Novel Snapshot-Based Hyperspectral Conversion for Dermatological Lesion Detection via YOLO Object Detection Models
Abstract
1. Introduction
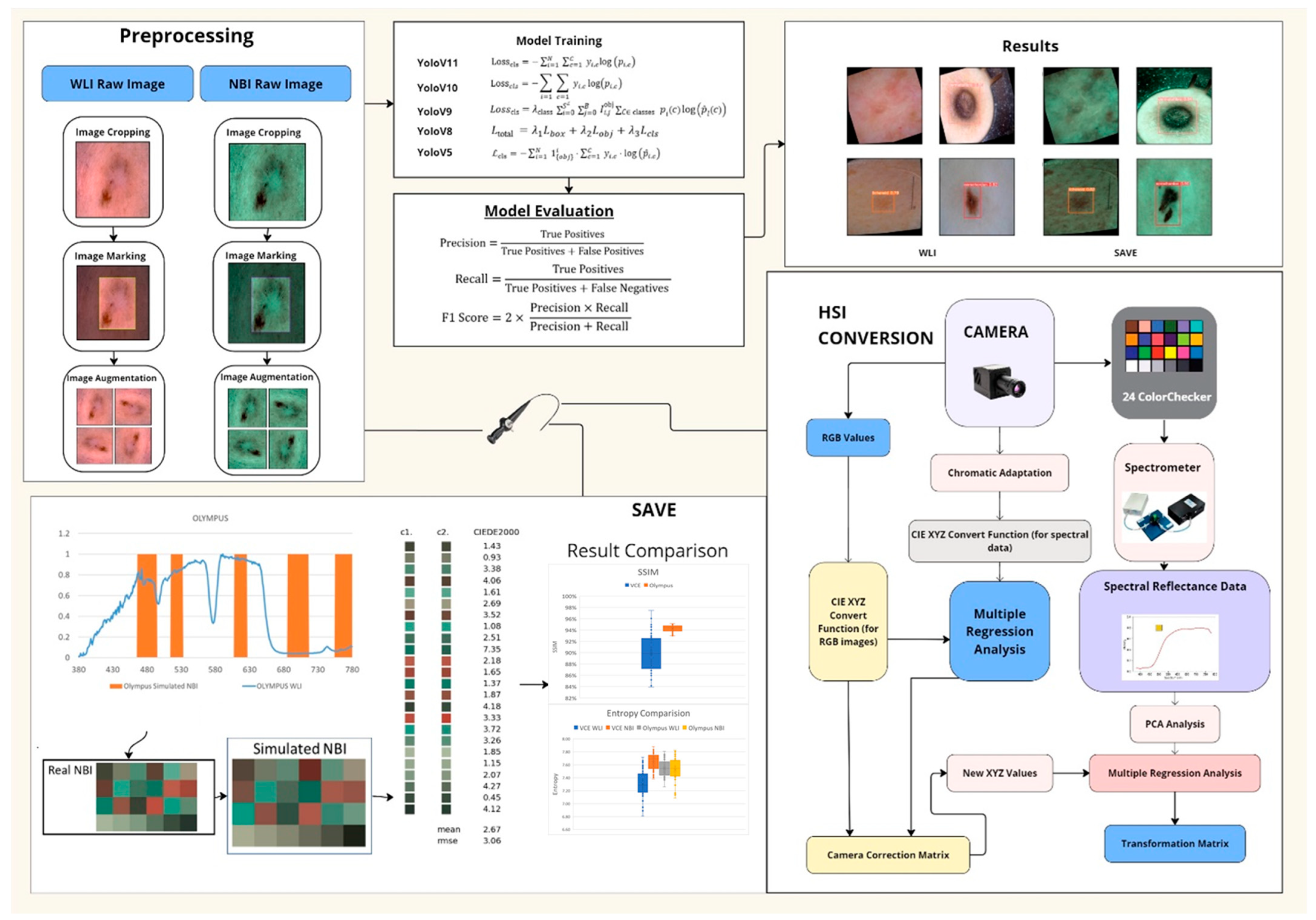
- SAVE: a snapshot-based technique that transforms any RGB image into a narrowband representation aligned with hemoglobin absorption peaks.
- Integration of SAVE with YOLO: adaptation of YOLOv5–YOLOv11 by incorporating SAVE outputs for real-time skin lesion detection.
- Comprehensive Evaluation: an exhaustive comparison of five YOLO variants across WLI and SAVE modalities, reporting class-specific precision, recall, F1 score, and statistical significance.
2. Materials and Methods
2.1. Dataset
- Acrochordon: 577 images;
- Dermatofibroma: 821 images;
- Lichenoid lesions: 805 images.
2.2. SAVE
2.3. ML Algorithms
2.3.1. YoloV11
2.3.2. YoloV10
2.3.3. YoloV9
2.3.4. YOLOv8
2.3.5. YOLOV5
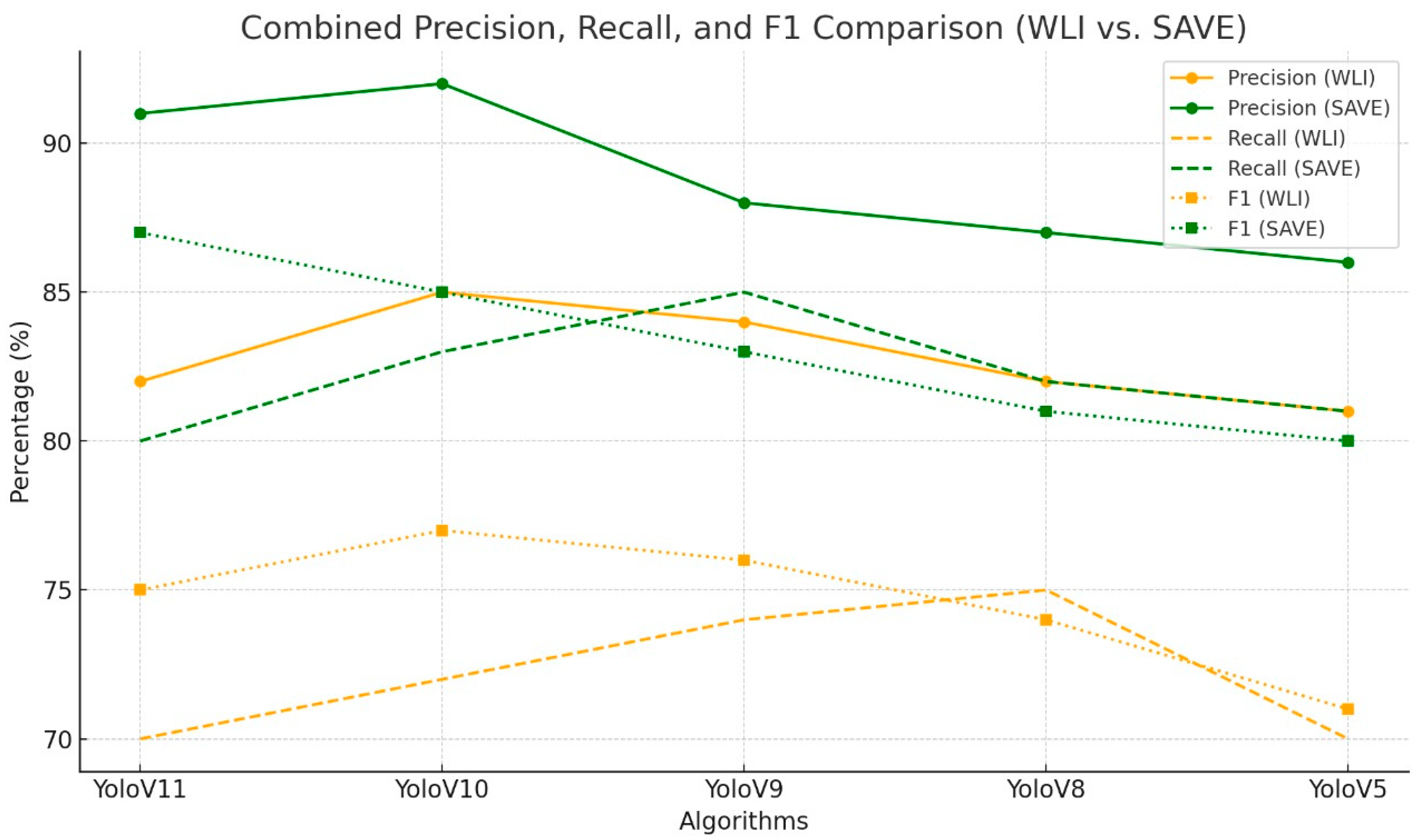
3. Results
4. Discussion
4.1. Uncertainty Analysis
4.2. Limitations and Computational Cost
4.3. Comparison to Alternative Spectral Modalities and Clinical Translation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Rundle, C.W.; Militello, M.; Barber, C.; Presley, C.L.; Rietcheck, H.R.; Dellavalle, R.P. Epidemiologic Burden of Skin Cancer in the Us and Worldwide. Curr. Dermatol. Rep. 2020, 9, 309–322. [Google Scholar] [CrossRef]
- LeBoit, P.E. Pathology and Genetics of Tumours of the Skin: Who Classification of Tumours; IARC: Lyon, France, 2006; Volume 6. [Google Scholar]
- Berklite, L.; Ranganathan, S.; John, I.; Picarsic, J.; Santoro, L.; Alaggio, R. Fibrous Histiocytoma/Dermatofibroma in Children: The Same as Adults? Hum. Pathol. 2020, 99, 107–115. [Google Scholar] [CrossRef]
- Gaufin, M.; Michaelis, T.; Duffy, K. Cellular Dermatofibroma: Clinicopathologic Review of 218 Cases of Cellular Dermatofibroma to Determine the Clinical Recurrence Rate. Dermatol. Surg. 2019, 45, 1359–1364. [Google Scholar] [CrossRef]
- Kim, J.M.; Cho, H.J.; Moon, S.-H. Rare Experience of Keloidal Dermatofibroma of Forehead. Arch. Craniofacial Surg. 2018, 19, 72. [Google Scholar] [CrossRef]
- Kadakia, S.; Chernobilsky, B.; Iacob, C. Dermatofibroma of the Auricle. J. Drugs Dermatol. 2016, 15, 1270–1272. [Google Scholar]
- Lehmer, L.M.; Ragsdale, B.D. Digital Dermatofibromas-Common Lesion, Uncommon Location: A Series of 26 Cases and Review of the Literature. Dermatol. Online J. 2011, 17, 2. [Google Scholar] [CrossRef]
- Orzan, O.A.; Dorobanțu, A.M.; Gurău, C.D.; Ali, S.; Mihai, M.M.; Popa, L.G.; Giurcăneanu, C.; Tudose, I.; Bălăceanu, B. Challenging Patterns of Atypical Dermatofibromas and Promising Diagnostic Tools for Differential Diagnosis of Malignant Lesions. Diagnostics 2023, 13, 671. [Google Scholar] [CrossRef]
- Cazzato, G.; Colagrande, A.; Cimmino, A.; Marrone, M.; Stellacci, A.; Arezzo, F.; Lettini, T.; Resta, L.; Ingravallo, G. Granular Cell Dermatofibroma: When Morphology Still Matters. Dermatopathology 2021, 8, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Tilly, J.J.; Drolet, B.A.; Esterly, N.B. Lichenoid Eruptions in Children. J. Am. Acad. Dermatol. 2004, 51, 606–624. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Jindal, A.; Patel, N.M. Acrochordons as a Cutaneous Sign of Metabolic Syndrome: A Case—Control Study. Ann. Med. Health Sci. Res. 2014, 4, 202–205. [Google Scholar] [PubMed]
- Faiz, S.M.; Bhargava, A.; Srivastava, S.; Singh, H.; Goswami, D.; Bhat, P. Giant Acrochordon of Left Side Neck, a Rare Case Finding. Era’s J. Med. Res. 2020, 7, 260–261. [Google Scholar] [CrossRef]
- Mohamed, T. A Comparative Study of Cutaneous Manifestations in Obese Patients and Non-Obese Controls at Vims, Ballari. Ph.D. Thesis, Rajiv Gandhi University of Health Sciences, Bengaluru, India, 2018. [Google Scholar]
- Shrestha, P.; Poudyal, Y.; Rajbhandari, S.L. Acrochordons and Diabetes Mellitus: A Case Control Study. Nepal J. Dermatol. Venereol. Leprol. 2016, 13, 32–37. [Google Scholar] [CrossRef]
- Sergi, C.M.; Consolato, M. Pathology of Childhood Sergi, and Adolescence: An Illustrated Guide. Soft Tissue; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1003–1094. [Google Scholar]
- Backer, E.L. Skin Tumors. In Family Medicine: Principles and Practice; Springer: Berlin/Heidelberg, Germany, 2022; pp. 1681–1705. [Google Scholar]
- Singh, B.S.; Tripathy, T.; Kar, B.R. Association of Skin Tag with Metabolic Syndrome and Its Components: A Case–Control Study from Eastern India. Indian Dermatol. Online J. 2019, 10, 284–287. [Google Scholar] [CrossRef]
- Sherin, N.; Khader, A.; Binitha, M.P.; George, B. Acrochordon as a Marker of Metabolic Syndrome—A Cross-Sectional Study from South India. J. Ski. Sex. Transm. Dis. 2023, 5, 40–46. [Google Scholar] [CrossRef]
- Johansen, T.H.; Møllersen, K.; Ortega, S.; Fabelo, H.; Garcia, A.; Callico, G.M.; Godtliebsen, F. Recent Advances in Hyperspectral Imaging for Melanoma Detection. Wiley Interdiscip. Rev. Comput. Stat. 2020, 12, e1465. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Nguyen, H.-T.; Lin, T.-L.; Saenprasarn, P.; Liu, P.-H.; Wang, H.-C. Identification of Skin Lesions by Snapshot Hyperspectral Imaging. Cancers 2024, 16, 217. [Google Scholar] [CrossRef]
- Leon, R.; Martinez-Vega, B.; Fabelo, H.; Ortega, S.; Melian, V.; Castaño, I.; Carretero, G.; Almeida, P.; Garcia, A.; Quevedo, E.; et al. Non-Invasive Skin Cancer Diagnosis Using Hyperspectral Imaging for In-Situ Clinical Support. J. Clin. Med. 2020, 9, 1662. [Google Scholar] [CrossRef]
- Pathan, S.; Prabhu, K.G.; Siddalingaswamy, P.C. Techniques and Algorithms for Computer Aided Diagnosis of Pigmented Skin Lesions—A Review. Biomed. Signal Process. Control 2018, 39, 237–262. [Google Scholar] [CrossRef]
- Yoon, J. Hyperspectral Imaging for Clinical Applications. BioChip J. 2022, 16, 1–12. [Google Scholar] [CrossRef]
- Adesokan, M.; Alamu, E.O.; Otegbayo, B.; Maziya-Dixon, B. A Review of the Use of Near-Infrared Hyperspectral Imaging (NIR-HSI) Techniques for the Non-Destructive Quality Assessment of Root and Tuber Crops. Appl. Sci. 2023, 13, 5226. [Google Scholar] [CrossRef]
- Guilloteau, C.; Oberlin, T.; Berne, O.; Dobigeon, N. Hyperspectral and Multispectral Image Fusion Under Spectrally Varying Spatial Blurs—Application to High Dimensional Infrared Astronomical Imaging. IEEE Trans. Comput. Imaging 2020, 6, 1362–1374. [Google Scholar] [CrossRef]
- Guerri, M.F.; Distante, C.; Spagnolo, P.; Bougourzi, F.; Taleb-Ahmed, A. Deep Learning Techniques for Hyperspectral Image Analysis in Agriculture: A Review. ISPRS Open J. Photogramm. Remote Sens. 2024, 12, 100062. [Google Scholar] [CrossRef]
- Rehman, A.; Qureshi, S.A. A Review of the Medical Hyperspectral Imaging Systems and Unmixing Algorithms’ in Biological Tissues. Photodiagnosis Photodyn. Ther. 2021, 33, 102165. [Google Scholar] [CrossRef]
- Khan, U.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V.K. Trends in Deep Learning for Medical Hyperspectral Image Analysis. IEEE Access 2021, 9, 79534–79548. [Google Scholar] [CrossRef]
- Gutiérrez-Gutiérrez, J.A.; Pardo, A.; Real, E.; López-Higuera, J.M.; Conde, O.M. Custom Scanning Hyperspectral Imaging System for Biomedical Applications: Modeling, Benchmarking, and Specifications. Sensors 2019, 19, 1692. [Google Scholar] [CrossRef]
- Thiele, S.T.; Bnoulkacem, Z.; Lorenz, S.; Bordenave, A.; Menegoni, N.; Madriz, Y.; Dujoncquoy, E.; Gloaguen, R.; Kenter, J. Mineralogical Mapping with Accurately Corrected Shortwave Infrared Hyperspectral Data Acquired Obliquely from UAVs. Remote Sens. 2021, 14, 5. [Google Scholar] [CrossRef]
- Cucci, C.; Picollo, M.; Chiarantini, L.; Uda, G.; Fiori, L.; De Nigris, B.; Osanna, M. Remote-Sensing Hyperspectral Imaging for Applications in Archaeological Areas: Non-Invasive Investigations on Wall Paintings and on Mural Inscriptions in the Pompeii site. Microchem. J. 2020, 158, 105082. [Google Scholar] [CrossRef]
- Aviara, N.A.; Liberty, J.T.; Olatunbosun, O.S.; Shoyombo, H.A.; Oyeniyi, S.K. Potential Application of Hyperspectral Imaging in Food Grain Quality Inspection, Evaluation and Control During Bulk Storage. J. Agric. Food Res. 2022, 8, 100288. [Google Scholar] [CrossRef]
- Faltynkova, A.; Johnsen, G.; Wagner, M. Hyperspectral Imaging as an Emerging Tool to Analyze Microplastics: A Systematic Review and Recommendations for Future Development. Microplastics Nanoplastics 2021, 1, 13. [Google Scholar] [CrossRef]
- He, Z.; Wang, P.; Liang, Y.; Fu, Z.; Ye, X.; Liu, A. Clinically Available Optical Imaging Technologies in Endoscopic Lesion Detection: Current Status and Future Perspective. J. Healthc. Eng. 2021, 2021, 7594513. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Liu, Z.; Sun, H.; Jiao, F.; Zhang, B.; Chen, J. A Narrative Review: Narrow-Band Imaging Endoscopic Classifications. Quant. Imaging Med. Surg. 2023, 13, 1138–1163. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.; Kong, L.; Zhao, Y. Contrast-Enhancing Snapshot Narrow-Band Imaging Method for Real-Time Computer-Aided Cervical Cancer Screening. J. Digit. Imaging 2019, 33, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Gono, K. Principles and History of Nbi. In Atlas of Endoscopy with Narrow Band Imaging; Springer: Berlin/Heidelberg, Germany, 2015; pp. 3–10. [Google Scholar]
- Goyal, M.; Knackstedt, T.; Yan, S.; Hassanpour, S. Artificial Intelligence-Based Image Classification Methods for Diagnosis of Skin Cancer: Challenges and Opportunities. Comput. Biol. Med. 2020, 127, 104065. [Google Scholar] [CrossRef]
- Tejada-Casado, M.; Herrera, L.J.; Carrillo-Perez, F.; Ruiz-López, J.; Ghinea, R.I.; Pérez, M.M. Exploring the CIEDE2000 Thresholds for Lightness, Chroma, and Hue Differences in Dentistry. J. Dent. 2024, 150, 105327. [Google Scholar] [CrossRef]
- Ultralytics. Ultralytics Yolo11; Ultralytics: Frederick, MD, USA, 2024. [Google Scholar]
- Wang, A.; Chen, H.; Liu, L.; Chen, K.; Lin, Z.; Han, J.; Ding, G. Yolov10: Real-Time End-to-End Object Detection. arXiv 2024, arXiv:2405.14458. [Google Scholar]
- Wang, C.-Y.; Yeh, I.-H.; Liao, H.-Y.M. Yolov9: Learning What You Want to Learn Using Programmable Gradient Information. arXiv 2024, arXiv:2402.13616. [Google Scholar]
- Yao, Q.; Zhuang, D.; Feng, Y.; Wang, Y.; Liu, J. Accurate Detection of Brain Tumor Lesions from Medical Images Based on Improved Yolov8 Algorithm. IEEE Access 2024, 12, 144260–144279. [Google Scholar] [CrossRef]
- Bashir, S.; Qureshi, R.; Shah, A.; Fan, X.; Alam, T. Yolov5-M: A Deep Neural Network for Medical Object Detection in Real-Time. In Proceedings of the 2023 IEEE Symposium on Industrial Electronics & Applications (ISIEA), Kuala Lumpur, Malaysia, 15–16 July 2023. [Google Scholar]
- Camastra, F.; Vinciarelli, A. Machine Learning for Audio, Image and Video Analysis: Theory and Applications; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Ravikumar, A.; Sriraman, H.; Saketh, P.M.S.; Lokesh, S.; Karanam, A. Effect of Neural Network Structure in Accelerating Performance and Accuracy of a Convolutional Neural Network with Gpu/Tpu for Image Analytics. PeerJ Comput. Sci. 2022, 8, e909. [Google Scholar] [CrossRef]
- Mahmud, T.; Barua, K.; Barua, A.; Das, S.; Basnin, N.; Hossain, M.S.; Andersson, K.; Kaiser, M.S.; Sharmen, N. Exploring Deep Transfer Learning Ensemble for Improved Diagnosis and Classification of Alzheimer’s Disease. In Proceedings of the 16th International Conference on Brain Informatics (BI2023), Hoboken, NJ, USA, 1–3 August 2023. [Google Scholar]
- Mayya, V.; Sowmya Kamath, S.; Kulkarni, U.; Surya, D.K.; Acharya, U.R. An Empirical Study of Preprocessing Techniques with Convolutional Neural Networks for Accurate Detection of Chronic Ocular Diseases Using Fundus Images. Appl. Intell. 2022, 53, 1548–1566. [Google Scholar] [CrossRef]
- Jiang, X.; Hu, Z.; Wang, S.; Zhang, Y. Deep Learning for Medical Image-Based Cancer Diagnosis. Cancers 2023, 15, 3608. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Hao, Y.; Liu, Q.; Liu, Y.; Zhu, M.; Xiao, L. Deep Learning for Hyperspectral Image Classification: A Critical Evaluation via Mutation Testing. Remote Sens. 2024, 16, 4695. [Google Scholar] [CrossRef]
- Roy, S.K.; Jamali, A.; Chanussot, J.; Ghamisi, P.; Ghaderpour, E.; Shahabi, H. SimPoolFormer: A Two-Stream Vision Transformer for Hyperspectral Image Classification. Remote Sens. Appl. Soc. Environ. 2025, 37, 101478. [Google Scholar] [CrossRef]


| Lesion Type | Total Images | Training (70%) | Validation (20%) | Test (10%) |
|---|---|---|---|---|
| Acrochordon | 577 | 404 (70%) | 116 (20%) | 57 (10%) |
| Dermatofibroma | 821 | 575 (70%) | 164 (20%) | 82 (10%) |
| Lichenoid | 805 | 563 (70%) | 161 (20%) | 81 (10%) |
| Total | 2203 | 1542 (70%) | 441 (20%) | 220 (10%) |
| Algorithm | Imaging Modality | Class | Precision in % | Recall in % | F1 in % |
|---|---|---|---|---|---|
| YoloV11 | WLI | All | 71 | 62.9 | 66.71 |
| Acrochordon | 62.8 | 40 | 48.87 | ||
| Dermatofibroma | 82.3 | 89.7 | 85.84 | ||
| Lichenoid | 67.9 | 59 | 63.14 | ||
| SAVE | All | 74.3 | 54.2 | 62.68 | |
| Acrochordon | 70.9 | 39.8 | 50.98 | ||
| Dermatofibroma | 86.1 | 76.3 | 80.9 | ||
| Lichenoid | 65.8 | 46.5 | 54.49 | ||
| YoloV10 | WLI | All | 79.2 | 58.6 | 67.36 |
| Acrochordon | 69.3 | 34.2 | 45.8 | ||
| Dermatofibroma | 90.9 | 83.3 | 86.93 | ||
| Lichenoid | 77.4 | 58.2 | 66.44 | ||
| SAVE | All | 79.2 | 58.3 | 67.16 | |
| Acrochordon | 72.3 | 39.2 | 50.84 | ||
| Dermatofibroma | 90.1 | 81.4 | 85.53 | ||
| Lichenoid | 75.1 | 54.4 | 63.1 | ||
| YoloV9 | WLI | All | 75.8 | 67 | 71.13 |
| Acrochordon | 62.8 | 48.8 | 54.92 | ||
| Dermatofibroma | 87.9 | 90.4 | 89.13 | ||
| Lichenoid | 76.8 | 61.9 | 68.55 | ||
| SAVE | All | 86.4 | 56.1 | 68.03 | |
| Acrochordon | 83.6 | 36.7 | 51.01 | ||
| Dermatofibroma | 87.8 | 79.2 | 83.28 | ||
| Lichenoid | 87.9 | 52.6 | 65.82 | ||
| YoloV8 | WLI | All | 78.2 | 60.8 | 68.41 |
| Acrochordon | 64.4 | 32.8 | 43.46 | ||
| Dermatofibroma | 89.5 | 87.5 | 88.49 | ||
| Lichenoid | 80.8 | 61.9 | 70.1 | ||
| SAVE | All | 81.2 | 57.9 | 67.6 | |
| Acrochordon | 74.8 | 36.7 | 49.24 | ||
| Dermatofibroma | 88.2 | 79.9 | 83.85 | ||
| Lichenoid | 88.5 | 57.2 | 69.49 | ||
| YoloV5 | WLI | All | 70.7 | 62.8 | 66.52 |
| Acrochordon | 57.8 | 40.5 | 47.63 | ||
| Dermatofibroma | 85.1 | 84.3 | 84.7 | ||
| Lichenoid | 69.1 | 63.4 | 66.13 | ||
| SAVE | All | 71.6 | 59.3 | 64.87 | |
| Acrochordon | 64.4 | 39.2 | 48.74 | ||
| Dermatofibroma | 81.8 | 81.5 | 81.65 | ||
| Lichenoid | 68.5 | 57.2 | 62.34 |
| Algorithm | Imaging Modality | Class | Precision in % | Recall in % | F1 in % |
|---|---|---|---|---|---|
| YoloV11 | WLI | All | 74.5 | 50.7 | 57.67 |
| Acrochordon | 72.6 | 37.9 | 49.91 | ||
| Dermatofibroma | 83.7 | 66.7 | 73.77 | ||
| Lichenoid | 67.2 | 47.4 | 55.71 | ||
| SAVE | All | 77.5 | 57.9 | 65.73 | |
| Acrochordon | 58.1 | 43.3 | 49.69 | ||
| Dermatofibroma | 93.4 | 82.7 | 87.04 | ||
| Lichenoid | 80.9 | 47.6 | 60.17 | ||
| YoloV10 | WLI | All | 70 | 52.9 | 60.3 |
| Acrochordon | 56.4 | 42.9 | 48.7 | ||
| Dermatofibroma | 85.4 | 68.5 | 76 | ||
| Lichenoid | 68 | 47.4 | 55.9 | ||
| SAVE | All | 85 | 60.3 | 70.6 | |
| Acrochordon | 75.7 | 49.7 | 60 | ||
| Dermatofibroma | 92 | 79.3 | 85.2 | ||
| Lichenoid | 87.3 | 52.1 | 65.3 | ||
| YoloV9 | WLI | All | 81.4 | 56.2 | 66.5 |
| Acrochordon | 85 | 47.6 | 61 | ||
| Dermatofibroma | 85 | 70.8 | 77.3 | ||
| Lichenoid | 74.1 | 50.2 | 59.9 | ||
| SAVE | All | 87.3 | 60.6 | 71.5 | |
| Acrochordon | 79.1 | 50 | 61.3 | ||
| Dermatofibroma | 92.3 | 81.7 | 86.7 | ||
| Lichenoid | 90.3 | 50 | 64.4 | ||
| YoloV8 | WLI | All | 80.4 | 55.9 | 65.9 |
| Acrochordon | 78.8 | 45.2 | 57.4 | ||
| Dermatofibroma | 87.5 | 71.9 | 78.9 | ||
| Lichenoid | 74.9 | 50.5 | 60.3 | ||
| SAVE | All | 83.3 | 64.3 | 72.6 | |
| Acrochordon | 80.5 | 56.8 | 66.6 | ||
| Dermatofibroma | 88.4 | 84.1 | 86.2 | ||
| Lichenoid | 80.9 | 52.1 | 63.4 | ||
| YoloV5 | WLI | All | 68 | 49.5 | 57.3 |
| Acrochordon | 60.8 | 33.3 | 43 | ||
| Dermatofibroma | 80.9 | 67.4 | 73.5 | ||
| Lichenoid | 62.3 | 47.8 | 54.1 | ||
| SAVE | All | 74.7 | 61.4 | 67.4 | |
| Acrochordon | 63.2 | 52.3 | 57.2 | ||
| Dermatofibroma | 87.5 | 82.9 | 85.1 | ||
| Lichenoid | 73.4 | 49 | 58.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, N.-C.; Mukundan, A.; Karmakar, R.; Syna, S.; Chang, W.-Y.; Wang, H.-C. Novel Snapshot-Based Hyperspectral Conversion for Dermatological Lesion Detection via YOLO Object Detection Models. Bioengineering 2025, 12, 714. https://doi.org/10.3390/bioengineering12070714
Huang N-C, Mukundan A, Karmakar R, Syna S, Chang W-Y, Wang H-C. Novel Snapshot-Based Hyperspectral Conversion for Dermatological Lesion Detection via YOLO Object Detection Models. Bioengineering. 2025; 12(7):714. https://doi.org/10.3390/bioengineering12070714
Chicago/Turabian StyleHuang, Nan-Chieh, Arvind Mukundan, Riya Karmakar, Syna Syna, Wen-Yen Chang, and Hsiang-Chen Wang. 2025. "Novel Snapshot-Based Hyperspectral Conversion for Dermatological Lesion Detection via YOLO Object Detection Models" Bioengineering 12, no. 7: 714. https://doi.org/10.3390/bioengineering12070714
APA StyleHuang, N.-C., Mukundan, A., Karmakar, R., Syna, S., Chang, W.-Y., & Wang, H.-C. (2025). Novel Snapshot-Based Hyperspectral Conversion for Dermatological Lesion Detection via YOLO Object Detection Models. Bioengineering, 12(7), 714. https://doi.org/10.3390/bioengineering12070714







