Active Contours Connected Component Analysis Segmentation Method of Cancerous Lesions in Unsupervised Breast Histology Images
Abstract
1. Introduction
- 1
- The use of the connected component analysis method to group components with similar characteristics into binary masks that assist in separating overlapping and non-overlapping objects, thus avoiding over-segmentation.
- 2
- The binary masks from the connected component analysis method further aid in addressing the inaccurate segmentation of the image boundaries of intersecting objects, which is common with the active contours method. The proposed method clearly distinguishes the different ROIs from each other, clearly isolating and segmenting the cancerous lesions as visually documented in Section 3 and Section 4.
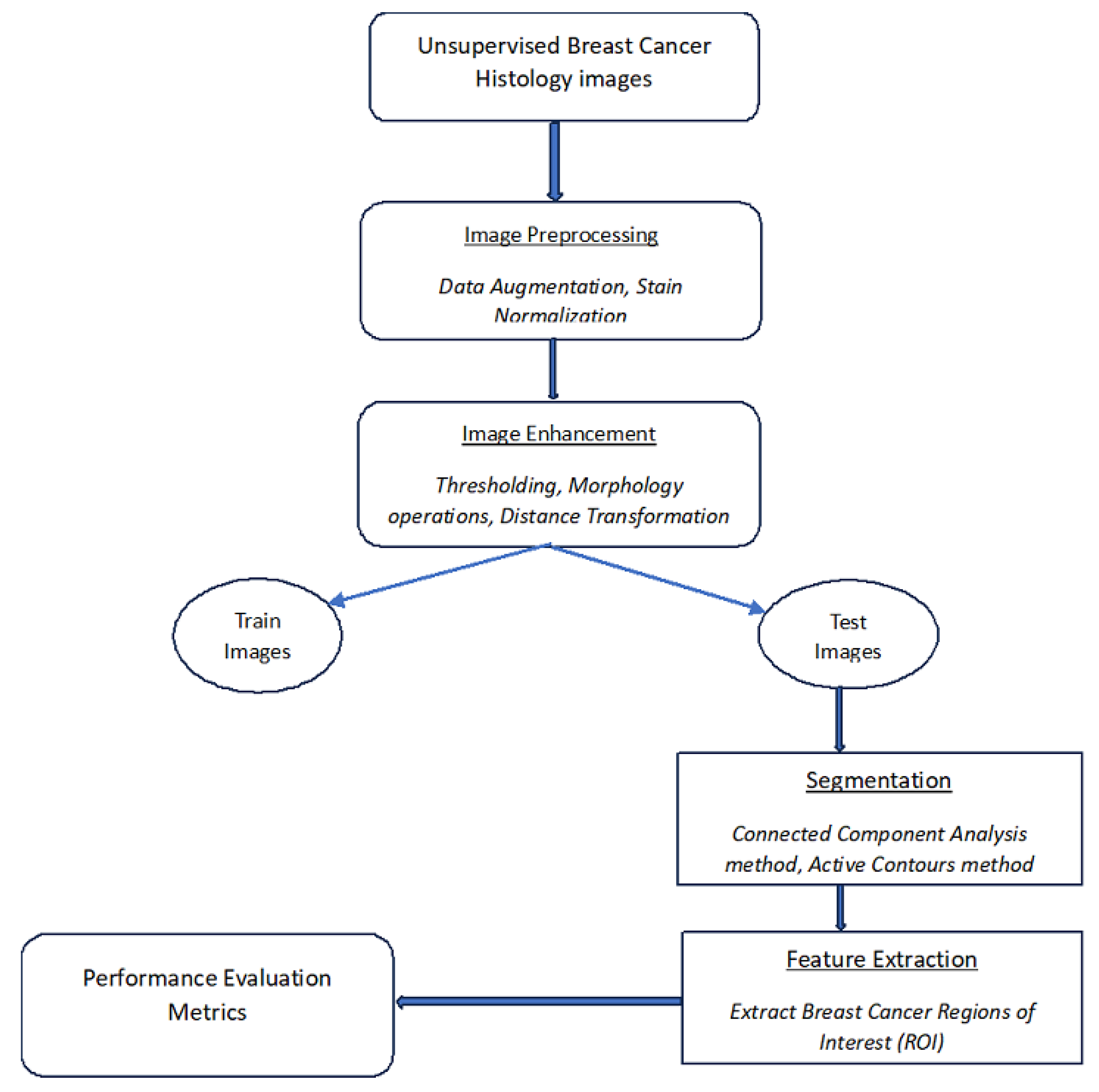
2. Proposed Segmentation Method
2.1. Dataset Pre-Processing
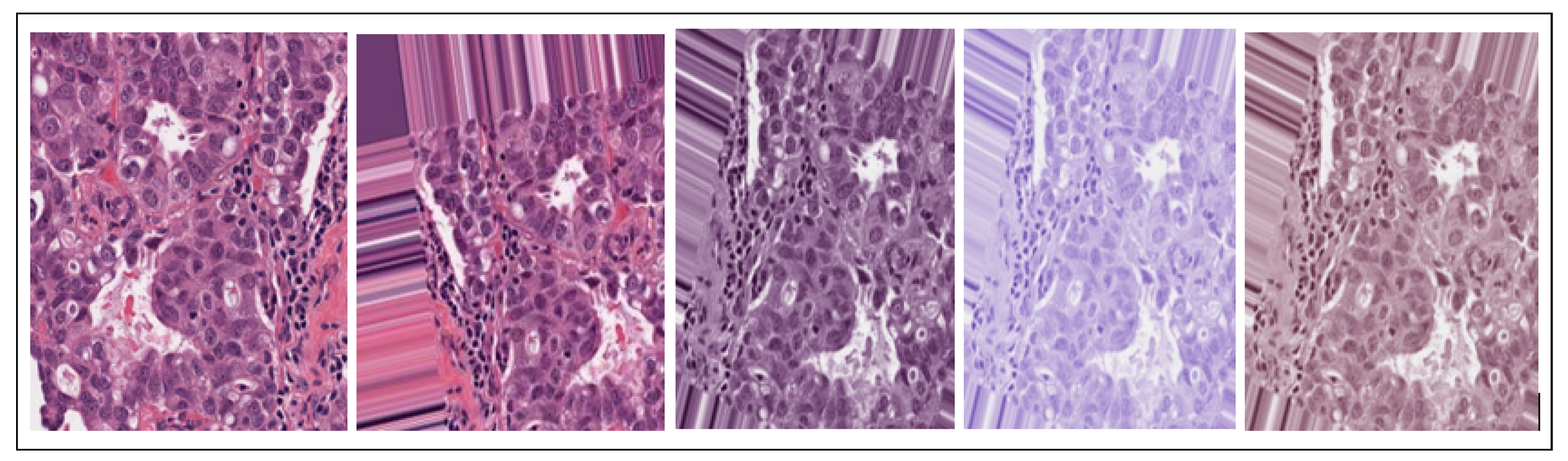
2.1.1. Dataset Augmentation
2.1.2. Data Stain Normalization
2.2. Image Enhancement
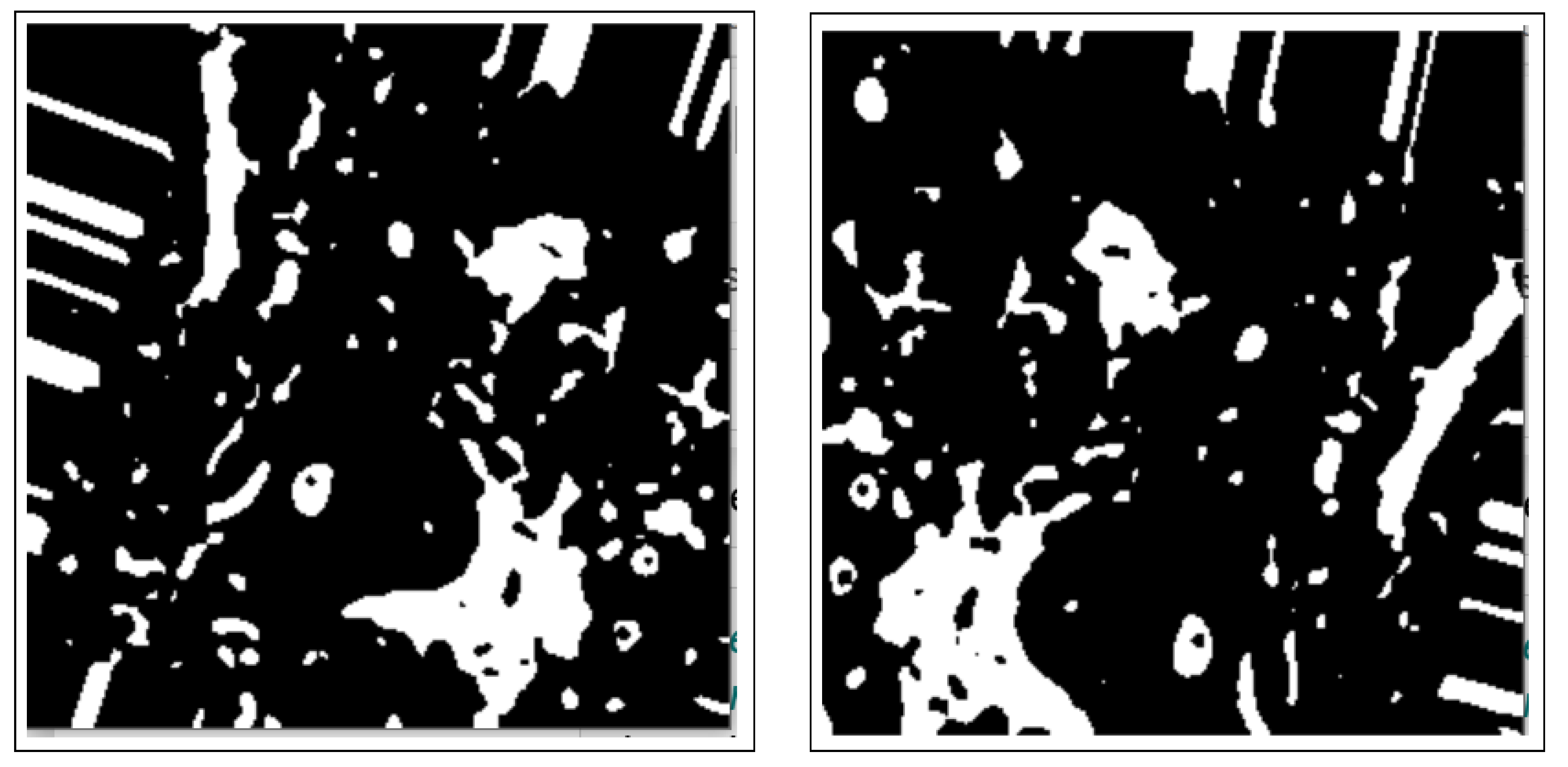
2.2.1. Thresholding
2.2.2. Morphology Operations
2.2.3. Distance Transform
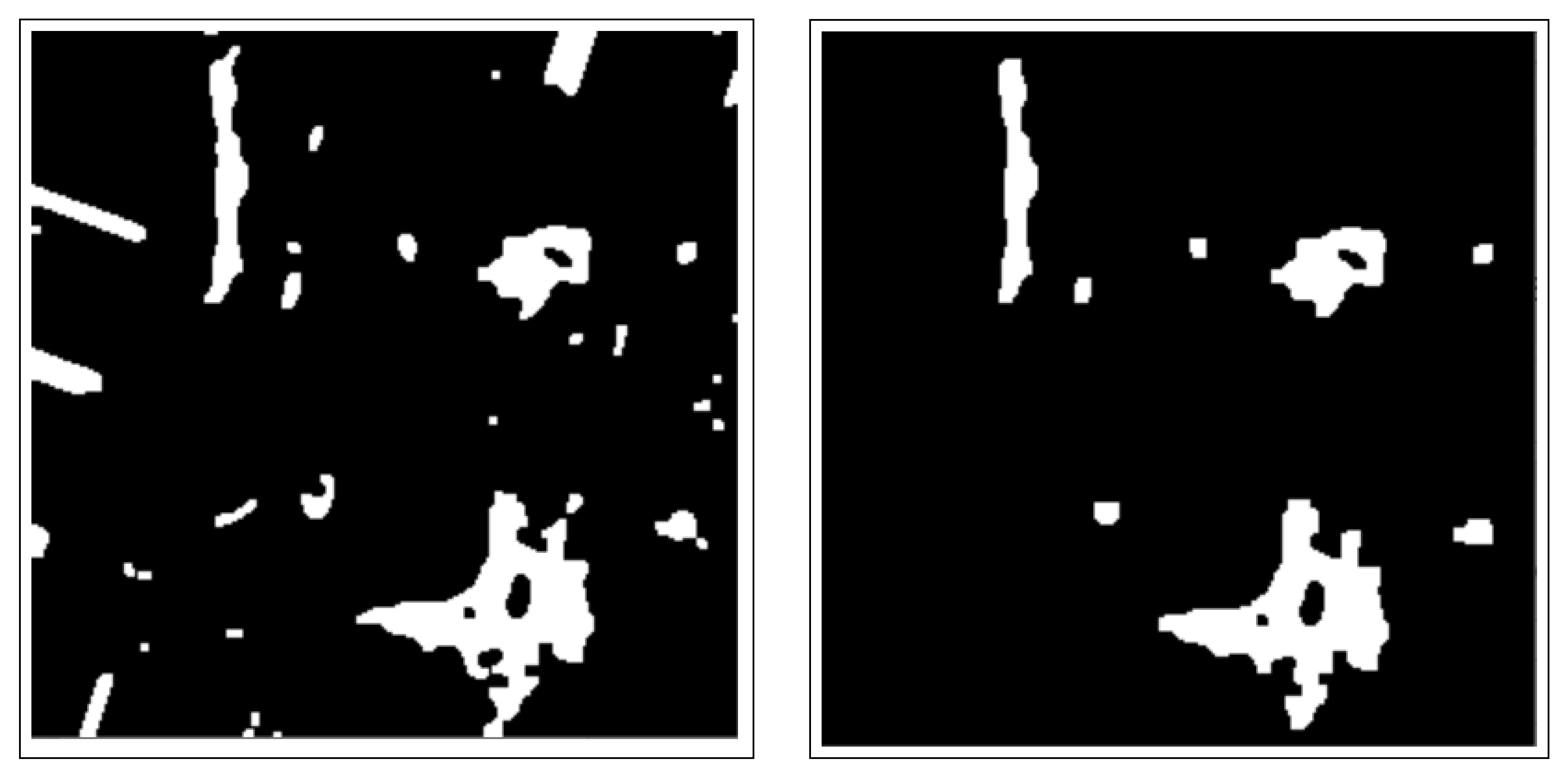
2.3. Segmentation
2.3.1. Connected Component Analysis
2.3.2. Active Contours Segmentation
3. Results and Discussion
| Algorithm 1 Proposed segmentation method for unsupervised breast cancer histology images segmentation |
|
Limitations
4. Conclusions
5. Future Work
- Data availability and integrity—most deep-learning approaches require significantly huge datasets to deduce meaningful and effective performance results. Therefore, it is necessary to access more publicly available BC histology image datasets, thus aiding deep learning. Additionally, the proposed model should be tested on other huge volume datasets to evaluate performance, not those specifically targeting breast cancer.
- Regularization methods—to improve the performance of models. This can be done through model hyperparameter tuning, such as optimizing learning rates, dropout, loss functions, activation functions, and early stopping methods.
- Blended approaches—combining various/several methods and their attributes to form hybrid methods, thus improving overall evaluation performance. This amalgamation can occur at any stage of the model architecture, namely, pre-processing, combining various attributes of different models to form one that will enhance the training, extraction, detection, and classification of nuclei objects. Additionally, in the future, our work can expand to infiltrate and diagnose image datasets of other human/animal gland histology images, not just limited to BC histology images.Consequently, the segmentation accuracy of these medical conditions could also be boosted by including attention-based models and other deep and machine-learning techniques. Results from the performance evaluation of these models could then be used to build clinical trust and thus the utilization of the proposed models. Furthermore, we plan to explore other models, including but not limited to CNN, LSTM, and UNet.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krithiga, R.; Geetha, P. Breast cancer detection, segmentation and classification on histopathology images analysis: A systematic review. Arch. Comput. Methods Eng. 2021, 28, 2607–2619. [Google Scholar] [CrossRef]
- Fox, H. Is H&E morphology coming to an end? J. Clin. Pathol. 2000, 53, 38–40. [Google Scholar]
- Wang, P.; Hu, X.; Li, Y.; Liu, Q.; Zhu, X. Automatic cell nuclei segmentation and classification of breast cancer histopathology images. Signal Process. 2016, 122, 1–13. [Google Scholar] [CrossRef]
- Ali, S.; Madabhushi, A. Segmenting multiple overlapping objects via a hybrid active contour model incorporating shape priors: Applications to digital pathology. In Medical Imaging 2011: Image Processing, Proceedings of the SPIE Medical Imaging, Lake Buena Vista, FL, USA, 14–16 February 2011; SPIE: Bellingham, WA, USA, 2011; Volume 7962, pp. 909–921. [Google Scholar]
- Venkataraman, G.; Rycyna, K.; Rabanser, A.; Heinze, G.; Baesens, B.M.; Ananthanarayanan, V.; Paner, G.P.; Barkan, G.A.; Flanigan, R.C.; Wojcik, E.M. Morphometric signature differences in nuclei of Gleason pattern 4 areas in Gleason 7 prostate cancer with differing primary grades on needle biopsy. J. Urol. 2009, 181, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Lal, S.; Desouza, R.; Maneesh, M.; Kanfade, A.; Kumar, A.; Perayil, G.; Alabhya, K.; Chanchal, A.K.; Kini, J. A robust method for nuclei segmentation of H&E stained histopathology images. In Proceedings of the 2020 7th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 27–28 February 2020; pp. 453–458. [Google Scholar]
- Kaushal, C.; Singla, A. Automated segmentation technique with self-driven post-processing for histopathological breast cancer images. CAAI Trans. Intell. Technol. 2020, 5, 294–300. [Google Scholar] [CrossRef]
- Zebari, D.A.; Zeebaree, D.Q.; Abdulazeez, A.M.; Haron, H.; Hamed, H.N.A. Improved threshold based and trainable fully automated segmentation for breast cancer boundary and pectoral muscle in mammogram images. IEEE Access 2020, 8, 203097–203116. [Google Scholar] [CrossRef]
- Kiran, I.; Raza, B.; Ijaz, A.; Khan, M.A. DenseRes-Unet: Segmentation of overlapped/clustered nuclei from multi organ histopathology images. Comput. Biol. Med. 2022, 143, 105267. [Google Scholar] [CrossRef]
- Fatakdawala, H.; Xu, J.; Basavanhally, A.; Bhanot, G.; Ganesan, S.; Feldman, M.; Tomaszewski, J.E.; Madabhushi, A. Expectation–maximization-driven geodesic active contour with overlap resolution (emagacor): Application to lymphocyte segmentation on breast cancer histopathology. IEEE Trans. Biomed. Eng. 2010, 57, 1676–1689. [Google Scholar] [CrossRef]
- Aswathy, M.; Jagannath, M. Performance analysis of segmentation algorithms for the detection of breast cancer. Procedia Comput. Sci. 2020, 167, 666–676. [Google Scholar] [CrossRef]
- Xu, J.; Gong, L.; Wang, G.; Lu, C.; Gilmore, H.; Zhang, S.; Madabhushi, A. Convolutional neural network initialized active contour model with adaptive ellipse fitting for nuclear segmentation on breast histopathological images. J. Med. Imaging 2019, 6, 017501. [Google Scholar] [CrossRef]
- Mouelhi, A.; Sayadi, M.; Fnaiech, F.; Mrad, K.; Romdhane, K.B. Automatic image segmentation of nuclear stained breast tissue sections using color active contour model and an improved watershed method. Biomed. Signal Process. Control 2013, 8, 421–436. [Google Scholar] [CrossRef]
- Ciecholewski, M. Malignant and benign mass segmentation in mammograms using active contour methods. Symmetry 2017, 9, 277. [Google Scholar] [CrossRef]
- Ciecholewski, M. An edge-based active contour model using an inflation/deflation force with a damping coefficient. Expert Syst. Appl. 2016, 44, 22–36. [Google Scholar] [CrossRef]
- Marquez-Neila, P.; Baumela, L.; Alvarez, L. A morphological approach to curvature-based evolution of curves and surfaces. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 36, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef]
- Niaz, A.; Memon, A.A.; Rana, K.; Joshi, A.; Soomro, S.; Kang, J.S.; Choi, K.N. Inhomogeneous image segmentation using hybrid active contours model with application to breast tumor detection. IEEE Access 2020, 8, 186851–186861. [Google Scholar] [CrossRef]
- Djekoune, O.; Benbelkacem, S. Segmentation of the Breast Masses in Mammograms Using Active Contour for Medical Practice: AR Based Surgery. In Proceedings of the Artificial Intelligence and Its Applications: Proceeding of the 2nd International Conference on Artificial Intelligence and Its Applications, Chongqing, China, 28–30 May 2021; Springer Nature: Berlin/Heidelberg, Germany, 2022; Volume 413, p. 447. [Google Scholar]
- Feng, B.; Zhou, H.; Feng, J.; Chen, Y.; Liu, Y.; Yu, T.; Liu, Z.; Long, W. Active contour model of breast cancer DCE-MRI segmentation with an extreme learning machine and a fuzzy C-means cluster. IET Image Process. 2022, 16, 2947–2958. [Google Scholar] [CrossRef]
- Hu, H.; Qiao, S.; Hao, Y.; Bai, Y.; Cheng, R.; Zhang, W.; Zhang, G. Breast cancer histopathological images recognition based on two-stage nuclei segmentation strategy. PLoS ONE 2022, 17, e0266973. [Google Scholar] [CrossRef]
- Michael, E.; Ma, H.; Li, H.; Kulwa, F.; Li, J. Breast cancer segmentation methods: Current status and future potentials. BioMed Res. Int. 2021, 2021, 9962109. [Google Scholar] [CrossRef]
- Paramanandam, M.; O’Byrne, M.; Ghosh, B.; Mammen, J.J.; Manipadam, M.T.; Thamburaj, R.; Pakrashi, V. Automated segmentation of nuclei in breast cancer histopathology images. PLoS ONE 2016, 11, e0162053. [Google Scholar] [CrossRef]
- Zhao, L.; Wan, T.; Feng, H.; Qin, Z. Improved nuclear segmentation on histopathology images using a combination of deep learning and active contour model. In Proceedings of the Neural Information Processing: 25th International Conference, ICONIP 2018, Siem Reap, Cambodia, 13–16 December 2018; Proceedings, Part VI 25; Springer: Berlin/Heidelberg, Germany, 2018; pp. 307–317. [Google Scholar]
- Nelson, A.D.; Krishna, S. An effective approach for the nuclei segmentation from breast histopathological images using star-convex polygon. Procedia Comput. Sci. 2023, 218, 1778–1790. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Schmidt, U.; Weigert, M.; Broaddus, C.; Myers, G. Cell detection with star-convex polygons. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; Proceedings, Part II 11; Springer: Berlin/Heidelberg, Germany, 2018; pp. 265–273. [Google Scholar]
- Xu, Y.; Dang, H.; Tang, L. KACM: A KIS-awared active contour model for low-contrast image segmentation. Expert Syst. Appl. 2024, 255, 124767. [Google Scholar] [CrossRef]
- Hajjami, J.; Napoléon, T.; Alfalou, A. Adaptation of Koschmieder dehazing model for underwater marker detection. In Proceedings of the Pattern Recognition and Tracking XXXI; SPIE: Bellingham, WA, USA, 2020; Volume 11400, p. 1140003. [Google Scholar]
- Mahanta, L.B.; Hussain, E.; Das, N.; Kakoti, L.; Chowdhury, M. IHC-Net: A fully convolutional neural network for automated nuclear segmentation and ensemble classification for Allred scoring in breast pathology. Appl. Soft Comput. 2021, 103, 107136. [Google Scholar] [CrossRef]
- Kaladevi, P.; Kanimozhi, N.; Nirmala, B.; Sivasankari, R. Morpho-contour exponential estimation algorithm for predicting breast tumor growth from MRI imagery. Int. J. Inf. Technol. 2024, 1–16. [Google Scholar] [CrossRef]
- Majanga, V.; Viriri, S. Dental images’ segmentation using threshold connected component analysis. Comput. Intell. Neurosci. 2021, 2021, 2921508. [Google Scholar] [CrossRef]
- Hussain, Z.; Gimenez, F.; Yi, D.; Rubin, D. Differential data augmentation techniques for medical imaging classification tasks. In Proceedings of the AMIA Annual Symposium Proceedings, Washington, DC, USA, 4–8 November 2017; American Medical Informatics Association: Washington, DC, USA, 2017; Volume 2017, p. 979. [Google Scholar]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Garcea, F.; Serra, A.; Lamberti, F.; Morra, L. Data augmentation for medical imaging: A systematic literature review. Comput. Biol. Med. 2023, 152, 106391. [Google Scholar] [CrossRef]
- Araújo, T.; Aresta, G.; Castro, E.; Rouco, J.; Aguiar, P.; Eloy, C.; Polónia, A.; Campilho, A. Classification of breast cancer histology images using convolutional neural networks. PLoS ONE 2017, 12, e0177544. [Google Scholar] [CrossRef]
- Veta, M.; Pluim, J.P.; Van Diest, P.J.; Viergever, M.A. Breast cancer histopathology image analysis: A review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411. [Google Scholar] [CrossRef]
- Macenko, M.; Niethammer, M.; Marron, J.S.; Borland, D.; Woosley, J.T.; Guan, X.; Schmitt, C.; Thomas, N.E. A method for normalizing histology slides for quantitative analysis. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 1107–1110. [Google Scholar]
- Elmoataz, A.; Schüpp, S.; Clouard, R.; Herlin, P.; Bloyet, D. Using active contours and mathematical morphology tools for quantification of immunohistochemical images. Signal Process. 1998, 71, 215–226. [Google Scholar] [CrossRef]
- Osher, S.; Sethian, J.A. Fronts propagating with curvature-dependent speed: Algorithms based on Hamilton-Jacobi formulations. J. Comput. Phys. 1988, 79, 12–49. [Google Scholar] [CrossRef]
- Sethian, J.A. Level Set Methods: Evolving Interfaces in Geometry, Fluid Mechanics, Computer Vision, and Materials Science. In Cambridge Monographs on Applied and Computational Mathematics; Cambridge University Press: Cambridge, UK, 1996; Volume 3. [Google Scholar]
- Kichenassamy, S.; Kumar, A.; Olver, P.; Tannenbaum, A.; Yezzi, A. Gradient flows and geometric active contour models. In Proceedings of the IEEE International Conference on Computer Vision, Cambridge, MA, USA, 20–23 June 1995; pp. 810–815. [Google Scholar]
- Caselles, V.; Kimmel, R.; Sapiro, G. Geodesic active contours. Int. J. Comput. Vis. 1997, 22, 61–79. [Google Scholar] [CrossRef]
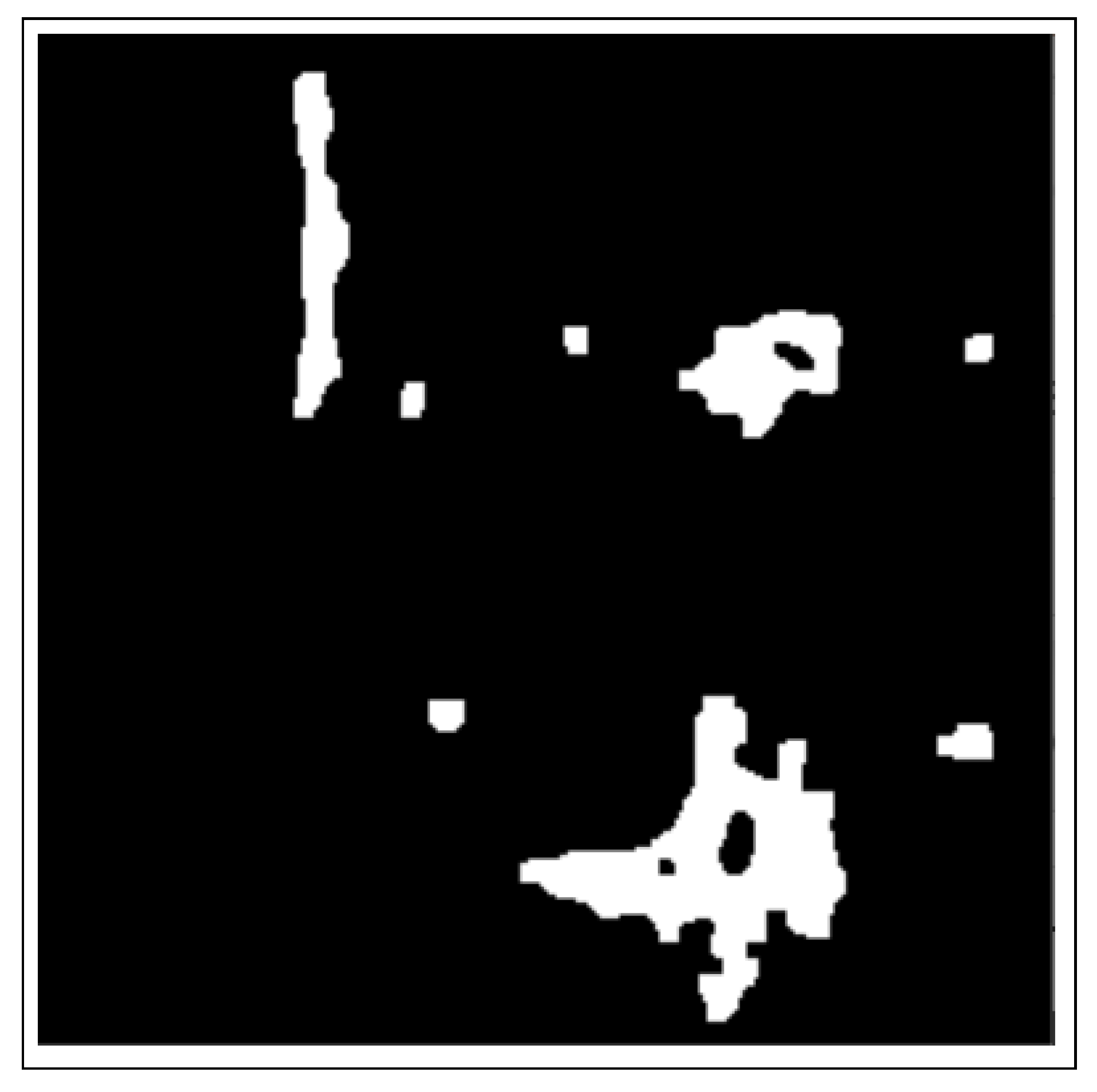

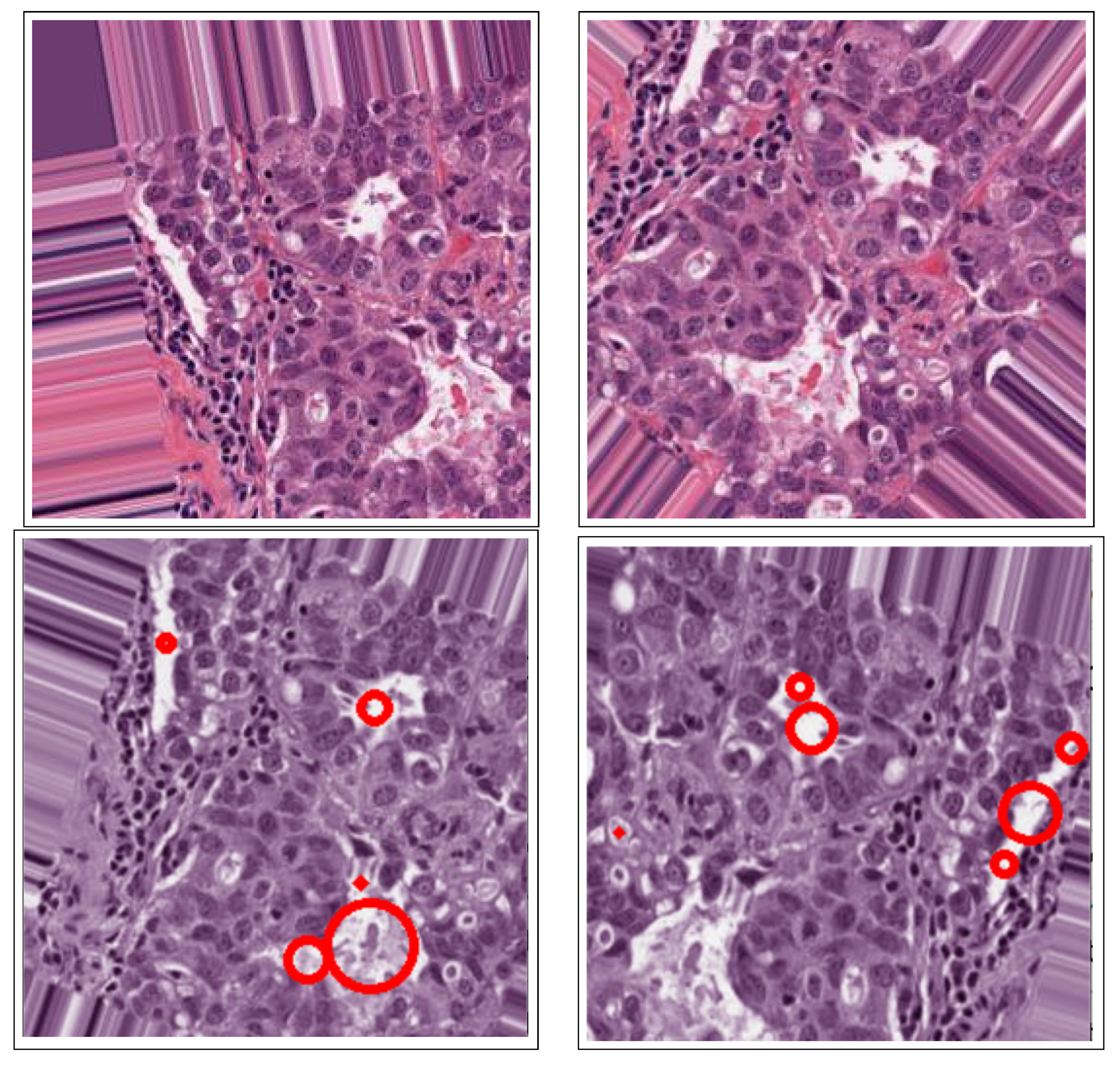
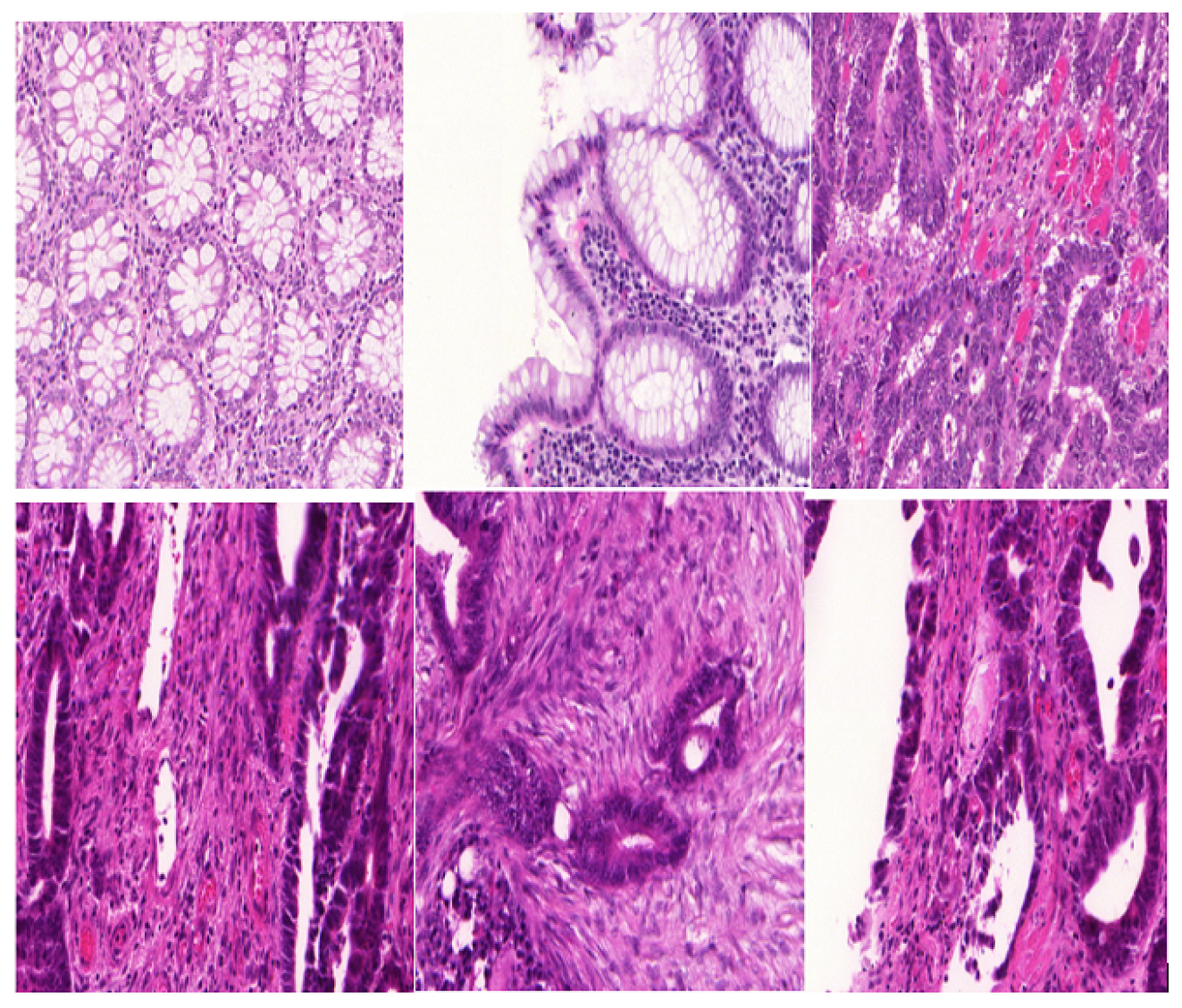
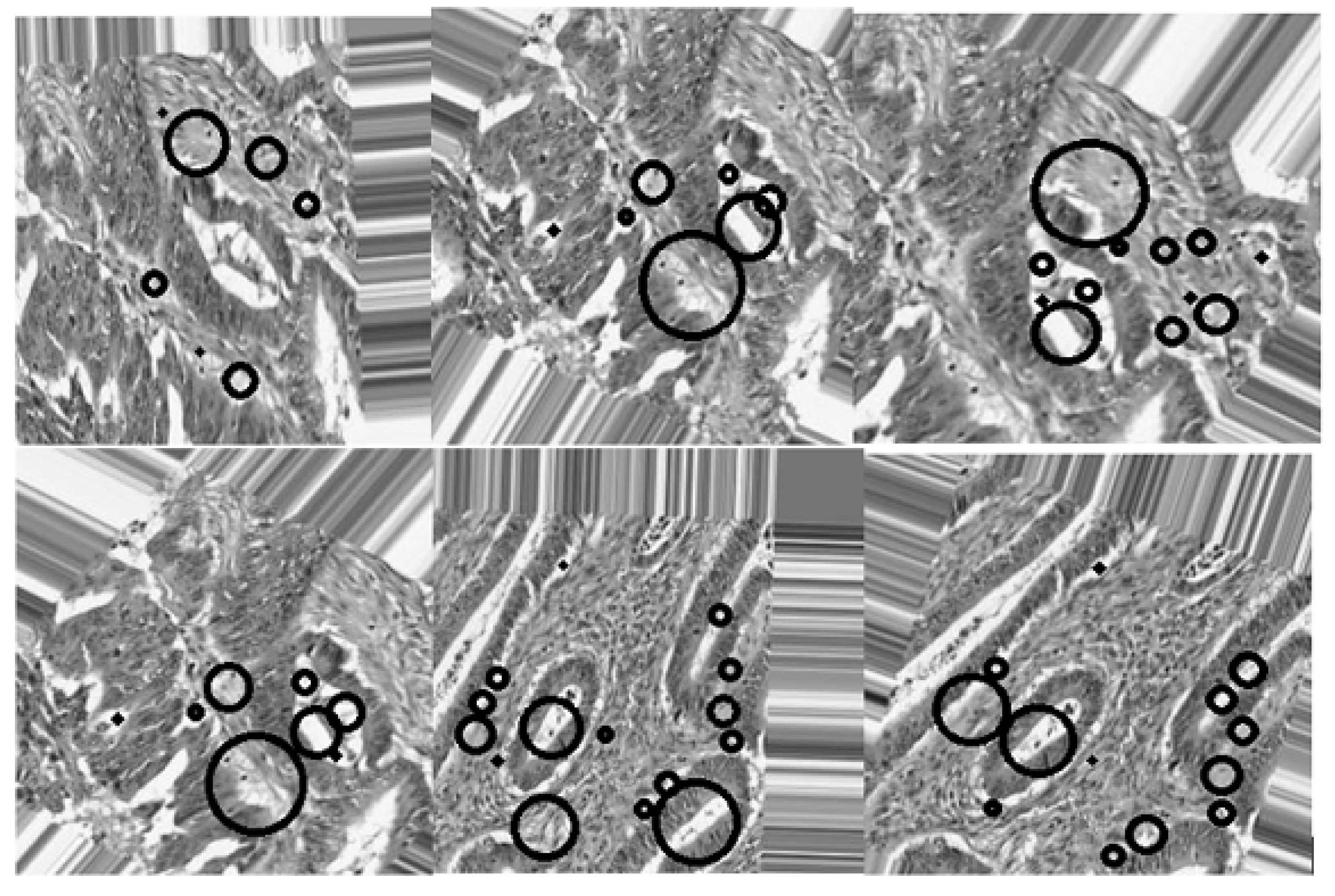
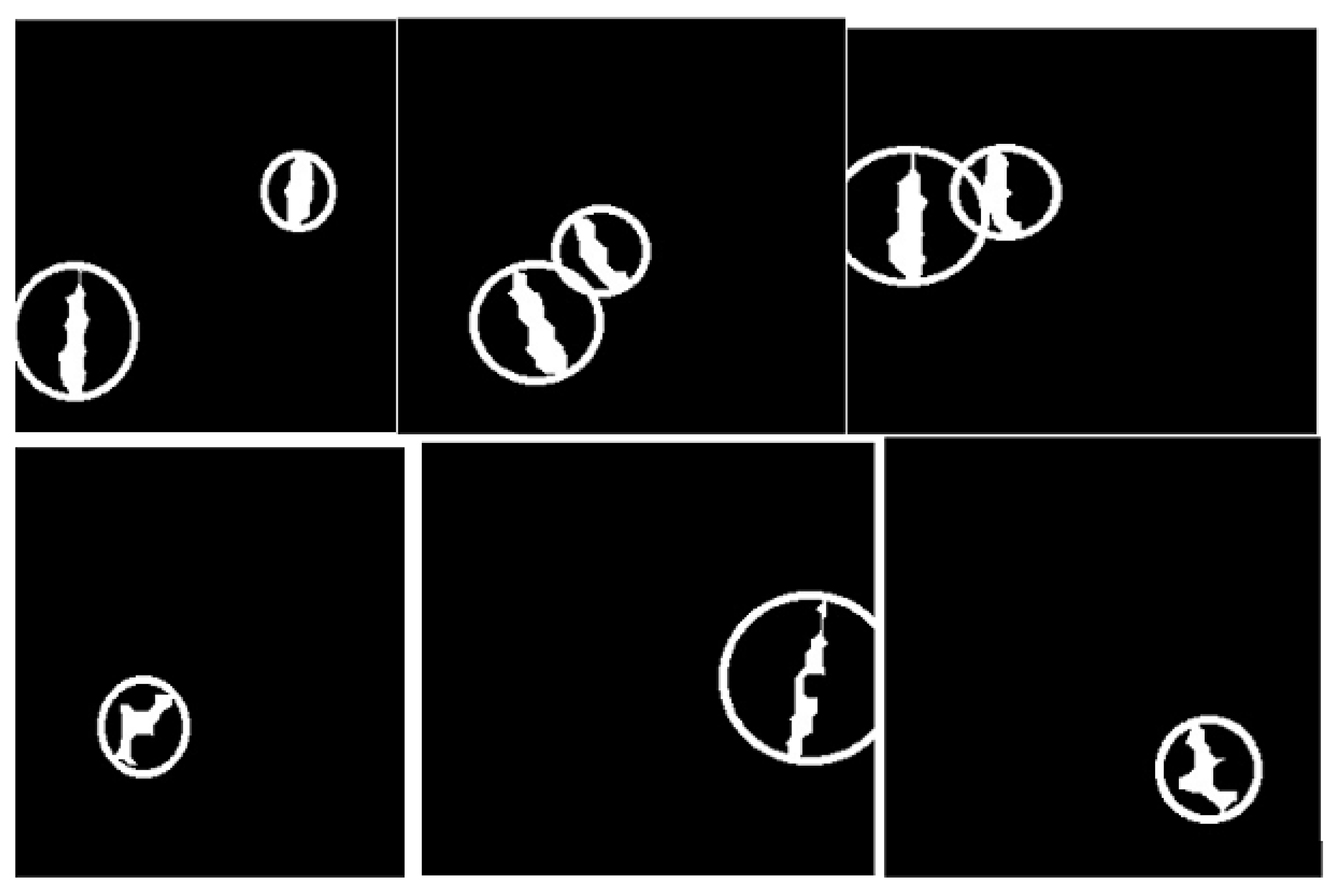
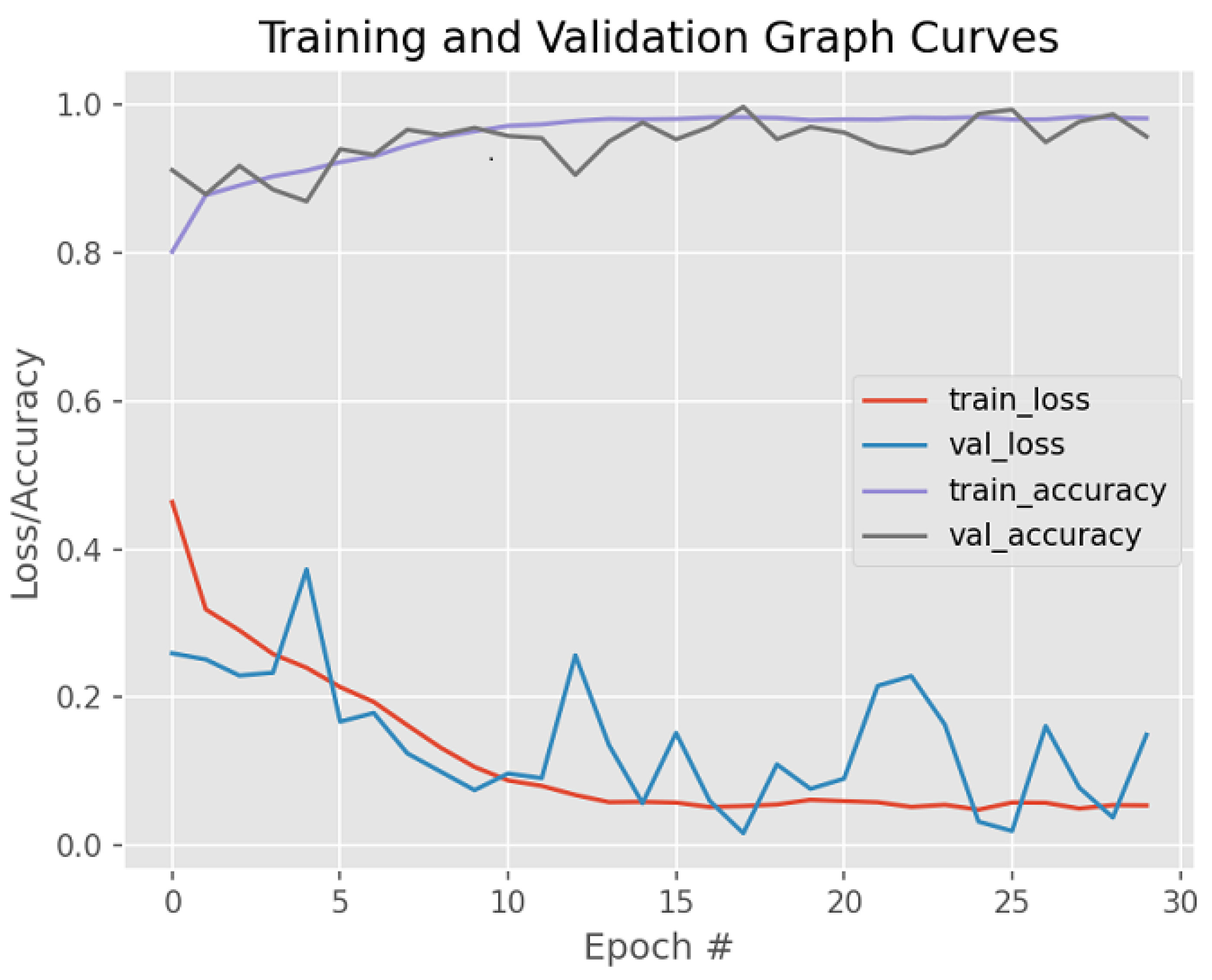
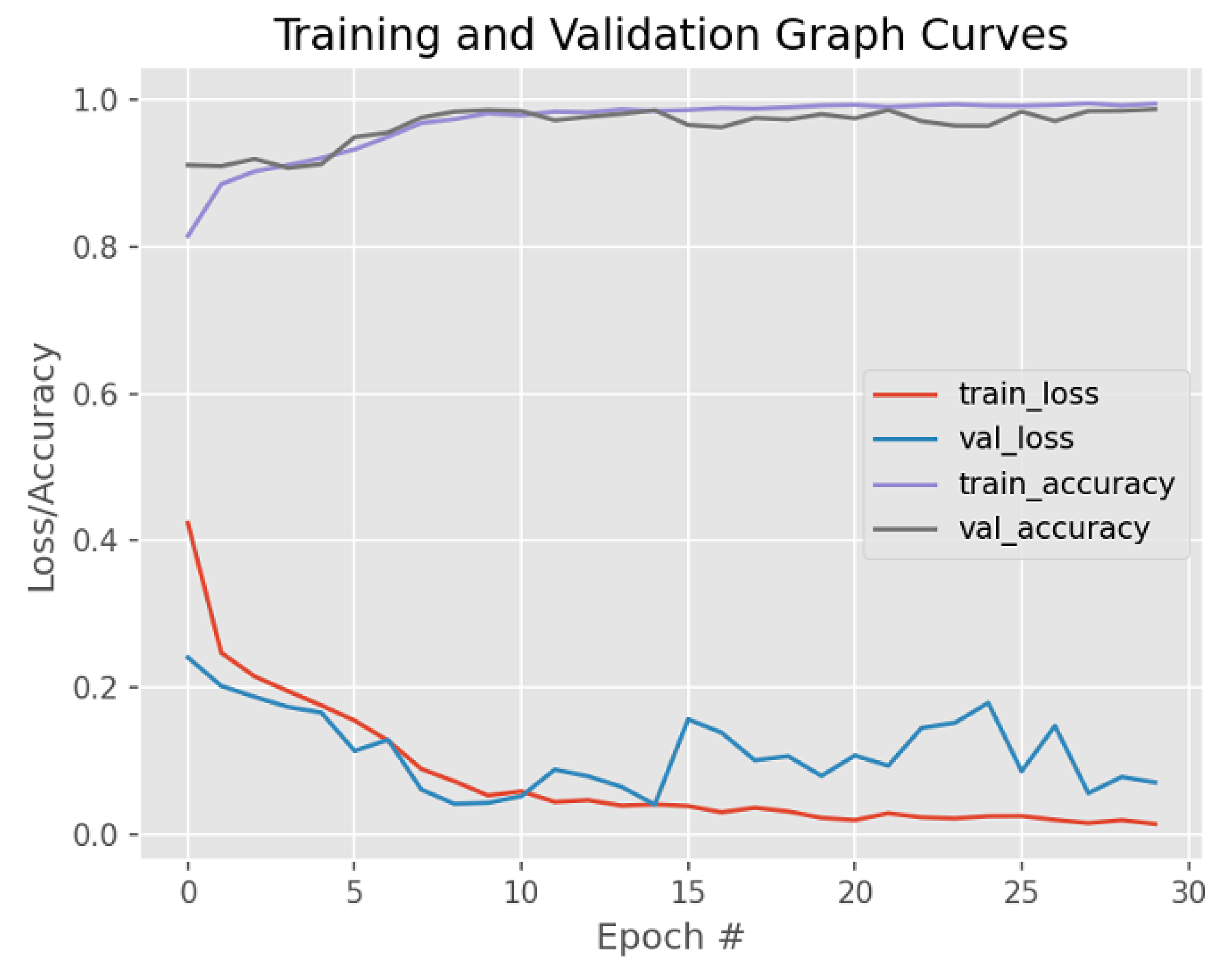
| Dataset | Augmented Images | After Implementation of ACCCA Method |
|---|---|---|
| Accuracy | 94.0% | 98.71% |
| Reference Authors | Methods | Accuracy |
|---|---|---|
| Mahanta et al. [30] | Deep-learning-based nuclei segmentation + ensemble classification scheme | 98.24% |
| Ronneberger et al. [26] | U-Net | 92.03% |
| Fatakdawala et al. [10] | Expectation max driven geodesic active contours with overlap resolution | 86% |
| Xu et al. [12] | CNN active contour model with adaptive ellipse fitting | 85.71% |
| Mouelhi et al. [13] | Colour active contour model + improved watershed method | 97% |
| Niaz et al. [18] | Inhomogeneous image segmentation using hybrid active contour model | 98.3% |
| Kaladevi et al. [31] | Morpho-contour exponential estimation algorithm | 93.12% |
| Hu et al. [21] | Two-stage nuclei segmentation strategy | 92.5% |
| Paramanandam et al. [23] | Tensor voting + Loopy Back Propagation algorithm | 93.0% |
| Zhao et al. [24] | Deep CNN + active contour method | 85.0% |
| Nelson et al. [25] | Star-convex polygon approach + non-maximum suppression technique | 66% |
| Proposed Method | Active contours + connected components analyis (ACCCA) | 98.71% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majanga, V.; Mnkandla, E.; Wang, Z.; Moulla, D.K. Active Contours Connected Component Analysis Segmentation Method of Cancerous Lesions in Unsupervised Breast Histology Images. Bioengineering 2025, 12, 642. https://doi.org/10.3390/bioengineering12060642
Majanga V, Mnkandla E, Wang Z, Moulla DK. Active Contours Connected Component Analysis Segmentation Method of Cancerous Lesions in Unsupervised Breast Histology Images. Bioengineering. 2025; 12(6):642. https://doi.org/10.3390/bioengineering12060642
Chicago/Turabian StyleMajanga, Vincent, Ernest Mnkandla, Zenghui Wang, and Donatien Koulla Moulla. 2025. "Active Contours Connected Component Analysis Segmentation Method of Cancerous Lesions in Unsupervised Breast Histology Images" Bioengineering 12, no. 6: 642. https://doi.org/10.3390/bioengineering12060642
APA StyleMajanga, V., Mnkandla, E., Wang, Z., & Moulla, D. K. (2025). Active Contours Connected Component Analysis Segmentation Method of Cancerous Lesions in Unsupervised Breast Histology Images. Bioengineering, 12(6), 642. https://doi.org/10.3390/bioengineering12060642







