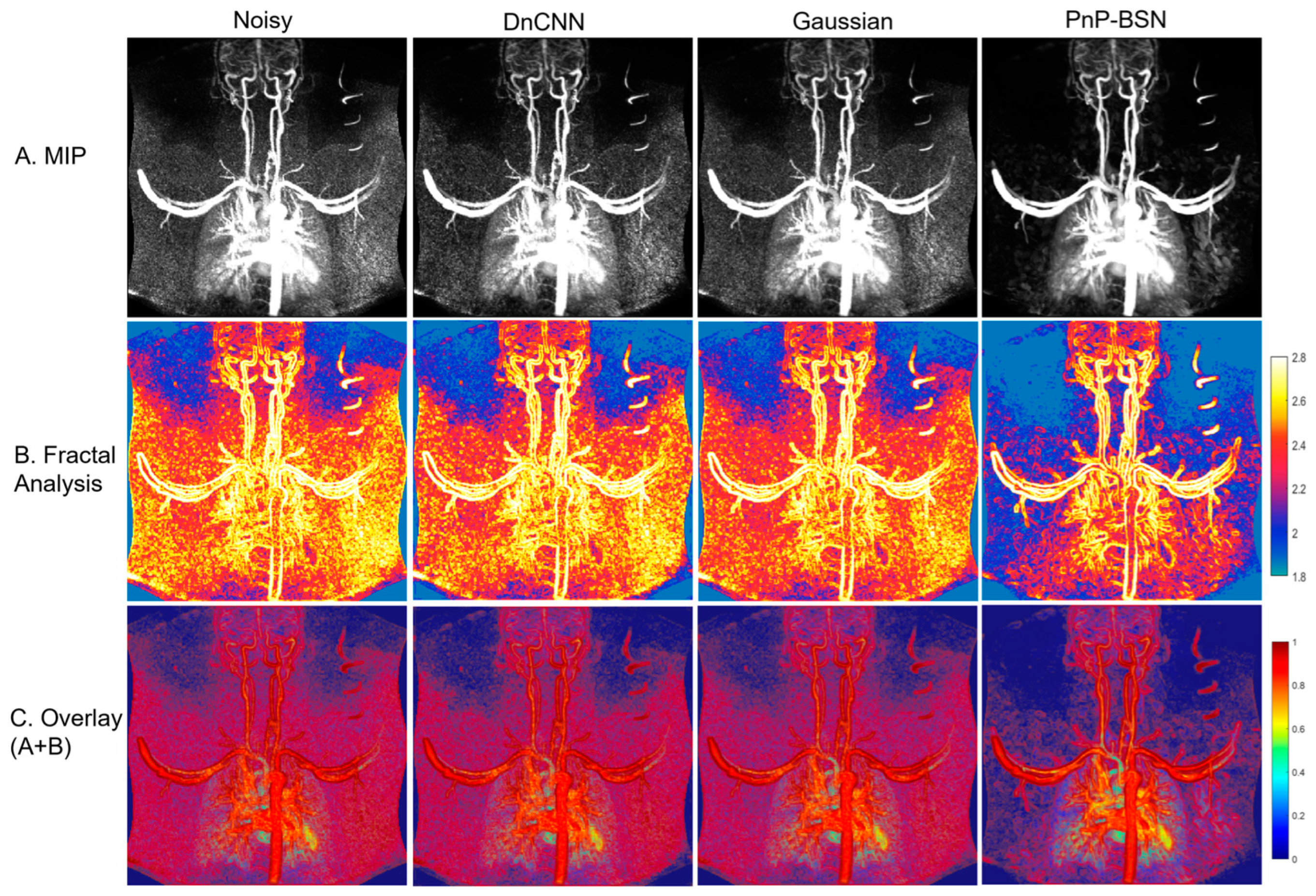
3.1. Denoising Performance and Fractal Analysis
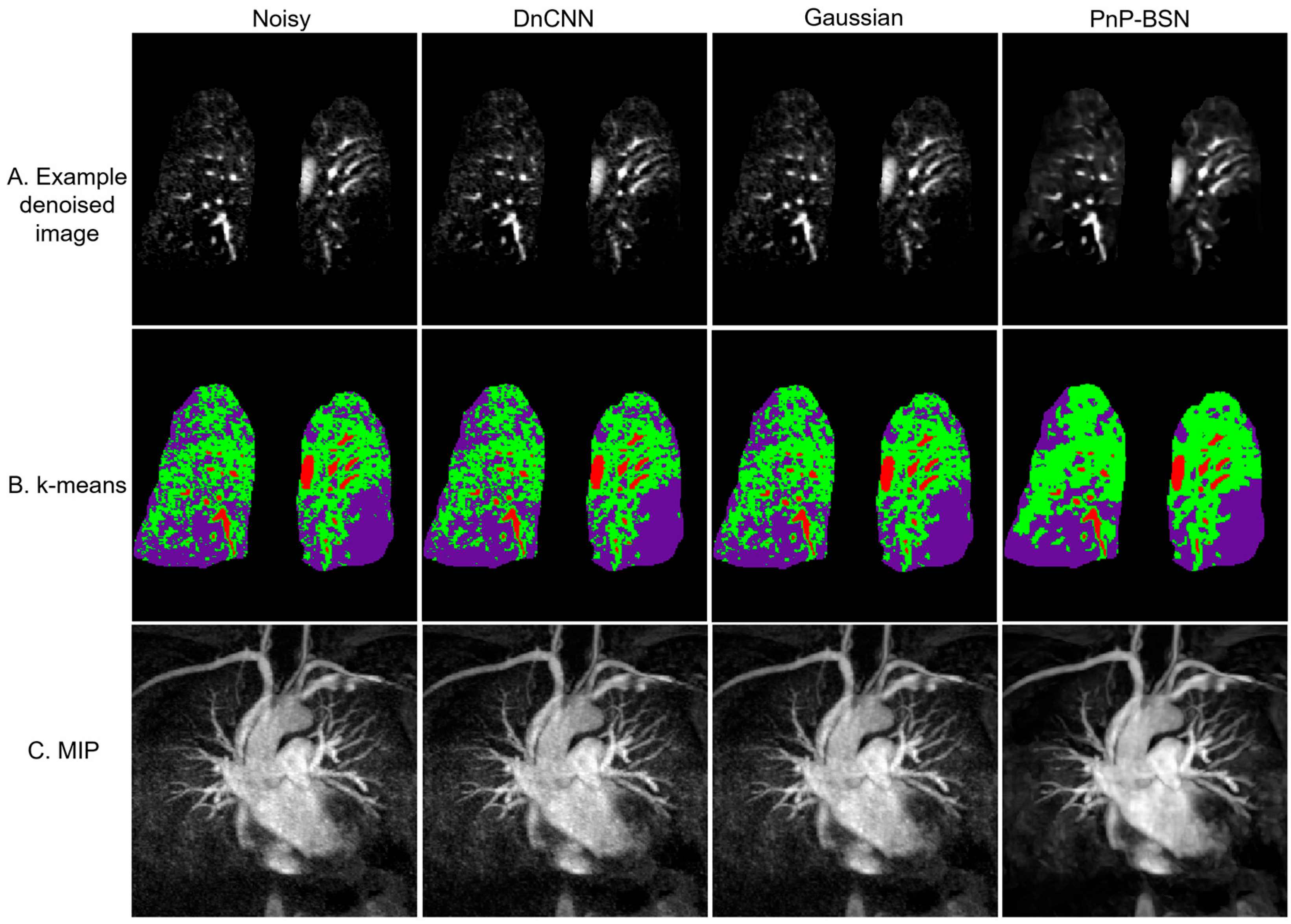
3.2. Patient Example: Pulmonary Embolism Case Study
3.3. Comparison with Reference Clinical Imaging
3.4. Evaluation of Image Quality
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D | two-dimensional |
| 3D | three-dimensional |
| ADMM | alternating direction method of multipliers |
| AP-BSN | asymmetric pixel-shuffle downsampling blind-spot network |
| BSN | blind-spot network |
| CT | computed tomography |
| DCE | dynamic contrast-enhanced |
| DL | deep learning |
| DnCNN | denoising convolutional neural network |
| FD | fractal dimension |
| MIP | maximum intensity projection |
| MRI | magnetic resonance imaging |
| PD | pixel-shuffle downsampling |
| PnP | plug-and-play |
| PnP-BSN | plug-and-play blind-spot network |
| SNR | signal-to-noise ratio |
| TWIST | time-resolved imaging with stochastic trajectories acquisition |
References
- Ley, S.; Ley-Zaporozhan, J. Pulmonary perfusion imaging using MRI: Clinical application. Insights Imaging 2012, 3, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Torres, L.A.; Lee, K.E.; Barton, G.P.; Hahn, A.D.; Sandbo, N.; Schiebler, M.L.; Fain, S.B. Dynamic contrast enhanced MRI for the evaluation of lung perfusion in idiopathic pulmonary fibrosis. Eur. Respir. J. 2022, 60, 2102058. [Google Scholar] [CrossRef] [PubMed]
- Lacharie, M.; Villa, A.; Milidonis, X.; Hasaneen, H.; Chiribiri, A.; Benedetti, G. Role of pulmonary perfusion magnetic resonance imaging for the diagnosis of pulmonary hypertension: A review. World J. Radiol. 2023, 15, 256–273. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, N.; Sato, S.; Suki, B.; Hirai, T. Fractal Analysis of Lung Structure in Chronic Obstructive Pulmonary Disease. Front. Physiol. 2020, 11, 603197. [Google Scholar] [CrossRef] [PubMed]
- Michallek, F.; Dewey, M. Fractal analysis of the ischemic transition region in chronic ischemic heart disease using magnetic resonance imaging. Eur. Radiol. 2017, 27, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Schiwek, M.; Triphan, S.M.F.; Biederer, J.; Weinheimer, O.; Eichinger, M.; Vogelmeier, C.F.; Jorres, R.A.; Kauczor, H.U.; Heussel, C.P.; Konietzke, P.; et al. Quantification of pulmonary perfusion abnormalities using DCE-MRI in COPD: Comparison with quantitative CT and pulmonary function. Eur. Radiol. 2022, 32, 1879–1890. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zuo, W.; Chen, Y.; Meng, D.; Zhang, L. Beyond a Gaussian Denoiser: Residual Learning of Deep CNN for Image Denoising. IEEE Trans. Image Process 2017, 26, 3142–3155. [Google Scholar] [CrossRef] [PubMed]
- Manjon, J.V.; Coupe, P.; Marti-Bonmati, L.; Collins, D.L.; Robles, M. Adaptive non-local means denoising of MR images with spatially varying noise levels. J. Magn. Reson. Imaging 2010, 31, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Wild, J.M.; Marshall, H.; Bock, M.; Schad, L.R.; Jakob, P.M.; Puderbach, M.; Molinari, F.; Van Beek, E.J.; Biederer, J. MRI of the lung (1/3): Methods. Insights Imaging 2012, 3, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Jiao, J.; Huang, H.; Wang, Y.; Wang, J.; Shi, H.; Huang, T. When awgn-based denoiser meets real noises. Proc. AAAI Conf. Artif. Intell. 2020, 34, 13074–13081. [Google Scholar] [CrossRef]
- Lee, W.; Son, S.; Lee, K. Self-supervised denoising for real-world images via asymmetric pd and blind-spot network. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 17704–17713. [Google Scholar]
- Ahmad, R.; Bouman, C.A.; Buzzard, G.T.; Chan, S.; Liu, S.; Reehorst, E.T.; Schniter, P. Plug-and-Play Methods for Magnetic Resonance Imaging: Using Denoisers for Image Recovery. IEEE Signal Process Mag. 2020, 37, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Senthil, K.; Altes, T.A. Phase-Specific Spatiotemporal Fractal Analysis and Radiomics of Free-breathing Stress Myocardial Perfusion. In Proceedings of the ISMRM 32nd Annual Meeting, Singapore, 4–9 May 2024. [Google Scholar]
- Peleg, S.; Naor, J.; Hartley, R.; Avnir, D. Multiple resolution texture analysis and classification. IEEE Trans. Pattern Anal. Mach. Intell. 1984, 6, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Biederer, J.; Mirsadraee, S.; Beer, M.; Molinari, F.; Hintze, C.; Bauman, G.; Both, M.; Van Beek, E.J.; Wild, J.; Puderbach, M. MRI of the lung (3/3)-current applications and future perspectives. Insights Imaging 2012, 3, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Krull, A.; Buchholz, T.-O.; Jug, F. Noise2void-learning denoising from single noisy images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2129–2137. [Google Scholar]
- Lehtinen, J.; Munkberg, J.; Hasselgren, J.; Laine, S.; Karras, T.; Aittala, M.; Aila, T. Noise2Noise: Learning image restoration without clean data. arXiv 2018, arXiv:1803.04189. [Google Scholar]
- Esteban, F.J.; Sepulcre, J.; de Mendizabal, N.V.; Goni, J.; Navas, J.; de Miras, J.R.; Bejarano, B.; Masdeu, J.C.; Villoslada, P. Fractal dimension and white matter changes in multiple sclerosis. Neuroimage 2007, 36, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Reishofer, G.; Studencnik, F.; Koschutnig, K.; Deutschmann, H.; Ahammer, H.; Wood, G. Age is reflected in the Fractal Dimensionality of MRI Diffusion Based Tractography. Sci. Rep. 2018, 8, 5431. [Google Scholar] [CrossRef] [PubMed]
- Cherstvy, A.G.; Safdari, H.; Metzler, R. Anomalous diffusion, nonergodicity, and ageing for exponentially and logarithmically time-dependent diffusivity: Striking differences for massive versus massless particles. J. Phys. D Appl. Phys. 2021, 54, 195401. [Google Scholar] [CrossRef]
- Dabov, K.; Foi, A.; Katkovnik, V.; Egiazarian, K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans. Image Process 2007, 16, 2080–2095. [Google Scholar] [CrossRef] [PubMed]
- Ulyanov, D.; Vedaldi, A.; Lempitsky, V. Deep Image Prior. Int. J. Comput. Vis. 2020, 128, 1867–1888. [Google Scholar] [CrossRef]
- Nazir, N.; Sarwar, A.; Saini, B.S. Recent developments in denoising medical images using deep learning: An overview of models, techniques, and challenges. Micron 2024, 180, 103615. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, C.; Wang, Y.; Thornburgh, C.; Lin, A.-L.; Qing, K.; Mugler, J.P., III; Altes, T.A. Plug-and-Play Self-Supervised Denoising for Pulmonary Perfusion MRI. Bioengineering 2025, 12, 724. https://doi.org/10.3390/bioengineering12070724
Sun C, Wang Y, Thornburgh C, Lin A-L, Qing K, Mugler JP III, Altes TA. Plug-and-Play Self-Supervised Denoising for Pulmonary Perfusion MRI. Bioengineering. 2025; 12(7):724. https://doi.org/10.3390/bioengineering12070724
Chicago/Turabian StyleSun, Changyu, Yu Wang, Cody Thornburgh, Ai-Ling Lin, Kun Qing, John P. Mugler, III, and Talissa A. Altes. 2025. "Plug-and-Play Self-Supervised Denoising for Pulmonary Perfusion MRI" Bioengineering 12, no. 7: 724. https://doi.org/10.3390/bioengineering12070724
APA StyleSun, C., Wang, Y., Thornburgh, C., Lin, A.-L., Qing, K., Mugler, J. P., III, & Altes, T. A. (2025). Plug-and-Play Self-Supervised Denoising for Pulmonary Perfusion MRI. Bioengineering, 12(7), 724. https://doi.org/10.3390/bioengineering12070724





