ResST-SEUNet++: Deep Model for Accurate Segmentation of Left Ventricle and Myocardium in Magnetic Resonance Imaging (MRI) Images
Abstract
1. Introduction
- A comprehensive end-to-end approach for the segmentation of left ventricular and myocardium structures is developed and implemented. The present work introduces a novel approach that incorporates a dual-stage image pre-processing pipeline, combining CLAHE and bilateral filtering. By taking this pre-processing stage, the quality of the input data is greatly enhanced, thereby leading to increased precision and reliability in future segmentations.
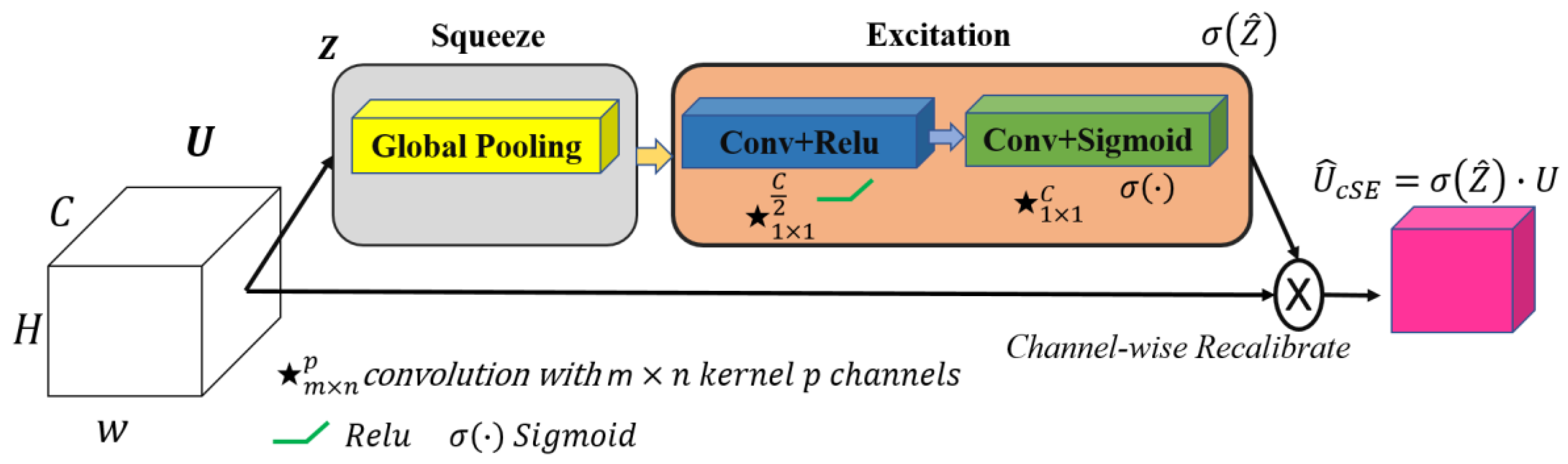
- Our work introduces ResST-SEUNet++, a DL model for segmenting cardiac and medical images, which incorporates ResNetSt (split-attention networks) as an encoder and an SE attention module in the Unet++’s decoder. To our knowledge, no prior work has employed this combination for cardiac segmentation. ResST-SEUNet++ was subjected to a thorough performance assessment and comparison to other advanced models like PAN, ResUnet, ResUnet++, Manet, Linknet, and PSPnet. Our findings demonstrated that the suggested model had a greatly superior performance when compared to existing methods in crucial rates like the IoU, Recall, Dice, accuracy, and F1-score.
- The verification of the proposed model’s exceptional precision and reliability utilizing experimental data demonstrated that the reported model exhibits an outstanding performance in cardiac LV and Myo structural segmentation tasks, as proven by the high metric values obtained from the experimental findings. These findings validate the theoretical relevance and practical applicability of the proposed methodologies in real medical situations.
- This study verified the generalization capabilities of the proposed model on five additional databases, namely the ACDC 2017 [11], Wound Closure Database [17], Kvasir-SEG [18], CVC-ClinicDB [19], and ISIC 2017 [20]. This affirmed the model’s adaptability and efficacy in a variety of datasets, illustrating its capacity to maintain a high segmentation accuracy under varying conditions.
- We incorporated an explainability scheme into the ResST-SEUNet++ model. This feature creates visual maps to show which parts of the image the model focuses on for each class. This makes the model easier to understand and interpret, helps spot mistakes, and makes its decisions more transparent and reliable.
2. Related Works
2.1. Left Ventricular and Myocardium Segmentation
2.2. Deep Learning Models for Segmentation
2.2.1. U-Net
2.2.2. LinkNet
2.2.3. PSPNet
2.2.4. PAN
2.2.5. Manet
2.2.6. ResUNet
2.2.7. ResUNet++
2.2.8. Unet++
3. Materials and Methodology
3.1. Datasets Description
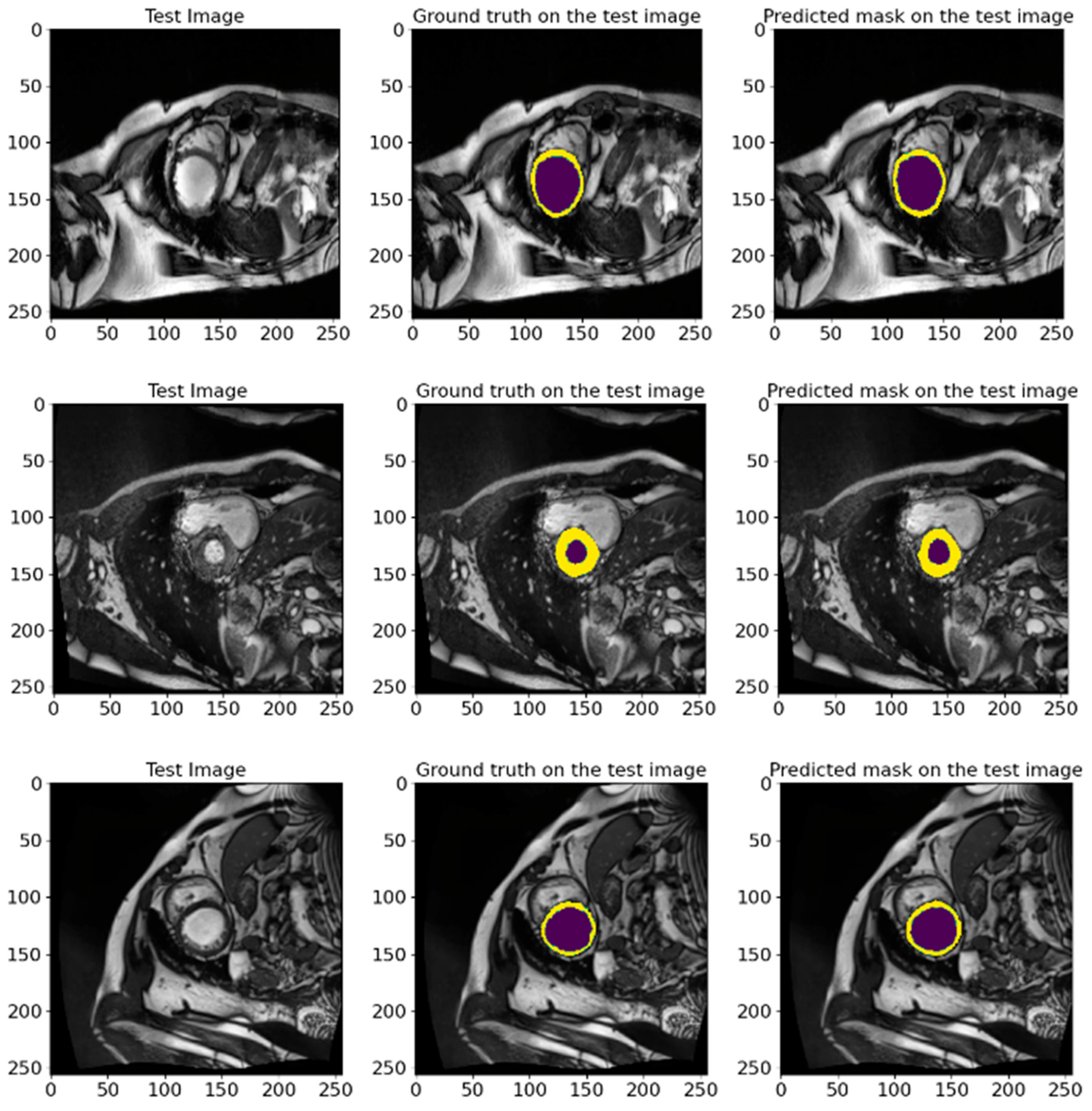
3.1.1. EMIDEC 2020
3.1.2. ACDC 2017
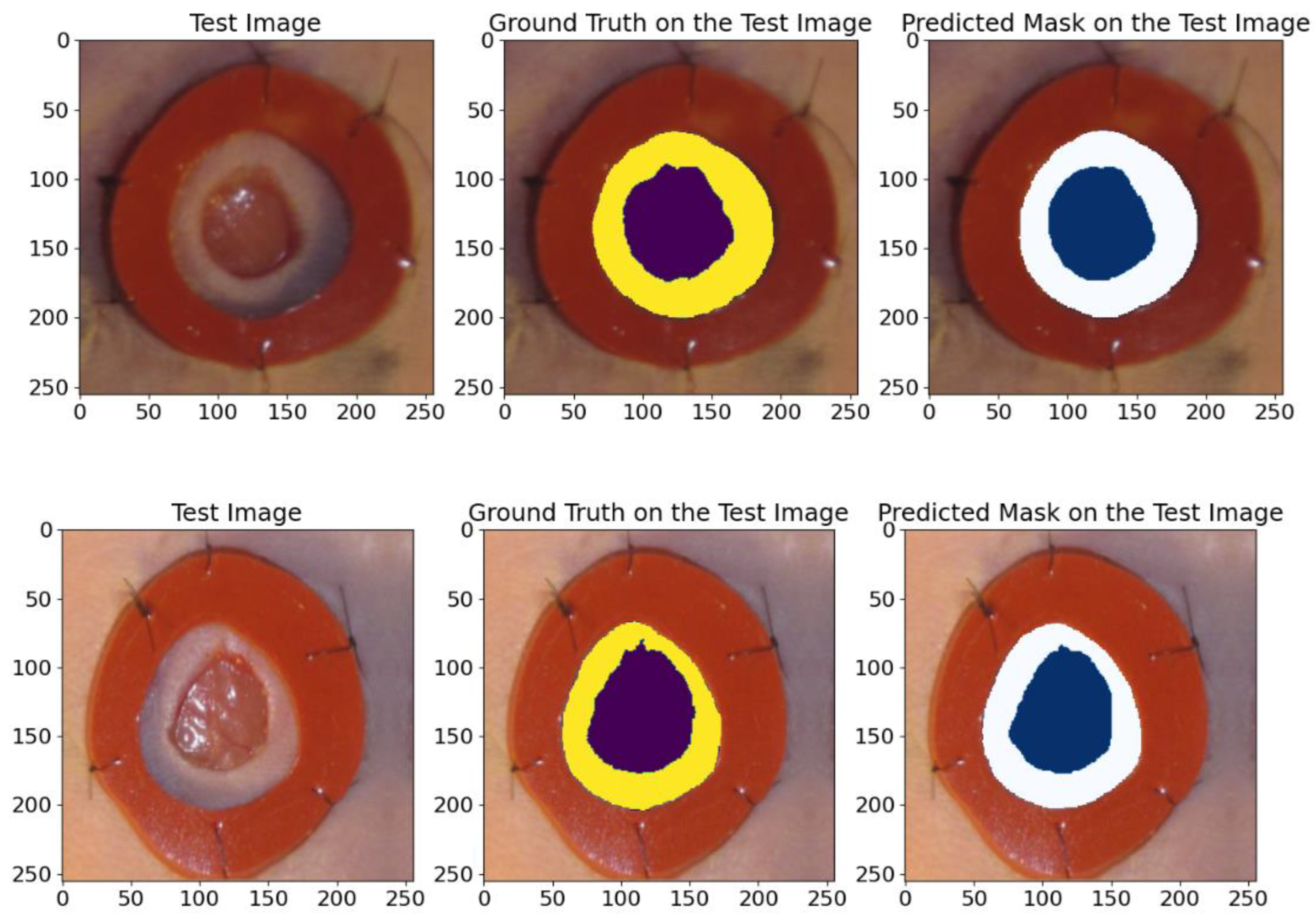
3.1.3. Wound Closure Database
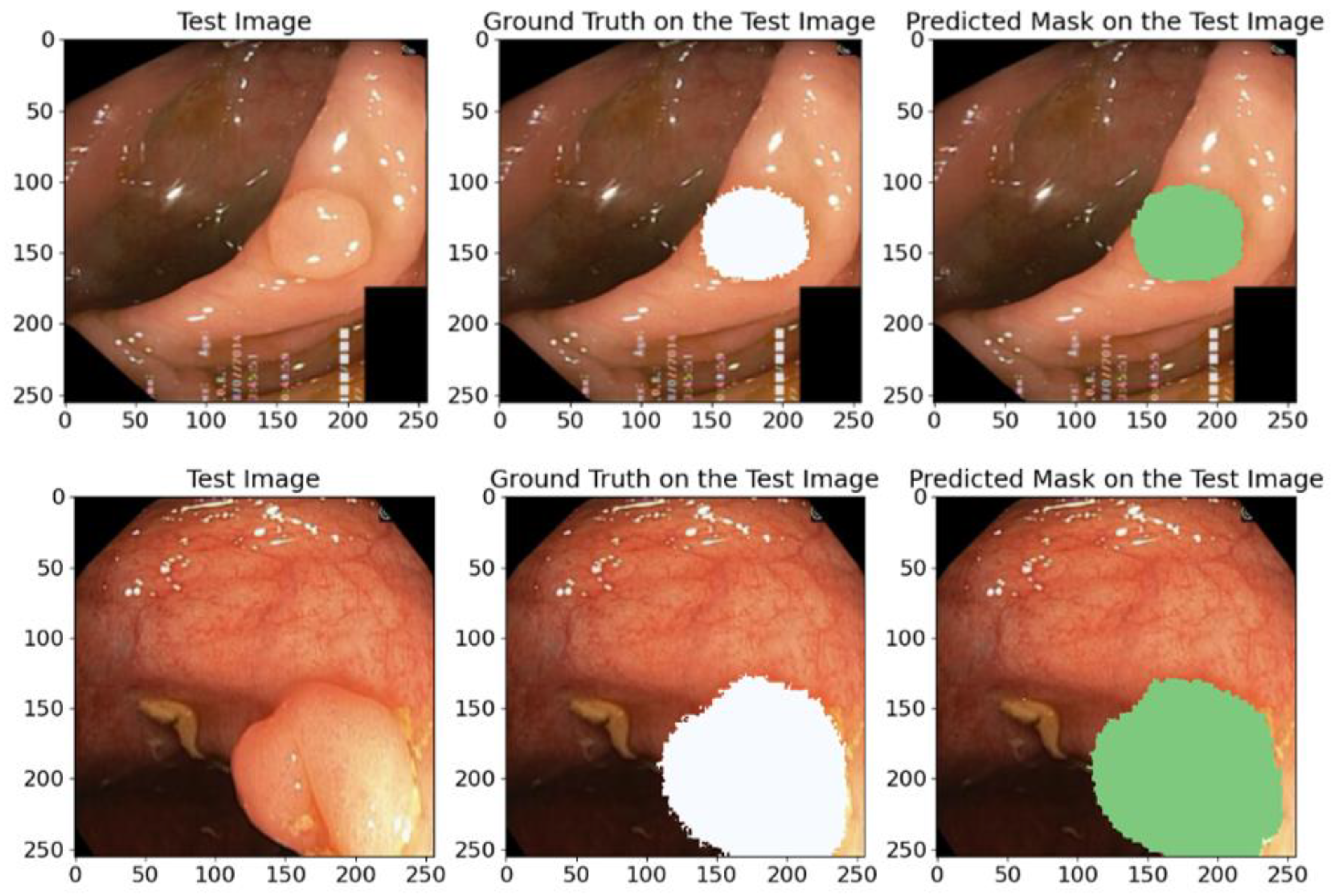
3.1.4. Kvasir-SEG
3.1.5. CVC-ClinicDB
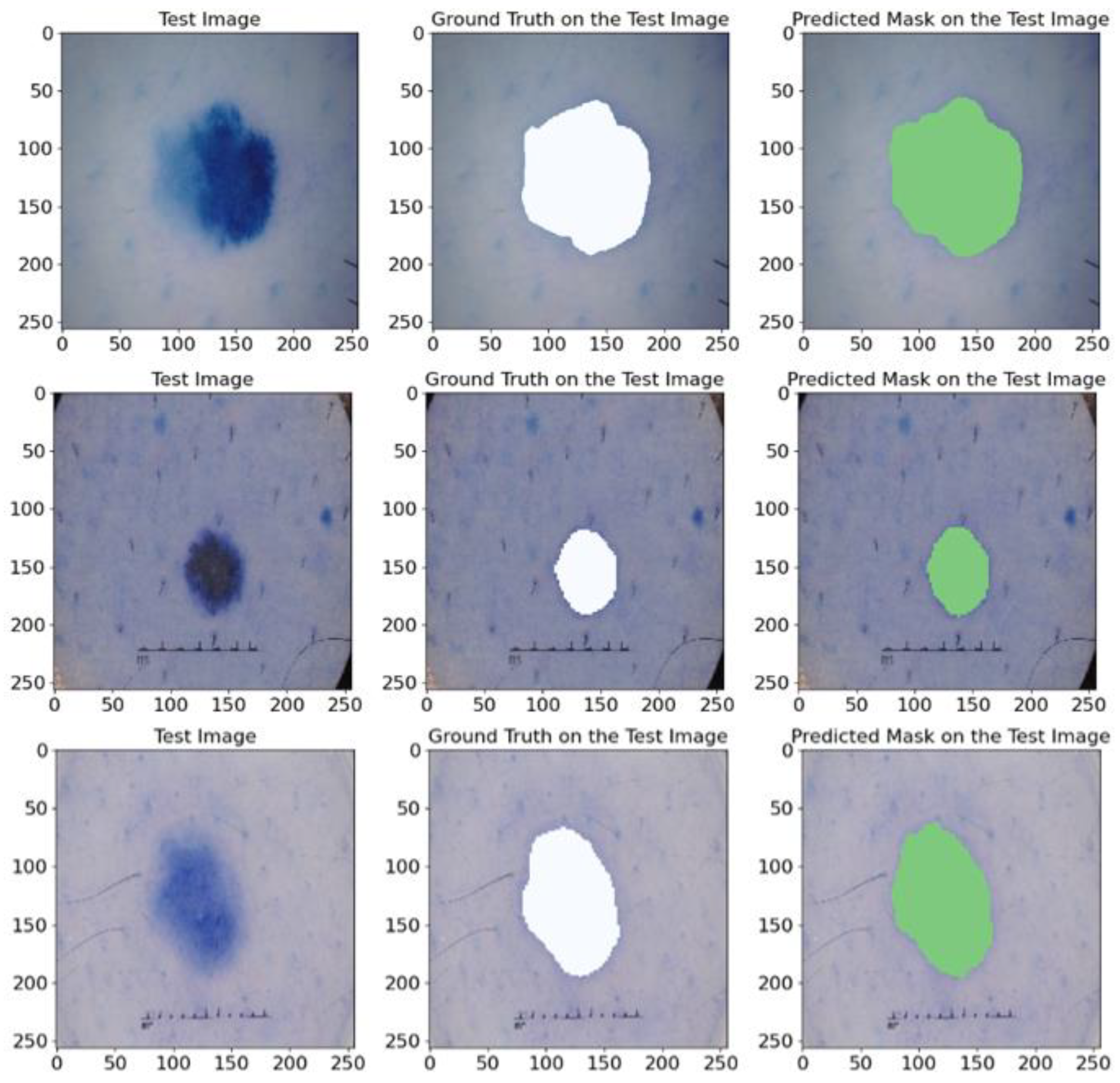
3.1.6. ISIC 2017
3.2. Pre-Processing
3.2.1. Cardiac MRI Enhancement
3.2.2. Cardiac MRI Augmentation
3.3. Proposed Model
3.4. Loss Function
3.5. Evaluation Metrics
4. Experimental Results
4.1. Training Environment and Hyperparameters
4.2. Results of Medical MRI Image Enhancement and Analysis for Cardiac Segmentation
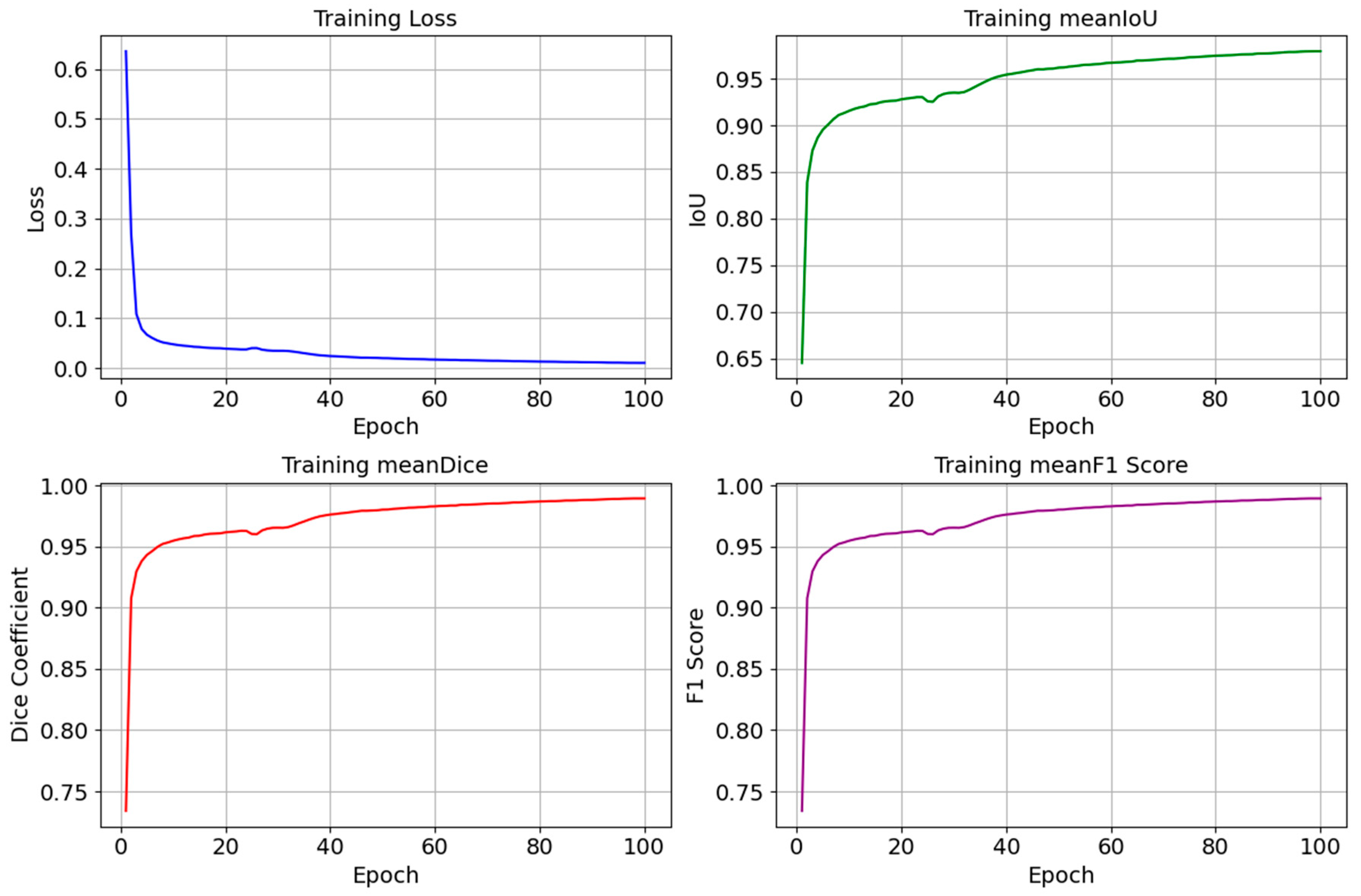
4.3. The Performance of the Proposed Model
4.4. Comparison with Other State-of-the-Art Models
4.5. Ablation Study
5. Discussion
5.1. Generalization Results
5.1.1. On Wound Images Database
5.1.2. On the Kavsir-SEG Database
5.1.3. On the CVC-Clinic Database
5.1.4. On ISIC 2017 Database
5.2. Validation Results on ACDC 2017 Database
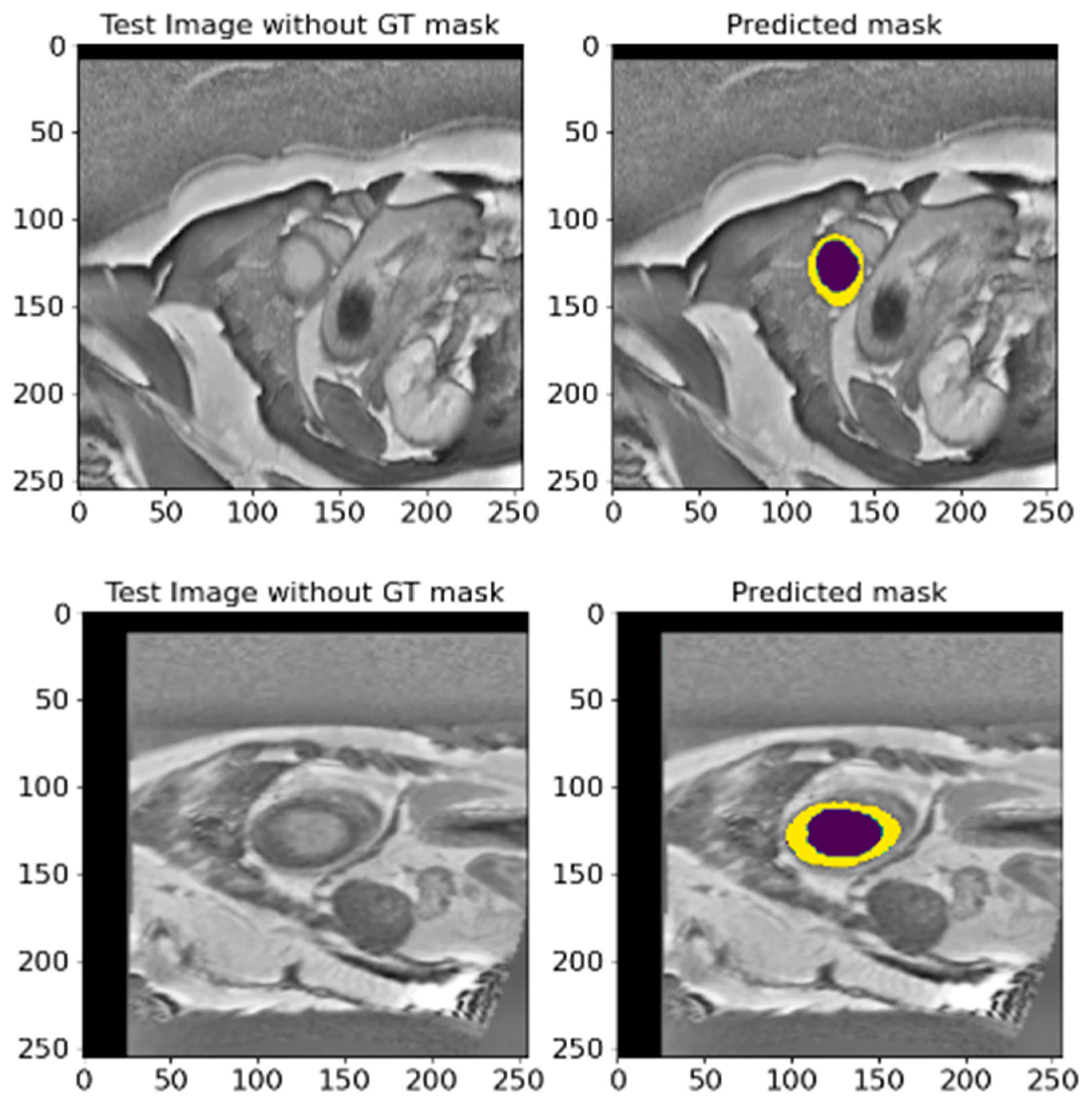
5.3. A Validation on an Unlabeled EMIDEC 2020 Database
5.4. Cross-Dataset Generalization: A Comparative Analysis
5.5. Comparison with Previous Studies
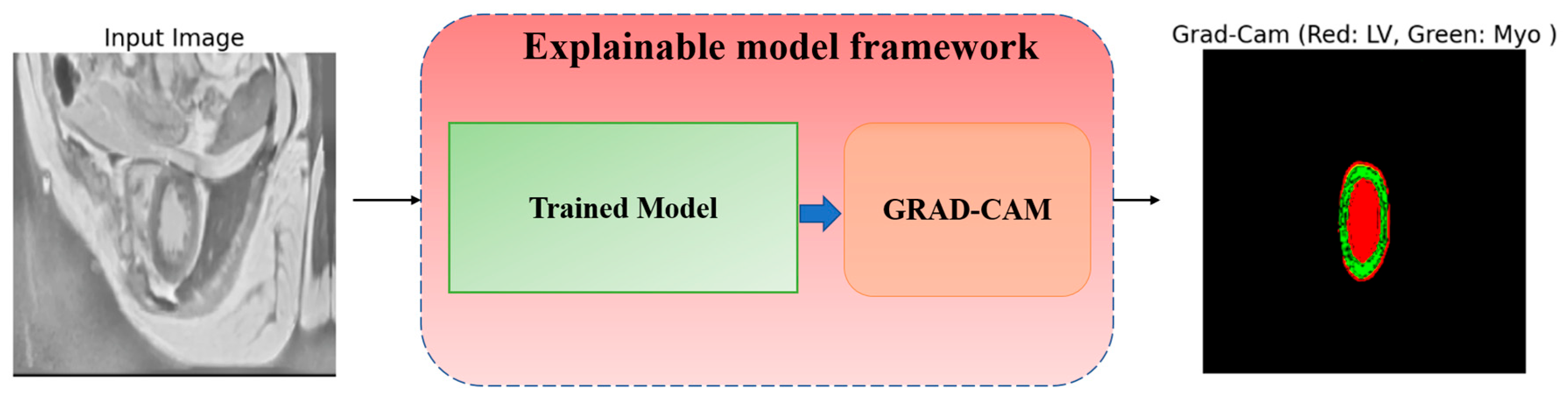
5.6. Explaining Model Decisions
- The increased intensity of overlays, to see crucial areas, confirms that the model confidently works with key structures.
- The model clearly distinguishes the boundaries between the LV and Myo, even if they are located close to each other.
5.7. Limitations
5.8. Future Work
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Heart Disease Facts and Statistics; Centers for Disease Control and Prevention, 15 May 2024. Available online: https://www.cdc.gov/heart-disease/data-research/facts-stats/index.html (accessed on 6 July 2024).
- Flachskampf, F.A.; Biering-Sørensen, T.; Solomon, S.D.; Duvernoy, O.; Bjerner, T.; Smiseth, O.A. Cardiac Imaging to Evaluate Left Ventricular Diastolic Function. JACC Cardiovasc. Imaging 2015, 8, 1071–1093. [Google Scholar] [CrossRef]
- Zhu, F.; Li, L.; Zhao, J.; Zhao, C.; Tang, S.; Nan, J.; Li, Y.; Zhao, Z.; Shi, J.; Chen, Z.; et al. A new method incorporating deep learning with shape priors for left ventricular segmentation in myocardial perfusion SPECT images. Comput. Biol. Med. 2023, 160, 106954. [Google Scholar] [CrossRef] [PubMed]
- Kar, J.; Cohen, M.V.; McQuiston, S.P.; Malozzi, C.M. A deep-learning semantic segmentation approach to fully automated MRI-based left-ventricular deformation analysis in cardiotoxicity. Magn. Reson. Imaging 2021, 78, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Leiner, T.; Rueckert, D.; Suinesiaputra, A.; Baeßler, B.; Nezafat, R.; Išgum, I.; Young, A.A. Machine learning in cardiovascular magnetic resonance: Basic concepts and applications. J. Cardiovasc. Magn. Reson. 2019, 21, 61. [Google Scholar] [CrossRef] [PubMed]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef]
- Zheng, R.; Zhao, X.; Zhao, X.; Wang, H. Deep Learning Based Multi-modal Cardiac MR Image Segmentation. In Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges, Proceedings of the STACOM 2019, Shenzhen, China, 13 October 2019; Lecture Notes in Computer Science; Pop, M., Sermesant, M., Camara, O., Zhuang, X., Li, S., Young, A., Mansi, T., Suinesiaputra, A., Eds.; Springer: Cham, Switzerland, 2020; Volume 12009. [Google Scholar] [CrossRef]
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep Learning for Cardiac Image Segmentation: A Review. Front. Cardiovasc. Med. 2020, 7, 25. [Google Scholar] [CrossRef]
- Ye, Y.; Chen, Y.; Wang, R.; Zhu, D.; Huang, Y.; Liu, J.; Chen, Y.; Shi, J.; Ding, B.; Xiahou, J. Image segmentation using improved U-Net model and convolutional block attention module based on cardiac magnetic resonance imaging. J. Radiat. Res. Appl. Sci. 2024, 17, 100816. [Google Scholar] [CrossRef]
- Singh, K.R.; Sharma, A.; Singh, G.K. Attention-guided residual W-Net for supervised cardiac magnetic resonance imaging segmentation. Biomed. Signal Process. Control 2023, 86, 105177. [Google Scholar] [CrossRef]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.-A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Chen, Z.; Lalande, A.; Salomon, M.; Decourselle, T.; Pommier, T.; Qayyum, A.; Shi, J.; Perrot, G.; Couturier, R. Automatic deep learning-based myocardial infarction segmentation from delayed enhancement MRI. Comput. Med. Imaging Graph. 2021, 95, 102014. [Google Scholar] [CrossRef]
- WHO. Cardiovascular Diseases. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 13 May 2024).
- Wang, R.; Lei, T.; Cui, R.; Zhang, B.; Meng, H.; Nandi, A.K. Medical image segmentation using deep learning: A survey. IET Image Process. 2022, 16, 1243–1267. [Google Scholar] [CrossRef]
- Bai, W.; Sinclair, M.; Tarroni, G.; Oktay, O.; Rajchl, M.; Vaillant, G.; Lee, A.M.; Aung, N.; Lukaschuk, E.; Sanghvi, M.M.; et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J. Cardiovasc. Magn. Reson. 2018, 20, 65. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Yang, H.-Y.; Bagood, M.; Carrion, H.; Isseroff, R. Photographs of 15-Day Wound Closure Progress in C57BL/6J Mice. 2022. Available online: https://datadryad.org/dataset/doi:10.25338/B84W8Q (accessed on 6 June 2024).
- Jha, D.; Smedsrud, P.H.; Riegler, M.; Halvorsen, P.; de Lange, T.; Johansen, D.; Johansen, H. Kvasir-Seg: A Segmented Polyp Dataset. Available online: https://datasets.simula.no/kvasir-seg/ (accessed on 10 June 2024).
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef]
- Codella, N.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2017, arXiv:1710.05006. [Google Scholar]
- Brahim, K.; Arega, T.W.; Boucher, A.; Bricq, S.; Sakly, A.; Meriaudeau, F. An Improved 3D Deep Learning-Based Segmentation of Left Ventricular Myocardial Diseases from Delayed-Enhancement MRI with Inclusion and Classification Prior Information U-Net (ICPIU-Net). Sensors 2022, 22, 2084. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, M.A.; Lai, K.W.; Chuah, J.H.; Hum, Y.C.; Ali, R.; Dhanalakshmi, S.; Wang, H.; Wu, X. Comparative studies of deep learning segmentation models for left ventricle segmentation. Front. Public Health 2022, 10, 981019. [Google Scholar] [CrossRef]
- Whig, P.; Sharma, P.; Nadikattu, R.R.; Bhatia, A.B.; Alkali, Y.J. GAN for Augmenting Cardiac MRI Segmentation. In GANs for Data Augmentation in Healthcare; Solanki, A., Naved, M., Eds.; Springer: Cham, Switzerland, 2023. [Google Scholar] [CrossRef]
- Tao, X.; Wei, H.; Xue, W.; Ni, D. Segmentation of Multi-modal Myocardial Images Using Shape-Transfer GAN. In Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges, Proceedings of the STACOM 2019, Shenzhen, China, 13 October 2019; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2020; Volume 12009. [Google Scholar] [CrossRef]
- Wang, Z.; Peng, Y.; Li, D.; Guo, Y.; Zhang, B. MMNet: A multi-scale deep learning network for the left ventricular segmentation of cardiac MRI images. Appl. Intell. 2021, 52, 5225–5240. [Google Scholar] [CrossRef]
- Yang, Y.; Shah, Z.; Jacob, A.J.; Hair, J.; Chitiboi, T.; Passerini, T.; Yerly, J.; Di Sopra, L.; Piccini, D.; Hosseini, Z.; et al. Deep learning-based left ventricular segmentation demonstrates improved performance on respiratory motion-resolved whole-heart reconstructions. Front. Radiol. 2023, 3, 1144004. [Google Scholar] [CrossRef]
- Nguyen, M.H.; Altintas, I.; Muse, E.D.; Quer, G.; Steinhubl, S.; Abdelmaguid, E.; Huang, J.; Kenchareddy, S.; Singla, D.; Wilke, L.; et al. Analytics pipeline for left ventricle segmentation and volume estimation on cardiac MRI using deep learning. In Proceedings of the 2018 IEEE 14th International Conference on e-Science (e-Science), Amsterdam, The Netherlands, 29 October–1 November 2018; pp. 305–306. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Chaurasia, A.; Culurciello, E. LinkNet: Exploiting encoder representations for efficient semantic segmentation. In Proceedings of the 2017 IEEE Visual Communications and Image Processing (VCIP), St. Petersburg, FL, USA, 10–13 December 2018; pp. 1–4. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Li, H.; Xiong, P.; An, J.; Wang, L. Pyramid Attention Network for Semantic Segmentation. arXiv 2018, arXiv:1805.10180. [Google Scholar]
- Fan, T.; Wang, G.; Li, Y.; Wang, H. MA-Net: A Multi-Scale Attention Network for Liver and Tumor Segmentation. IEEE Access 2020, 8, 179656–179665. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Q.; Wang, Y. Road Extraction by Deep Residual U-Net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; De Lange, T.; Halvorsen, P.; Johansen, H.D. ResUNet++: An Advanced Architecture for Medical Image Segmentation. In Proceedings of the IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, 9–11 December 2019. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Lalande, A.; Chen, Z.; Decourselle, T.; Qayyum, A.; Pommier, T.; Lorgis, L.; de la Rosa, E.; Cochet, A.; Cottin, Y.; Ginhac, D.; et al. Emidec: A Database Usable for the Automatic Evaluation of Myocardial Infarction from Delayed-Enhancement Cardiac MRI. Data 2020, 5, 89. [Google Scholar] [CrossRef]
- Ma, J. Cascaded Framework for Automatic Evaluation of Myocardial Infarction from Delayed-Enhancement Cardiac MRI. arXiv 2020, arXiv:2012.14556. [Google Scholar]
- Wada, K. labelme: Image Polygonal Annotation with Python. [CrossRef]
- Buslaev, A.; Iglovikov, V.I.; Khvedchenya, E.; Parinov, A.; Druzhinin, M.; Kalinin, A.A. Albumentations: Fast and Flexible Image Augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef]
- Zuiderveld, K. Contrast Limited Adaptive Histogram Equalization; Heckbert, P.S., Ed.; Academic Press: Cambridge, MA, USA, 1994; Chapter VIII.5, Graphics Gems IV; pp. 474–485. [Google Scholar]
- Tomasi, C.; Manduchi, R. Bilateral filtering for gray and color images. In Proceedings of the Sixth International Conference on Computer Vision (IEEE Cat. No. 98CH36271), Bombay, India, 7 January 1998; pp. 839–846. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Alabdulhafith, M.; Mahel, A.S.B.; Samee, N.A.; Mahmoud, N.F.; Talaat, R.; Muthanna, M.S.A.; Nassef, T.M. Automated wound care by employing a reliable U-Net architecture combined with ResNet feature encoders for monitoring chronic wounds. Front. Med. 2024, 11, 1310137. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, C.; Zhang, Z.; Zhu, Y.; Lin, H.; Zhang, Z.; Sun, Y.; He, T.; Mueller, J.; Manmatha, R.; et al. ResNeSt: Split-Attention Networks. arXiv 2020, arXiv:2004.08955. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar] [CrossRef]
- Roy, A.G.; Navab, N.; Wachinger, C. Recalibrating Fully Convolutional Networks with Spatial and Channel ‘Squeeze & Excitation’ Blocks. arXiv 2018, arXiv:1808.08127. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Cui, H.; Yuwen, C.; Jiang, L.; Xia, Y.; Zhang, Y. Multiscale attention guided U-Net architecture for cardiac segmentation in short-axis MRI images. Comput. Methods Programs Biomed. 2021, 206, 106142. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.K.; Liew, Y.M.; Lim, E.; McLaughlin, R.A. Convolutional neural network regression for short-axis left ventricle segmentation in cardiac cine MR sequences. Med. Image Anal. 2017, 39, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.V. A Fully Convolutional Neural Network for Cardiac Segmentation in Short-Axis MRI. arXiv 2016, arXiv:1604.00494. [Google Scholar]
- Khened, M.; Kollerathu, V.A.; Krishnamurthi, G. Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. Med. Image Anal. 2019, 51, 21–45. [Google Scholar] [CrossRef]
- Wang, Z.; Xie, L.; Qi, J. Dynamic pixel-wise weighting-based fully convolutional neural networks for left ventricle segmentation in short-axis MRI. Magn. Reson. Imaging 2020, 66, 131–140. [Google Scholar] [CrossRef]
- Chen, N.; Wang, S.; Lu, R.; Li, W.; Shi, X. PHMamba: Preheating State Space Models with Context-Augmented Features for Medical Image Segmentation. In Proceedings of the ICASSP 2025—2025 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Hyderabad, India, 6–11 April 2025; pp. 1–5. [Google Scholar] [CrossRef]
- da Silva, I.F.S.; Silva, A.C.; de Paiva, A.C.; Gattass, M. A cascade approach for automatic segmentation of cardiac structures in short-axis cine-MR images using deep neural networks. Expert Syst. Appl. 2022, 197, 116704. [Google Scholar] [CrossRef]
- Al-Antari, M.A.; Shaaf, Z.F.; Jamil, M.M.A.; Samee, N.A.; Alkanhel, R.; Talo, M.; Al-Huda, Z. Deep learning myocardial infarction segmentation framework from cardiac magnetic resonance images. Biomed. Signal Process. Control 2023, 89, 105710. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. arXiv 2016, arXiv:1610.02391. [Google Scholar]
- Wang, Y.; Jin, P.; Meng, X.; Li, L.; Mao, Y.; Zheng, M.; Liu, L.; Liu, Y.; Yang, J. Treatment of Severe Pulmonary Regurgitation in Enlarged Native Right Ventricular Outflow Tracts: Transcatheter Pulmonary Valve Replacement with Three-Dimensional Printing Guidance. Bioengineering 2023, 10, 1136. [Google Scholar] [CrossRef]
- Zhang, W.; Peng, D.; Cheng, S.; Ni, R.; Yang, M.; Cai, Y.; Chen, J.; Liu, F.; Liu, Y. Inflammatory Cell-Targeted Delivery Systems for Myocardial Infarction Treatment. Bioengineering 2025, 12, 205. [Google Scholar] [CrossRef] [PubMed]

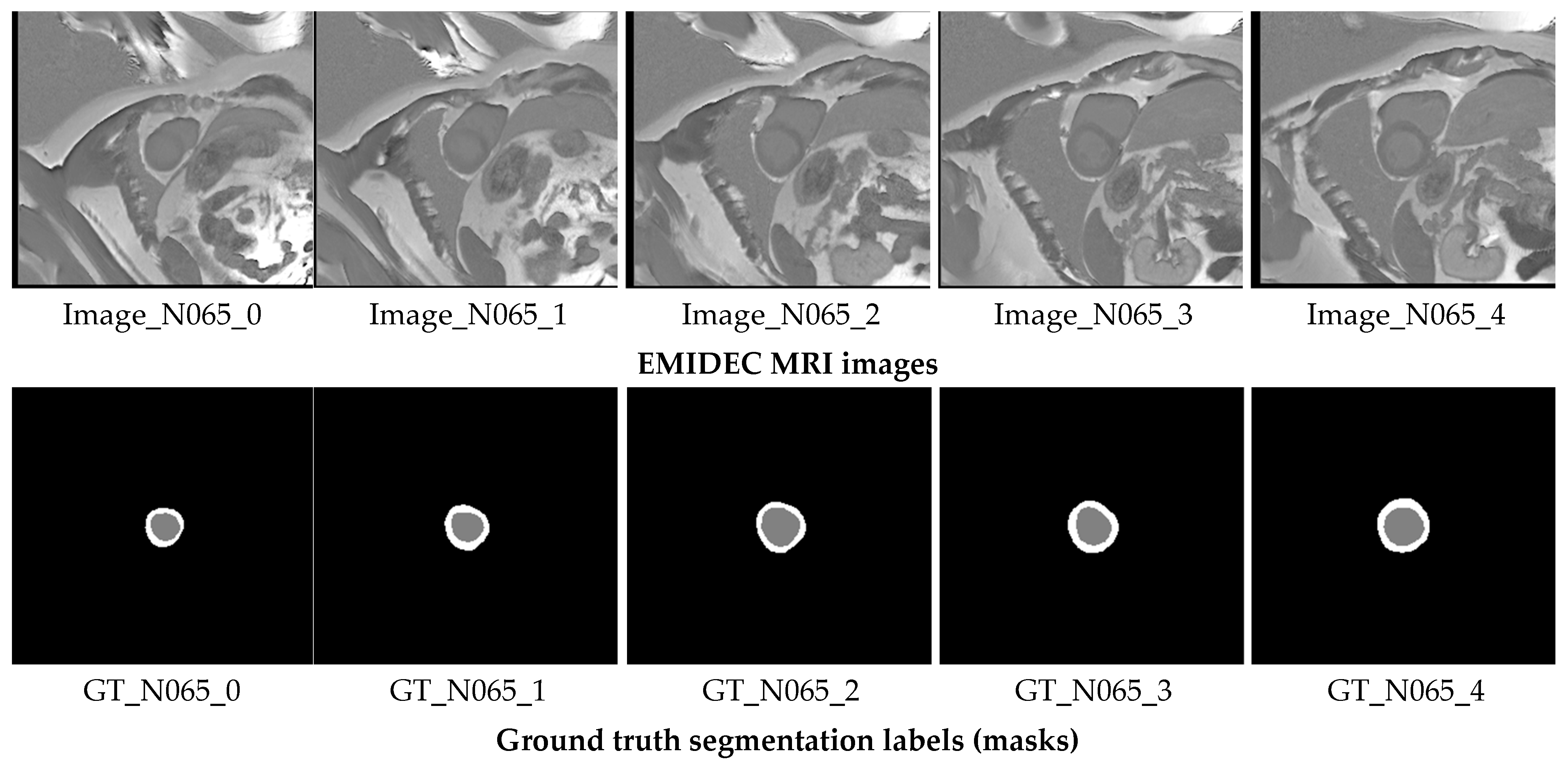
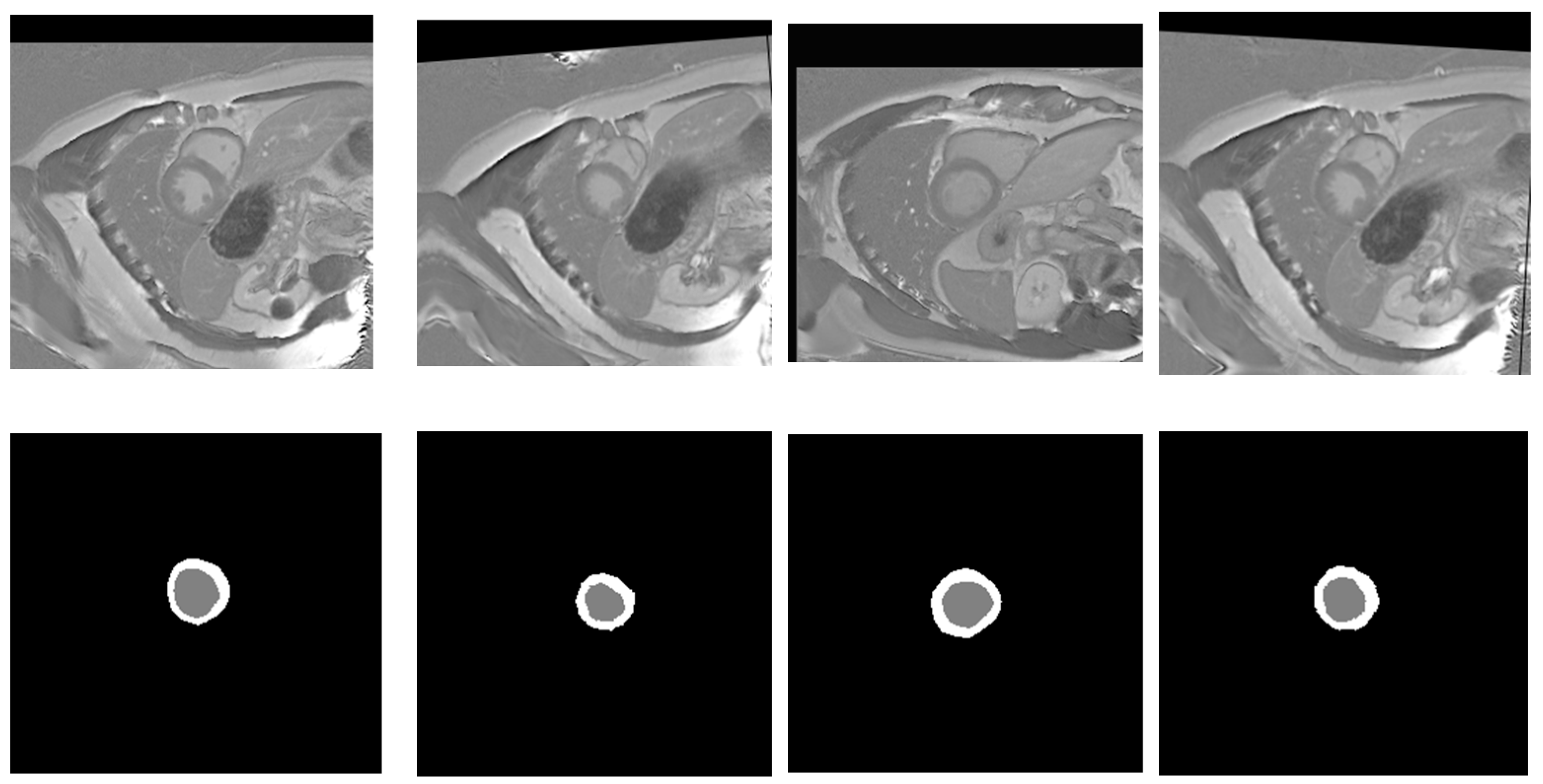








| Model | Mean IoU (%) ± SD | Mean Recall (%) ± SD | Mean Dice (%) ± SD | Mean Precision (%) ± SD | Mean F1-Score (%) ± SD | Epochs | Params (M) | LR/Batch |
|---|---|---|---|---|---|---|---|---|
| Proposed | 93.73 ± 0.0137 | 96.54 ± 0.0081 | 96.70 ± 0.0076 | 96.86 ± 0.0074 | 96.70 ± 0.0076 | 100 | 53 | 0.0001/8 |
| Model | Mean IoU (%) ± SD | Mean Recall (%) ± SD | Mean Dice (%) ± SD | Mean Precision (%) ± SD | Mean F1-Score (%) ± SD | LR/Batch |
|---|---|---|---|---|---|---|
| ResUnet++ [34] | 91.20 ± 0.0074 | 93.79 ± 0.0052 | 95.28 ± 0.0043 | 97.00 ± 0.0041 | 95.28 ± 0.043 | 0.0001/8 |
| PAN [31] | 90.90 ± 0.0095 | 93.49 ± 0.0066 | 95.10 ± 0.0055 | 96.98 ± 0.0054 | 95.10 ± 0.0055 | 0.0001/8 |
| ResUnet [33] | 91.33 ± 0.0091 | 93.49 ± 0.0073 | 95.35 ± 0.0053 | 97.47 ± 0.0045 | 95.35 ± 0.0053 | 0.0001/8 |
| LinkNet [29] | 91.29 ± 0.0099 | 93.75 ± 0.0069 | 95.33 ± 0.0057 | 97.12 ± 0.0054 | 95.33 ± 0.0057 | 0.0001/8 |
| MAnet [32] | 91.53 ± 0.0078 | 93.66 ± 0.0059 | 95.46 ± 0.0044 | 97.52 ± 0.0041 | 95.46 ± 0.0044 | 0.0001/8 |
| PSPNet [30] | 90.69 ± 0.0103 | 93.38 ± 0.0074 | 94.98 ± 0.0060 | 96.85 ± 0.0058 | 94.98 ± 0.0060 | 0.0001/8 |
| Proposed | 93.73 ± 0.0137 | 96.54 ± 0.0081 | 96.70 ± 0.0076 | 96.86 ± 0.0074 | 96.70 ± 0.0076 | 0.0001/8 |
| Model | Mean IoU (%) ± SD | Mean F1-Score (%) ± SD |
|---|---|---|
| Unet++ with Resnet34 (M1) | 91.21 ± 0.0082 | 95.28 ± 0.0047 |
| Unet++ with Resnet34 and SE decoder attention (M2) | 91.15 ± 0.0078 | 95.25 ± 0.0045 |
| Unet++ with Resnet 34 and scSE decoder attention (M3) | 90.92 ± 0.0092 | 95.11 ± 0.0053 |
| Unet++ alone (M4) | 88.83 ± 0.0130 | 93.90 ± 0.0078 |
| Unet++ with resnest50d and scSE decoder attention (M5) | 91.74 ± 0.0077 | 95.58 ± 0.0044 |
| Proposed without CLAHE (M6) | 91.34 ± 0.0070 | 95.36 ± 0.0040 |
| Proposed (Unet++ with resnest50d and SE decoder attention) | 93.73 ± 0.0137 | 96.70 ± 0.0076 |
| Model | Mean IoU (%) ± SD | Mean Recall (%) ± SD | Mean Dice (%) ± SD | Mean Precision (%) ± SD | Mean F1-Score (%) ± SD | Epochs | Params (M) | LR/Batch |
|---|---|---|---|---|---|---|---|---|
| Proposed | 92.95 ± 0.0101 | 95.45 ± 0.0085 | 96.27 ± 0.0057 | 97.14 ± 0.0042 | 96.27 ± 0.0057 | 100 | 53 | 0.0001/8 |
| Model | Mean IoU (%) ± SD | Mean Recall (%) ± SD | Mean Dice (%) ± SD | Precision (%) ± SD | Mean F1-Score (%) ± SD | Epochs | Params (M) | LR/Batch |
|---|---|---|---|---|---|---|---|---|
| Proposed | 86.04 ± 0.0737 | 91.26 ± 0.0625 | 92.32 ± 0.0451 | 93.74 ± 0.0493 | 92.32 ± 0.0451 | 100 | 53 | 0.0001/8 |
| Model | Mean IoU (%) ± SD | Mean Recall (%) ± SD | Mean Dice (%) ± SD | Mean Precision (%) ± SD | Mean F1-Score (%) ± SD | Epochs | Params (M) | LR/Batch |
|---|---|---|---|---|---|---|---|---|
| Proposed | 89.72 ± 0.0345 | 93.80 ± 0.0339 | 94.54 ± 0.0195 | 95.40 ± 0.0206 | 94.54 ± 0.0195 | 100 | 53 | 0.0001/8 |
| Model | Mean IoU (%) ± SD | Mean Recall (%) ± SD | Mean Dice (%) ± SD | Mean Precision (%) ± SD | Mean F1-Score (%) ± SD | Epochs | Params (M) | LR/Batch |
|---|---|---|---|---|---|---|---|---|
| Proposed | 85.37 ± 0.0689 | 91.63 ± 0.0529 | 91.91 ± 0.0430 | 92.65 ± 0.0629 | 91.91 ± 0.0430 | 100 | 53 | 0.0001/8 |
| Model | Mean IoU (%) ± SD | Mean Recall (%) ± SD | Mean Dice (%) ± SD | Mean Precision (%) ± SD | Mean F1-Score (%) ± SD | Epochs | Params (M) | LR |
|---|---|---|---|---|---|---|---|---|
| Proposed | 88.78 ± 0.0175 | 93.39 ± 0.0138 | 93.85 ± 0.0106 | 94.38 ± 0.0101 | 93.85 ± 0.0106 | 100 | 53 | 0.0001 |
| Database | Model | Mean IoU (%) | Mean Recall (%) | Mean Dice (%) | Mean Precision (%) | Mean F1-Score (%) |
|---|---|---|---|---|---|---|
| Wound Closure | MAnet | 92.20 ± 0.0180 | 94.77 ± 0.0178 | 95.83 ± 0.0232 | 97.01 ± 0.0010 | 95.83 ± 0.0232 |
| ResUnet | 92.06 ± 0.0213 | 94.69 ± 0.0185 | 95.75 ± 0.0133 | 96.92 ± 0.0067 | 95.75 ± 0.0133 | |
| LinkNET | 92.26 ± 0.0134 | 94.87 ± 0.0118 | 95.88 ± 0.0077 | 96.98 ± 0.0046 | 95.88 ± 0.0077 | |
| Proposed | 92.95 ± 0.0101 | 95.45 ± 0.0085 | 96.27 ± 0.0057 | 97.14 ± 0.0042 | 96.27 ± 0.0057 | |
| Kavsir-SEG | MAnet | 80.32 ± 0.0812 | 89.99 ± 0.0832 | 88.85 ± 0.0516 | 88.60 ± 0.0705 | 88.85 ± 0.0516 |
| ResUnet | 83.25 ± 0.0669 | 89.64 ± 0.0627 | 90.71 ± 0.0413 | 92.18 ± 0.0495 | 90.71 ± 0.0413 | |
| LinkNET | 83.30 ± 0.0823 | 89.63 ± 0.0757 | 90.65 ± 0.0520 | 92.27 ± 0.0594 | 90.65 ± 0.0520 | |
| Proposed | 86.04 ± 0.0737 | 91.26 ± 0.0625 | 92.32 ± 0.0451 | 93.74 ± 0.0493 | 92.32 ± 0.0451 | |
| CVC-Clinic | MAnet | 88.81 ± 0.0454 | 92.78 ± 0.0437 | 94.01 ± 0.0262 | 95.41 ± 0.0215 | 94.01 ± 0.0262 |
| ResUnet | 88.24 ± 0.0503 | 92.04 ± 0.0523 | 93.67 ± 0.0295 | 95.54 ± 0.0155 | 93.67 ± 0.0295 | |
| LinkNET | 88.79 ± 0.0527 | 93.42 ± 0.0367 | 93.97 ± 0.0327 | 94.63 ± 0.0401 | 93.97 ± 0.0327 | |
| Proposed | 89.72 ± 0.0345 | 93.80 ± 0.0339 | 94.54 ± 0.0195 | 95.40 ± 0.0206 | 94.54 ± 0.0195 | |
| ISIC 2017 | MAnet | 83.39 ± 0.0702 | 90.04 ± 0.0695 | 90.78 ± 0.0433 | 91.92 ± 0.0432 | 90.78 ± 0.0433 |
| ResUnet | 83.13 ± 0.0747 | 87.73 ± 0.0743 | 90.59 ± 0.0478 | 94.08 ± 0.0410 | 90.59 ± 0.0478 | |
| LinkNET | 83.30 ± 0.0801 | 87.97 ± 0.0907 | 90.67 ± 0.0518 | 94.21 ± 0.0296 | 90.67 ± 0.0518 | |
| Proposed | 85.37 ± 0.0689 | 91.63 ± 0.0529 | 91.91 ± 0.0430 | 92.65 ± 0.0629 | 91.91 ± 0.0430 | |
| ACDC 2017 | MAnet | 87.87 ± 0.0150 | 92.15 ± 0.0147 | 93.22 ± 0.0171 | 94.56 ± 0.046 | 93.22 ± 0.0171 |
| ResUnet | 87.56 ± 0.0682 | 91.95 ± 0.0713 | 92.73 ± 0.0704 | 93.65 ± 0.0711 | 92.73 ± 0.0704 | |
| LinkNET | 87.22 ± 0.0676 | 91.28 ± 0.0706 | 92.52 ± 0.0701 | 93.98 ± 0.0717 | 92.52 ± 0.0701 | |
| Proposed | 88.78 ± 0.0175 | 93.39 ± 0.0138 | 93.85 ± 0.0106 | 94.38 ± 0.0101 | 93.85 ± 0.0106 |
| Method | Dataset | Methods | Mean IoU (%) | Mean Dice (%) | Mean Recall (%) | Mean F1-Score (%) |
|---|---|---|---|---|---|---|
| Cui et al. [48] | LVSC | An Attention U-Net with image pyramids and Focal Tversky Loss for cardiac MRI segmentation. | 75 | – | – | – |
| Tan et al. [49] | LVSC | Dual-network model for LV segmentation via center localization and polar border regression. | 77 | – | – | – |
| Tran et al. [50] | LVSC | A fully convolutional network (FCN) for end-to-end, pixel-level segmentation of cardiac MRI images. | 74 | – | 83 | – |
| Khened et al. [51] | LV-2011 | DenseNet-based FCN using Inception modules and dual loss for efficient cardiac segmentation. | 74 | 84 | – | – |
| Wang et al. [52] | LV DETERMINE dataset | Dynamic pixel-wise weighted FCN with scale-invariant features for automatic LV MRI segmentation. | 70 | 80 | – | – |
| Chen et al. [53] | ACDC-2017 | PH-Mamba boosts segmentation by preheating state space models with contextual cues and compensation. | 83.89 | 91.04 | – | – |
| Silva et al. [54] | ACDC-2017 | A U-Net identifies regions of interest, a custom FCN segments cardiac structures, followed by refinement via a reconstruction module. | 80.89 | 86.88 | 87.48 | 87.43 |
| Al-antari et al. [55] | EMIDEC 2020 | Multi-class cardiac segmentation using enhanced MRI and an adapted ResUNet. | 84.23 | – | 85.24 | 85.35 |
| Proposed | EMIDEC 2020 | CLAHE-enhanced MRI and ResST-SEUNet++, a deep learning model integrating a ResNetSt encoder with SE-attention mechanisms in the UNet++ decoder, for improved cardiac image segmentation. | 93.73 | 96.70 | 96.54 | 96.70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ba Mahel, A.S.; Al-Gaashani, M.S.A.M.; Alotaibi, F.M.G.; Alkanhel, R.I. ResST-SEUNet++: Deep Model for Accurate Segmentation of Left Ventricle and Myocardium in Magnetic Resonance Imaging (MRI) Images. Bioengineering 2025, 12, 665. https://doi.org/10.3390/bioengineering12060665
Ba Mahel AS, Al-Gaashani MSAM, Alotaibi FMG, Alkanhel RI. ResST-SEUNet++: Deep Model for Accurate Segmentation of Left Ventricle and Myocardium in Magnetic Resonance Imaging (MRI) Images. Bioengineering. 2025; 12(6):665. https://doi.org/10.3390/bioengineering12060665
Chicago/Turabian StyleBa Mahel, Abduljabbar S., Mehdhar S. A. M. Al-Gaashani, Fahad Mushabbab G. Alotaibi, and Reem Ibrahim Alkanhel. 2025. "ResST-SEUNet++: Deep Model for Accurate Segmentation of Left Ventricle and Myocardium in Magnetic Resonance Imaging (MRI) Images" Bioengineering 12, no. 6: 665. https://doi.org/10.3390/bioengineering12060665
APA StyleBa Mahel, A. S., Al-Gaashani, M. S. A. M., Alotaibi, F. M. G., & Alkanhel, R. I. (2025). ResST-SEUNet++: Deep Model for Accurate Segmentation of Left Ventricle and Myocardium in Magnetic Resonance Imaging (MRI) Images. Bioengineering, 12(6), 665. https://doi.org/10.3390/bioengineering12060665








