WHA-Net: A Low-Complexity Hybrid Model for Accurate Pseudopapilledema Classification in Fundus Images
Abstract
1. Introduction
- Construction of a multi-source dataset comprising normal optic discs, true optic disc edema, and pseudopapilledema images.
- Proposal of the WHA-Net lightweight architecture, which delivers competitive classification accuracy without pretraining, despite limited pseudopapilledema data availability.
- To enhance vessel and optic disc detail feature extraction while mitigating image noise interference, we introduced a wavelet convolution module that performs multi-level wavelet transforms and convolution on images, capturing richer image information compared to standard convolutional operations. Additionally, the model integrates a HAIR module to strengthen the perception of critical features such as vascular morphology, hemorrhages, and exudates, while maintaining low computational complexity.
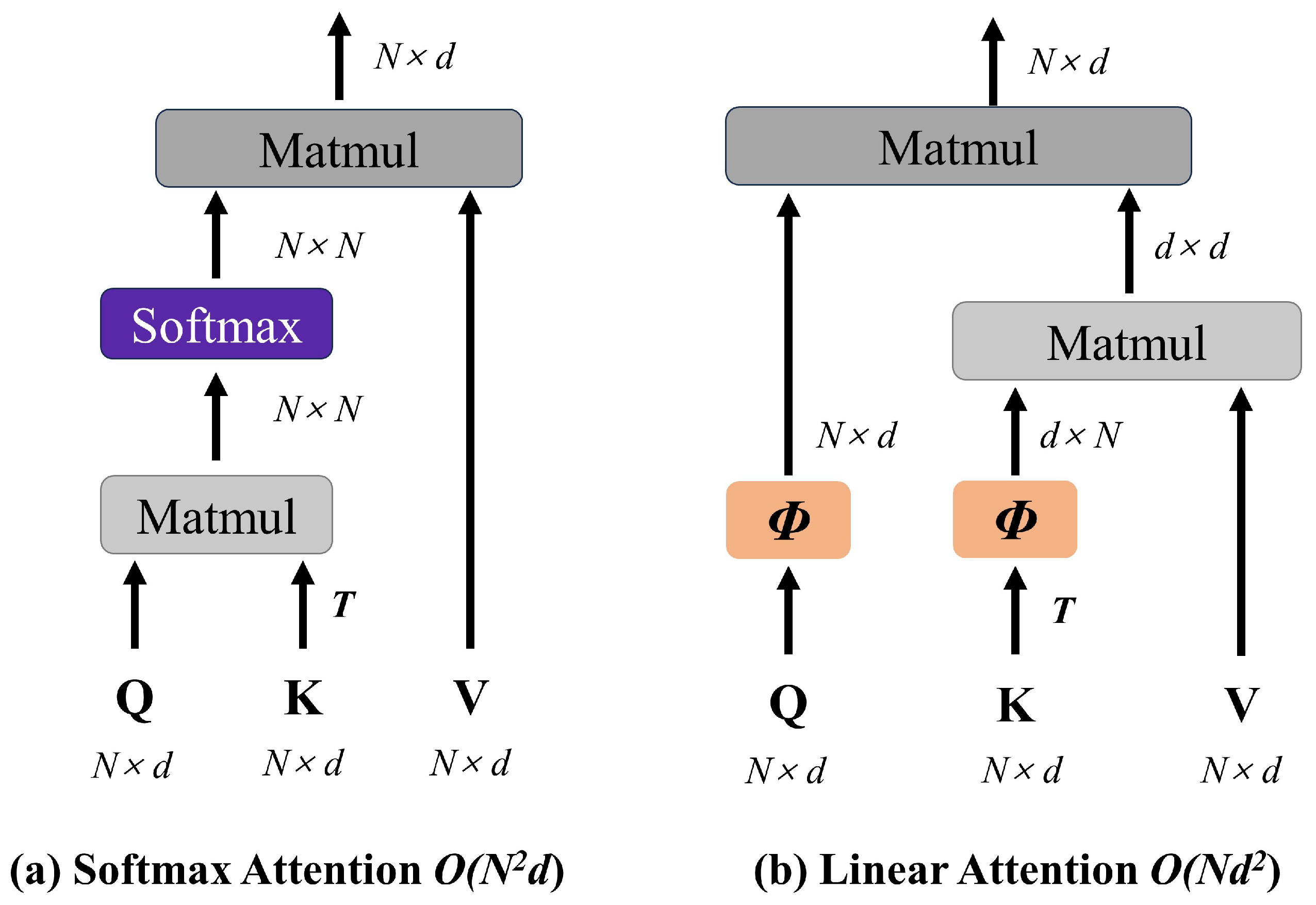
- To address the high computational complexity of self-attention mechanisms, we propose the Agent-MViT block, which replaces the Softmax attention in the original transformer module with agent attention. This achieves linear-complexity representation of global features, including vascular distribution patterns, optic disc color, and edema extent, while reducing computational overhead.
2. Related Works
2.1. Fundus Diseases Classification
2.2. CNN–Transformer Hybrid Models
2.3. Attention Mechanisms in CV
2.4. Wavelet Transforms
3. Methods
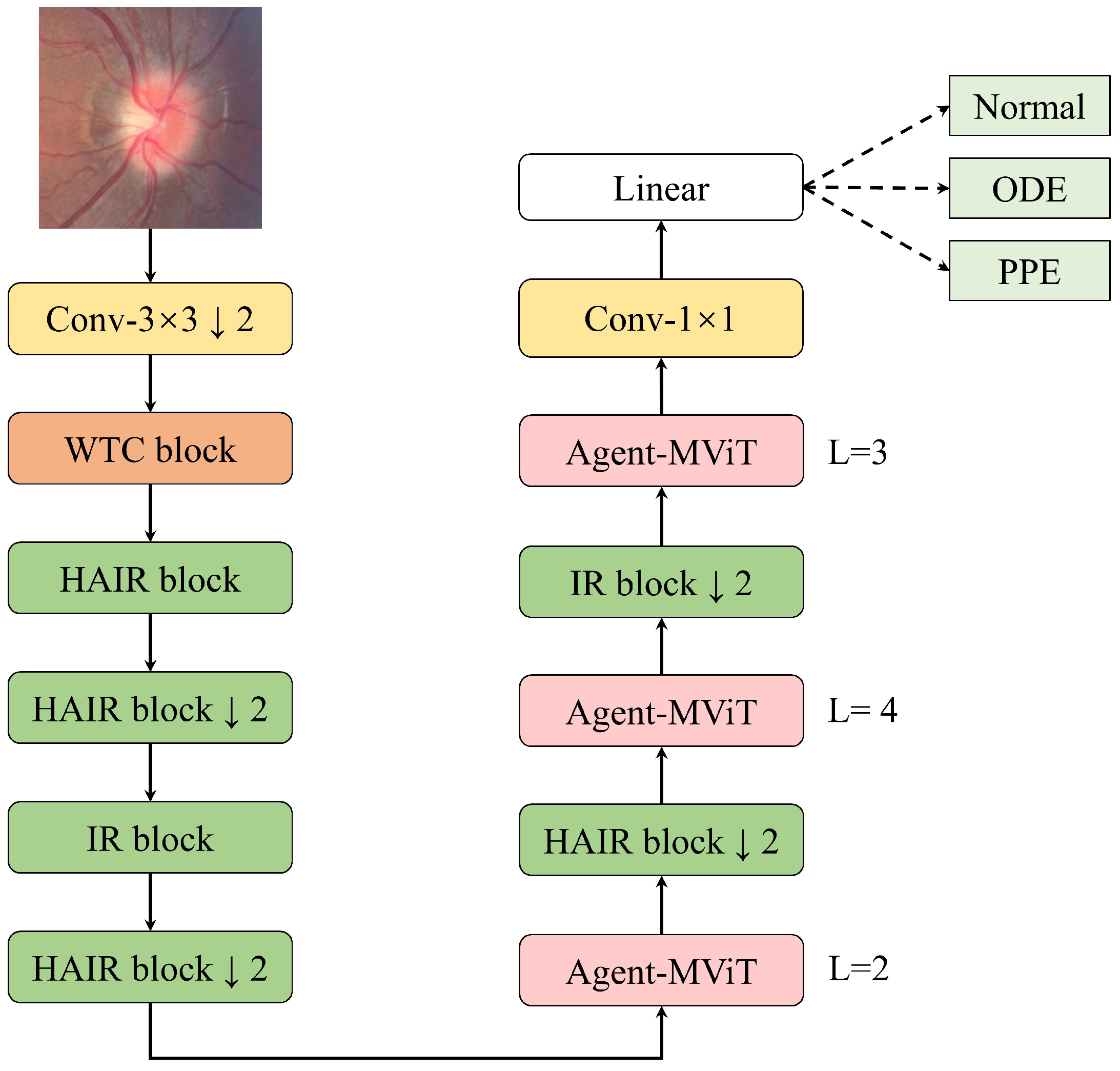
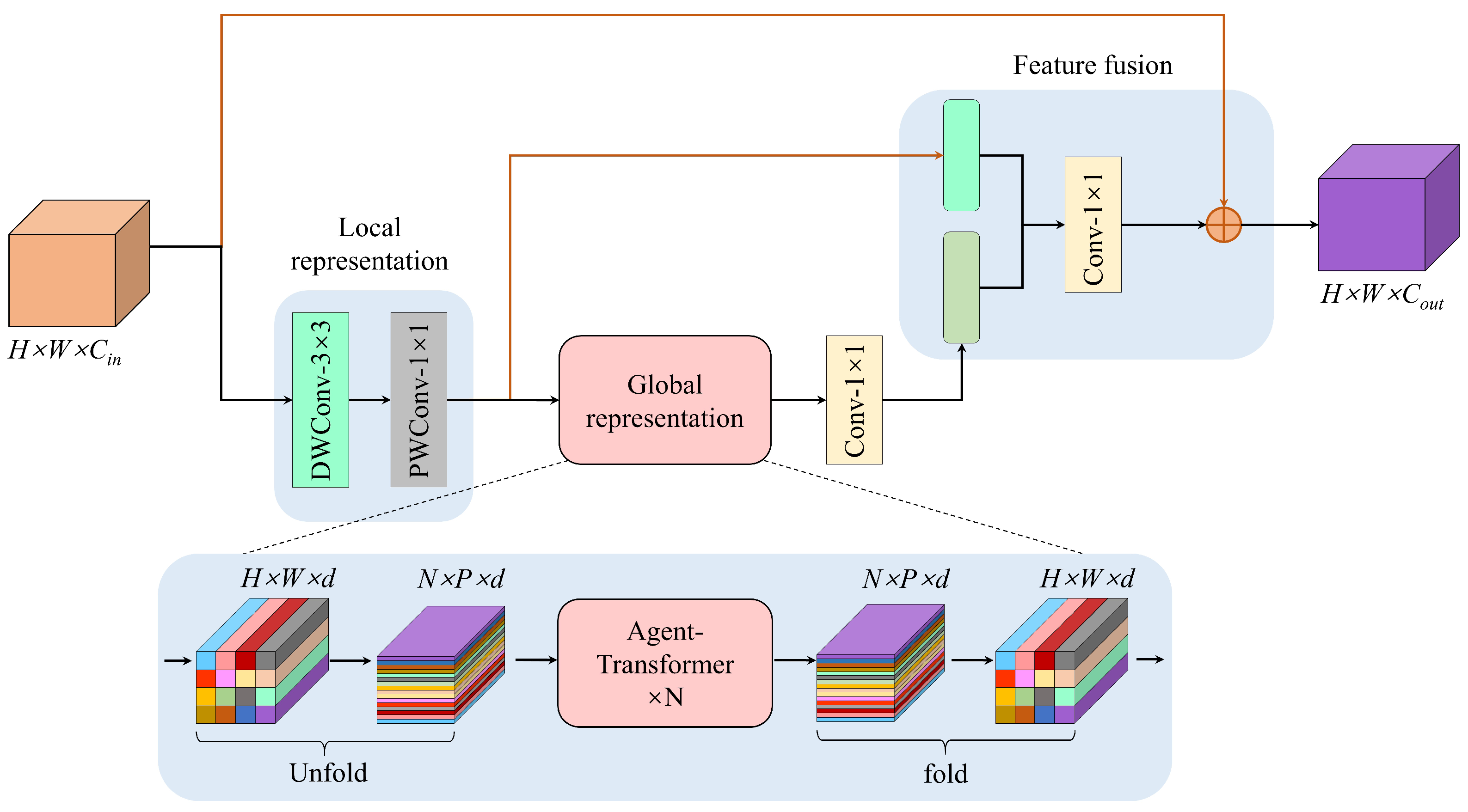
3.1. Architecture of the WHA-Net
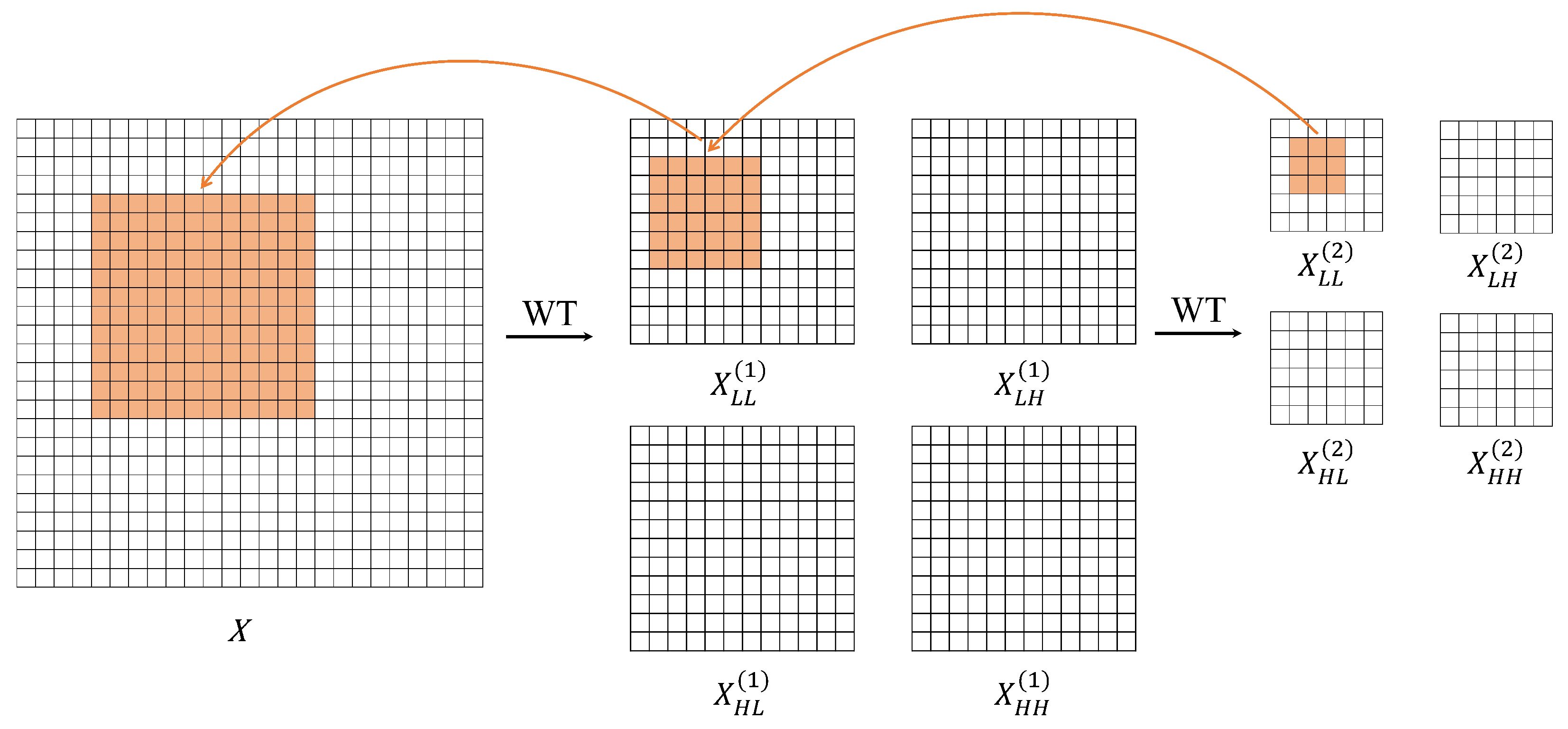
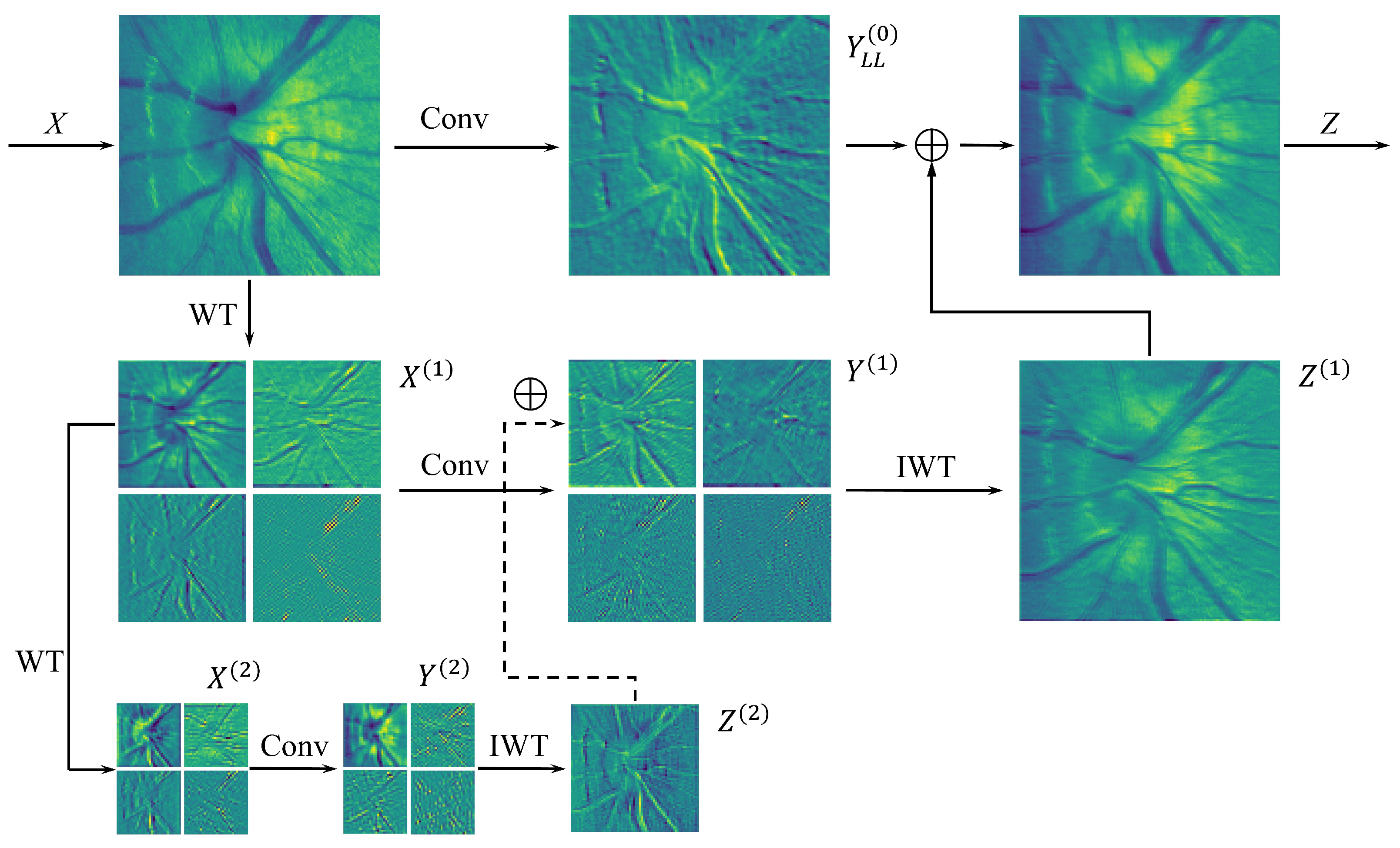
3.2. Wavelet Convolution (WTC) Block
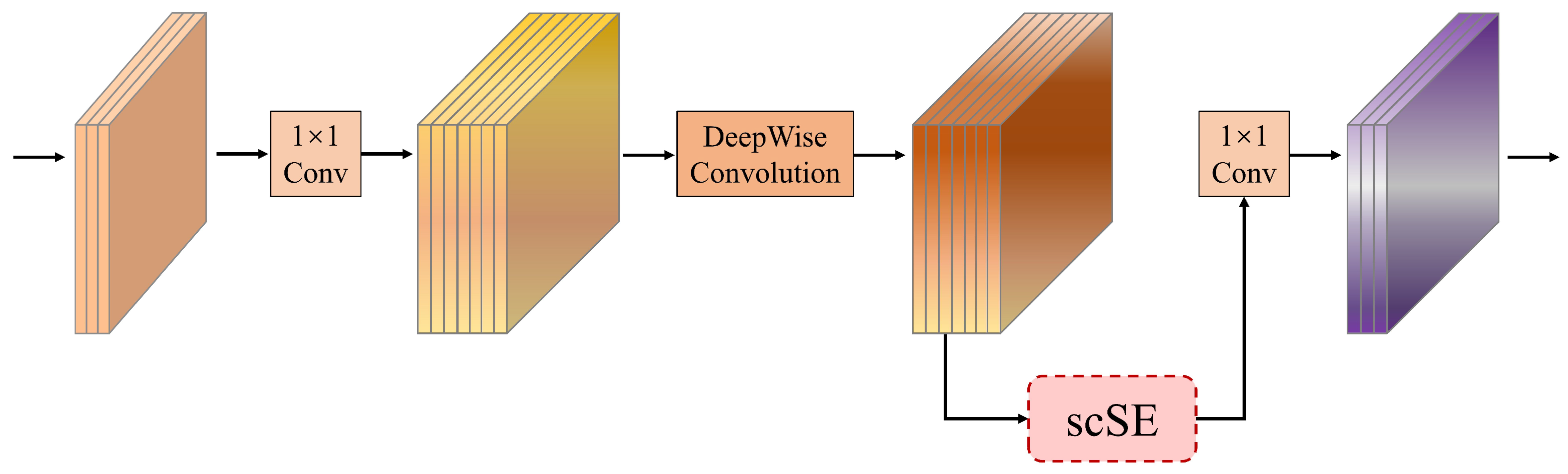
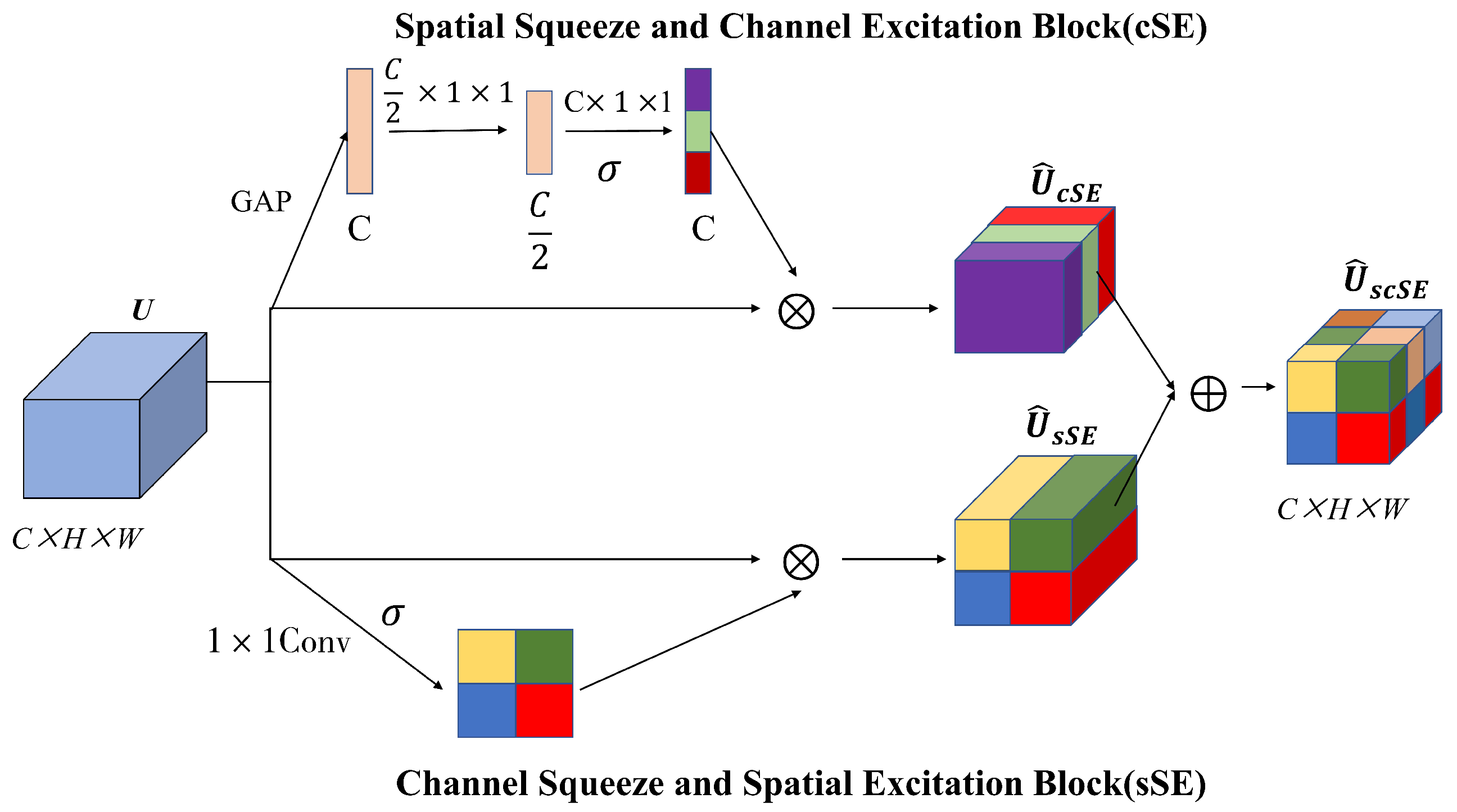
3.3. Hybrid Attention Inverted Residual (HAIR) Block
3.4. Agent-MViT Block
4. Experiments and Results
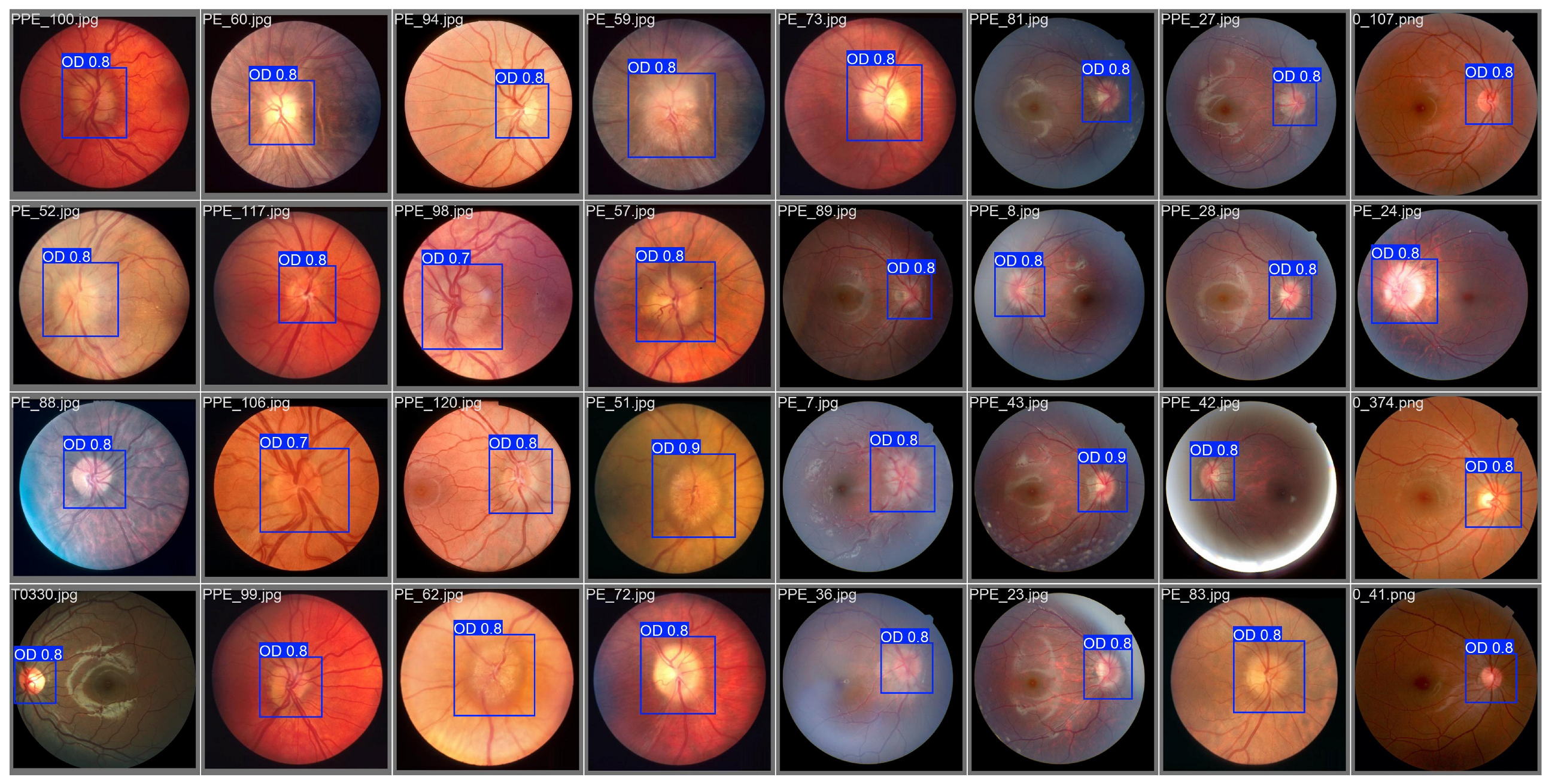
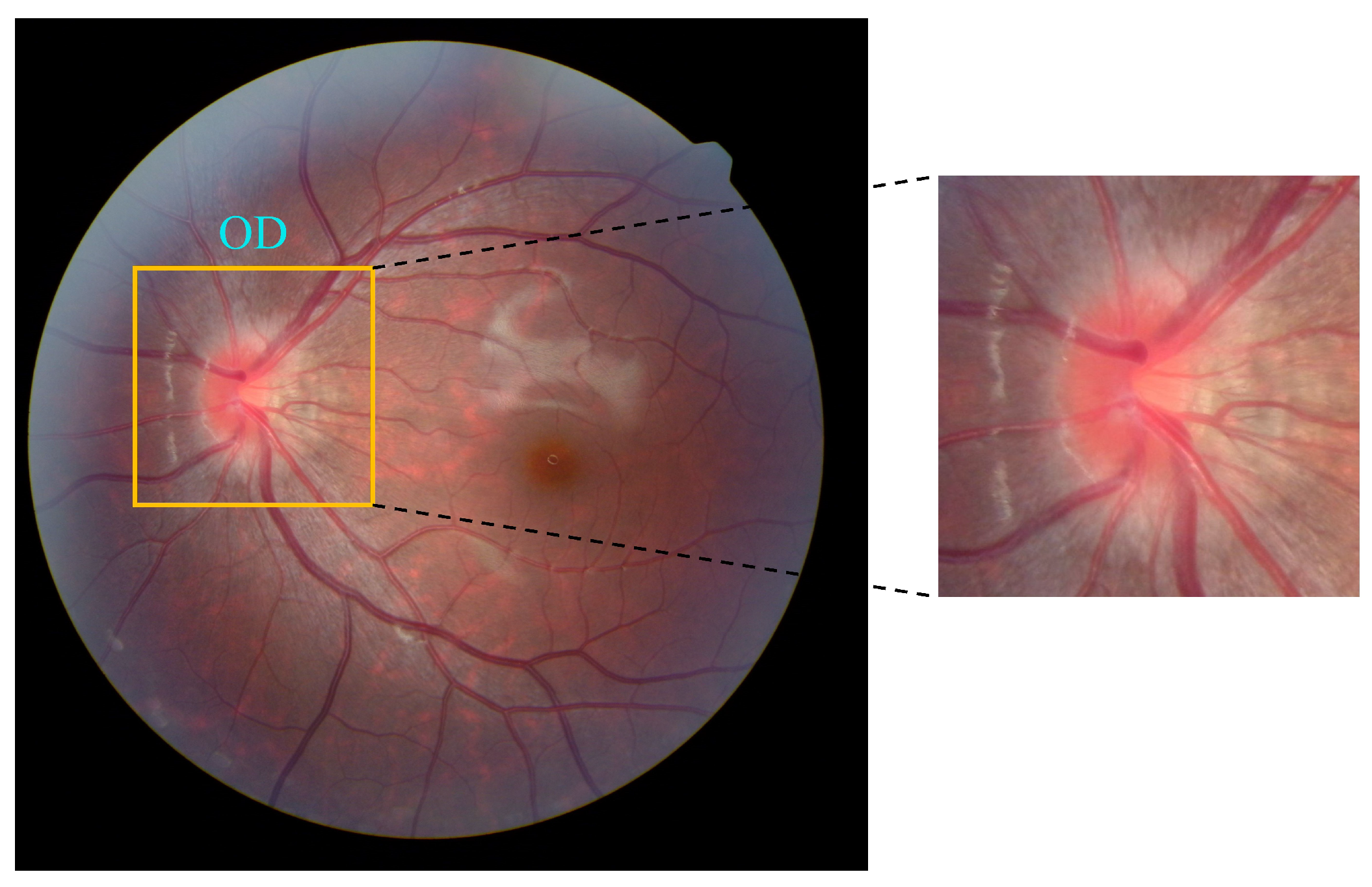
4.1. Dataset
4.2. Evaluation Metrics
4.3. Experimental Settings
4.4. Classification Performance of WHA-Net
4.5. Comparison with Other Methods
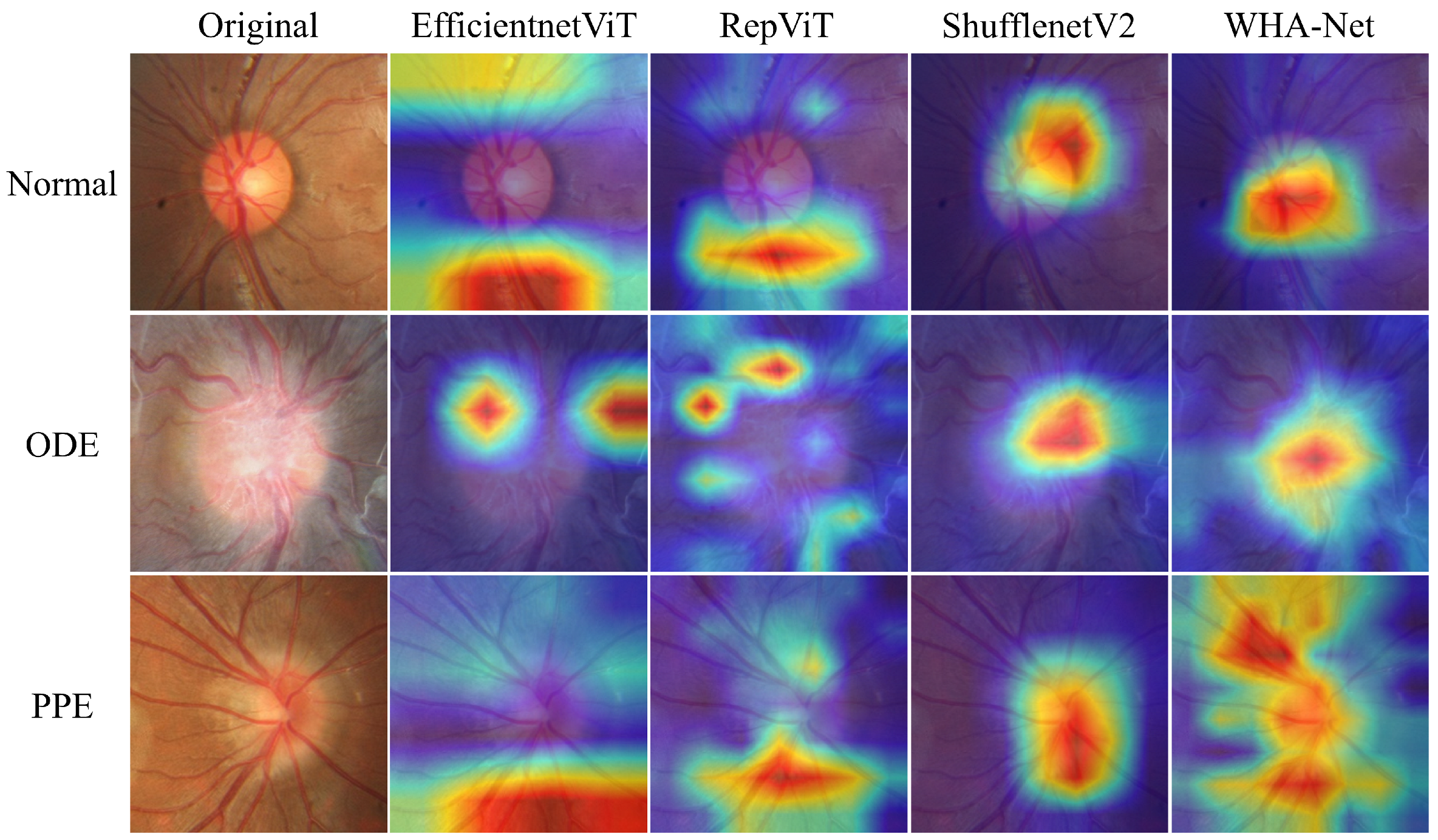
4.6. Explaining Model with Grad-CAM
4.7. Ablation Study
5. Discussion
- Expand the challenging case database: Systematically collect fundus images covering all ODE progression stages, with emphasis on early-stage cases exhibiting overlapping features with PPE to enhance the model’s sensitivity to subtle discriminative patterns.
- Confidence threshold alert: When prediction probability falls below 95%, activate a “Low Diagnostic Confidence - Clinical Review Recommended” prompt, guiding clinicians and patients toward supplementary diagnostic verification.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, G.T.; Volpe, N.J.; Galetta, S.L. CHAPTER 6—Optic disc swelling: Papilledema and other causes. In Neuro-Ophthalmology, 2nd ed.; Liu, G.T., Volpe, N.J., Galetta, S.L., Eds.; W.B. Saunders: London, UK, 2010; pp. 199–236. [Google Scholar]
- Yan, Y.; Liao, Y.J. Updates on ophthalmic imaging features of optic disc drusen, papilledema, and optic disc edema. Curr. Opin. Neurol. 2021, 34, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Sefi-Yurdakul, N. True or pseudo optic disc edema: Clinically-based approach to the differential diagnosis. Int. Ophthalmol. 2024, 44, 384. [Google Scholar] [CrossRef] [PubMed]
- Fard, M.A.; Sahraiyan, A.; Jalili, J.; Hejazi, M.; Suwan, Y.; Ritch, R.; Subramanian, P.S. Optical Coherence Tomography Angiography in Papilledema Compared with Pseudopapilledema. Investig. Ophthalmol. Vis. Sci. 2019, 60, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.N.; Diehl, M.L.; Hamm, C.W.; Sommerville, D.N.; Petroski, G.F. Differentiating Optic Disc Edema from Optic Nerve Head Drusen on Optical Coherence Tomography. Arch. Ophthalmol. 2009, 127, 45–49. [Google Scholar] [CrossRef]
- Kohane, I. AI for the Eye - Automated Assistance for Clinicians Screening for Papilledema. N. Engl. J. Med. 2020, 382, 1760–1761. [Google Scholar] [CrossRef]
- Rambabu, L.; Smith, B.G.; Tumpa, S.; Kohler, K.; Kolias, A.G.; Hutchinson, P.J.; Bashford, T. Artificial intelligence-enabled ophthalmoscopy for papilledema: A systematic review protocol. Int. J. Surg. Protoc. 2024, 28, 27–30. [Google Scholar] [CrossRef]
- Chang, M.Y.; Heidary, G.; Beres, S.; Pineles, S.L.; Gaier, E.D.; Gise, R.; Reid, M.; Avramidis, K.; Rostami, M.; Narayanan, S. Artificial Intelligence to Differentiate Pediatric Pseudopapilledema and True Papilledema on Fundus Photographs. Ophthalmol. Sci. 2024, 4, 100496. [Google Scholar] [CrossRef]
- Chen, C.; Mat Isa, N.A.; Liu, X. A review of convolutional neural network based methods for medical image classification. Comput. Biol. Med. 2025, 185, 109507. [Google Scholar] [CrossRef]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in medical imaging: A survey. Med. Image Anal. 2023, 88, 102802. [Google Scholar] [CrossRef]
- Farahat, Z.; Zrira, N.; Souissi, N.; Bennani, Y.; Bencherif, S.; Benamar, S.; Belmekki, M.; Ngote, M.N.; Megdiche, K. Diabetic retinopathy screening through artificial intelligence algorithms: A systematic review. Surv. Ophthalmol. 2024, 69, 707–721. [Google Scholar] [CrossRef]
- Ashtari-Majlan, M.; Dehshibi, M.M.; Masip, D. Glaucoma diagnosis in the era of deep learning: A survey. Expert Syst. Appl. 2024, 256, 124888. [Google Scholar] [CrossRef]
- Hassan, B.; Raja, H.; Hassan, T.; Akram, M.U.; Raja, H.; Abd-Alrazaq, A.A.; Yousefi, S.; Werghi, N. A comprehensive review of artificial intelligence models for screening major retinal diseases. Artif. Intell. Rev. 2024, 57, 111. [Google Scholar] [CrossRef]
- Cen, L.P.; Ji, J.; Lin, J.W.; Ju, S.T.; Lin, H.J.; Li, T.P.; Wang, Y.; Yang, J.F.; Liu, Y.F.; Tan, S.; et al. Automatic detection of 39 fundus diseases and conditions in retinal photographs using deep neural networks. Nat. Commun. 2021, 12, 4828. [Google Scholar] [CrossRef]
- Łajczak, P.M.; Sirek, S.; Wyględowska-Promieńska, D. Unveiling AI’s role in papilledema diagnosis from fundus images: A systematic review with diagnostic test accuracy meta-analysis and comparison of human expert performance. Comput. Biol. Med. 2025, 184, 109350. [Google Scholar] [CrossRef]
- Ahn, J.M.; Kim, S.; Ahn, K.S.; Cho, S.H.; Kim, U.S. Accuracy of machine learning for differentiation between optic neuropathies and pseudopapilledema. BMC Ophthalmol. 2019, 19, 178. [Google Scholar] [CrossRef]
- Milea, D.; Najjar, R.P.; Zhubo, J.; Ting, D.; Suwan, Y. Artificial Intelligence to Detect Papilledema from Ocular Fundus Photographs. New Engl. J. Med. 2020, 382, 1687–1695. [Google Scholar] [CrossRef]
- Biousse, V.; Newman, N.J.; Najjar, R.P.; Vasseneix, C.; Xu, X.; Ting, D.S.; Milea, L.B.; Hwang, J.M.; Kim, D.H.; Yang, H.K.; et al. Optic Disc Classification by Deep Learning versus Expert Neuro-Ophthalmologists. Ann. Neurol. 2020, 88, 785–795. [Google Scholar] [CrossRef]
- Vasseneix, C.; Najjar, R.P.; Xu, X.; Tang, Z.; Milea, D. Accuracy of a Deep Learning System for Classification of Papilledema Severity on Ocular Fundus Photographs. Neurology 2021, 97, e369–e377. [Google Scholar] [CrossRef]
- Saba, T.; Akbar, S.; Kolivand, H.; Ali Bahaj, S. Automatic detection of papilledema through fundus retinal images using deep learning. Microsc. Res. Tech. 2021, 84, 3066–3077. [Google Scholar] [CrossRef]
- Liu, K.; Liu, S.; Tan, X.; Li, W.; Wang, L.; Li, X.; Xu, X.; Fu, Y.; Liu, X.; Hong, J.; et al. Deep learning system for distinguishing optic neuritis from non-arteritic anterior ischemic optic neuropathy at acute phase based on fundus photographs. Front. Med. 2023, 10, 1188542. [Google Scholar] [CrossRef]
- Salaheldin, A.; Abdelwahed, M.; Talaat, M.; Saleh, N. Deep Learning-Based Automated Detection and Grading of Papilledema From OCT Images: A Promising Approach for Improved Clinical Diagnosis and Management. Int. J. Imaging Syst. Technol. 2024, 34, e23133. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Haruna, Y.; Qin, S.; Adama Chukkol, A.H.; Yusuf, A.A.; Bello, I.; Lawan, A. Exploring the synergies of hybrid convolutional neural network and Vision Transformer architectures for computer vision: A survey. Eng. Appl. Artif. Intell. 2025, 144, 110057. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Howard, A.; Sandler, M.; Chen, B.; Wang, W.; Chen, L.C.; Tan, M.; Chu, G.; Vasudevan, V.; Zhu, Y.; Pang, R.; et al. Searching for mobileNetV3. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar]
- Ma, N.; Zhang, X.; Zheng, H.T.; Sun, J. ShuffleNet V2: Practical Guidelines for Efficient CNN Architecture Design. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Mehta, S.; Rastegari, M. Mobilevit: Light-Weight, General-Purpose, and Mobile-Friendly Vision Transformer. In Proceedings of the ICLR 2022—10th International Conference on Learning Representations, Virtual, Online, 25 April 2022. [Google Scholar]
- Liu, X.; Peng, H.; Zheng, N.; Yang, Y.; Hu, H.; Yuan, Y. EfficientViT: Memory Efficient Vision Transformer with Cascaded Group Attention. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 14420–14430. [Google Scholar]
- Zhang, H.; Hu, W.; Wang, X. ParC-Net: Position Aware Circular Convolution with Merits from ConvNets and Transformer. In Proceedings of the Computer Vision–ECCV 2022, Tel Aviv, Israel, 23–27 October 22022; Avidan, S., Brostow, G., Cissé, M., Farinella, G.M., Hassner, T., Eds.; Spring: Cham, Switzerland, 2022; pp. 613–630. [Google Scholar]
- Guo, M.H.; Xu, T.X.; Liu, J.J.; Liu, Z.N.; Jiang, P.T.; Mu, T.J.; Zhang, S.H.; Martin, R.R.; Cheng, M.M.; Hu, S.M. Attention mechanisms in computer vision: A survey. Comput. Vis. Media 2022, 8, 331–368. [Google Scholar] [CrossRef]
- Bastola, A.; Luitel, N.; Wang, H.; Paudel, D.P.; Poudel, R.; Razi, A. RobustFormer: Noise-Robust Pre-training for images and videos. arXiv 2024, arXiv:2411.13040. [Google Scholar]
- Wu, Z.; Sun, C.; Xuan, H.; Liu, G.; Yan, Y. WaveFormer: Wavelet Transformer for Noise-Robust Video Inpainting. In Proceedings of the AAAI Conference on Artificial Intelligence, Vancouver, BC, Canada, 26–27 February 2024; Volume 38, pp. 6180–6188. [Google Scholar]
- Finder, S.E.; Amoyal, R.; Treister, E.; Freifeld, O. Wavelet Convolutions for Large Receptive Fields. In Proceedings of the European Conference on Computer Vision, Malmö, Sweden, 29 September–4 October 2024. [Google Scholar]
- Roy, A.G.; Navab, N.; Wachinger, C. Concurrent Spatial and Channel Squeeze & Excitation in Fully Convolutional Networks. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: Proceedings of the 21st International Conference, Granada, Spain, 16–20 September 2018; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Wadekar, S.N.; Chaurasia, A. MobileViTv3: Mobile-Friendly Vision Transformer with Simple and Effective Fusion of Local, Global and Input Features. arXiv 2022, arXiv:2209.15159. [Google Scholar]
- Katharopoulos, A.; Vyas, A.; Pappas, N.; Fleuret, F. Transformers are RNNs: Fast Autoregressive Transformers with Linear Attention. In Proceedings of the 37th International Conference on Machine Learning, ICML 2020, Virtual, Online, 13–18 July 2020; Volume Part F168147-7, pp. 5112–5121. [Google Scholar]
- Han, D.; Ye, T.; Han, Y.; Xia, Z.; Pan, S.; Wan, P.; Song, S.; Huang, G. Agent attention: On the integration of Softmax and linear attention. In Proceedings of the European Conference on Computer Vision, Milan, Italy, 29 September–4 October 2024. [Google Scholar]
- Hoyt, W.F. Collection-Novel. Available online: https://novel.utah.edu/collection/william-f-hoyt/#tabcollection (accessed on 15 June 2024).
- Kim, U. Machine Learning for Pseudopapilledema. 2018. Available online: https://doi.org/10.17605/OSF.IO/2W5CE (accessed on 18 December 2023).
- Pachade, S.; Porwal, P.; Thulkar, D.; Kokare, M.; Deshmukh, G.; Sahasrabuddhe, V.; Giancardo, L. Retinal Fundus Multi-Disease Image Dataset (RFMiD): A Dataset for Multi-Disease Detection Research. Data 2021, 6, 14. [Google Scholar] [CrossRef]
- Li, N.; Li, T.; Hu, C.; Wang, K.; Kang, H. A Benchmark of Ocular Disease Intelligent Recognition: One Shot for Multi-disease Detection. In Benchmarking, Measuring, and Optimizing: Third BenchCouncil International Symposium, Bench 2020, Virtual Event, 15–16 November 2020; Wolf, F., Gao, W., Eds.; Springer: Cham, Switzerland, 2021; pp. 177–193. [Google Scholar]
- Bajwa, M.N.; Singh, G.A.P.; Neumeier, W.; Malik, M.I.; Dengel, A.; Ahmed, S. G1020: A benchmark retinal fundus Image dataset for computer-aided glaucoma detection. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–7. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017; Volume 2017-January, pp. 2261–2269. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNetV2: Smaller Models and Faster Training. In Proceedings of the 38th International Conference on Machine Learning, ICML 2021, Virtual, Online, 18–24 July 2021; Volume 139, pp. 10096–10106. [Google Scholar]
- Qin, D.; Leichner, C.; Delakis, M.; Fornoni, M.; Luo, S.; Yang, F.; Wang, W.; Banbury, C.; Ye, C.; Akin, B. MobileNetV4: Universal Models fortheMobile Ecosystem. In Proceedings of the European Conference on Computer Vision, Milan, Italy, 29 September–4 October 2024. [Google Scholar]
- Wang, A.; Chen, H.; Lin, Z.; Han, J.; Ding, G. Rep ViT: Revisiting Mobile CNN From ViT Perspective. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 17–21 June 2024; pp. 15909–15920. [Google Scholar]
- Arrigo, A.; Aragona, E.; Battaglia Parodi, M.; Bandello, F. Quantitative approaches in multimodal fundus imaging: State of the art and future perspectives. Prog. Retin. Eye Res. 2023, 92, 101111. [Google Scholar] [CrossRef]
- Silva-Rodríguez, J.; Chakor, H.; Kobbi, R.; Dolz, J.; Ben Ayed, I. A Foundation Language-Image Model of the Retina (FLAIR): Encoding expert knowledge in text supervision. Med. Image Anal. 2025, 99, 103357. [Google Scholar] [CrossRef]
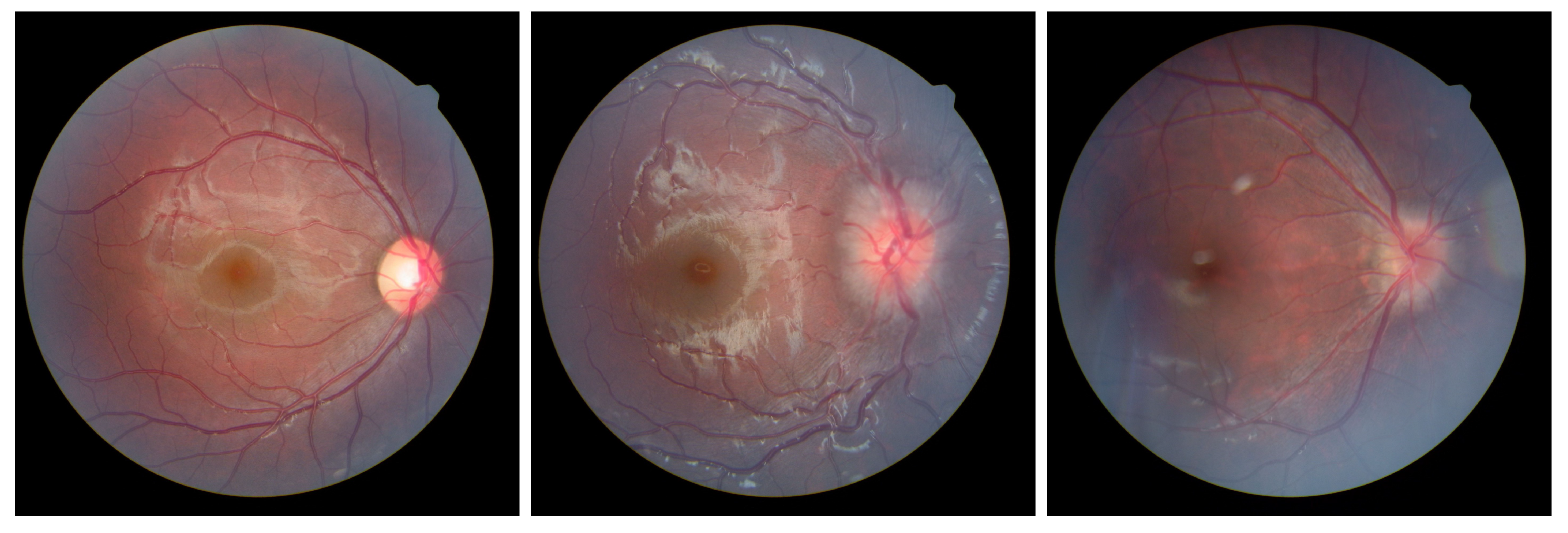




| Label | Normal | ODE | PPE |
|---|---|---|---|
| Training set | 616 | 330 | 263 |
| Validation set | 189 | 106 | 108 |
| Test set | 90 | 49 | 42 |
| Total | 895 | 485 | 413 |
| Label | Acccuracy | Precision | Recall | Specificity |
|---|---|---|---|---|
| Normal | 1.0 | 1.0 | 1.0 | 1.0 |
| ODE | 0.9669 | 0.9574 | 0.9184 | 0.9848 |
| PPE | 0.9669 | 0.9091 | 0.9524 | 0.9712 |
| Average | 0.9779 | 0.9555 | 0.9569 | 0.9853 |
| Model | Acccuracy | Precision | Recall | Specificity | Params (M) | Flops |
|---|---|---|---|---|---|---|
| DenseNet-169 | 0.9595 | 0.9238 | 0.9215 | 0.9716 | 12.49 | 3.44 G |
| EfficientNetV2-S | 0.9595 | 0.9301 | 0.9223 | 0.9713 | 20.18 | 6.74 G |
| ShuffleNetV2 (x1.0) | 0.9632 | 0.9329 | 0.9294 | 0.9730 | 1.26 | 197 M |
| MobileViTv3-xs | 0.9558 | 0.9130 | 0.9189 | 0.9706 | 1.85 | 1.0 G |
| EfficientViT-M4 | 0.9632 | 0.9290 | 0.9325 | 0.9752 | 8.42 | 300 M |
| MobileNetV4-M | 0.9668 | 0.9429 | 0.9362 | 0.9752 | 8.44 | 1.11 G |
| RepViT (m1.0) | 0.9632 | 0.9304 | 0.9294 | 0.9741 | 6.41 | 1.13 G |
| WHA-Net (Ours) | 0.9779 | 0.9555 | 0.9569 | 0.9853 | 2.15 | 810 M |
| Model | Valid_Acc (%) | Parameters (M) | Flops |
|---|---|---|---|
| MobileViTv3 | 97.27 | 1.85 | 1.0 G |
| Agent- MViTv3 | 97.02 | 1.96 (+5.95%) | 889 M (−11.10%) |
| Model | Valid_Acc (%) | Parameters (M) | Flops (M) |
|---|---|---|---|
| Agent (baseline) | 96.77 | 1.939 | 797.93 |
| Agent + WTC | 97.27 | 1.941 | 804.05 |
| Agent + HAIR | 97.02 | 2.145 | 803.69 |
| WHA-Net (ours) | 97.52 | 2.147 | 809.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, J.; Wang, Y.; Ge, M.; Li, J.; Li, Y.; Wang, W.; Zhou, X. WHA-Net: A Low-Complexity Hybrid Model for Accurate Pseudopapilledema Classification in Fundus Images. Bioengineering 2025, 12, 550. https://doi.org/10.3390/bioengineering12050550
Pei J, Wang Y, Ge M, Li J, Li Y, Wang W, Zhou X. WHA-Net: A Low-Complexity Hybrid Model for Accurate Pseudopapilledema Classification in Fundus Images. Bioengineering. 2025; 12(5):550. https://doi.org/10.3390/bioengineering12050550
Chicago/Turabian StylePei, Junpeng, Yousong Wang, Mingliang Ge, Jun Li, Yixing Li, Wei Wang, and Xiaohong Zhou. 2025. "WHA-Net: A Low-Complexity Hybrid Model for Accurate Pseudopapilledema Classification in Fundus Images" Bioengineering 12, no. 5: 550. https://doi.org/10.3390/bioengineering12050550
APA StylePei, J., Wang, Y., Ge, M., Li, J., Li, Y., Wang, W., & Zhou, X. (2025). WHA-Net: A Low-Complexity Hybrid Model for Accurate Pseudopapilledema Classification in Fundus Images. Bioengineering, 12(5), 550. https://doi.org/10.3390/bioengineering12050550







