Patch-Based Texture Feature Extraction Towards Improved Clinical Task Performance
Abstract
1. Introduction
Motivations and Contributions
- (1)
- We develop novel 3D patch-based texture features. Specifically, we construct patch patterns using k-means clustering and discretize images based on the labels of these patterns. The computation formulas of conventional texture features are modified to drive the extraction of patch-level (instead of voxel-level) texture features.
- (2)
- In our proposed method, we design a multi-resolution framework via resampling image volumes to different voxel sizes and introduce multi-scale 3D patches to capture inter-patch correlation at different spatial scales simultaneously, thereby enabling a more comprehensive quantification of tissue characteristics.
- (3)
- The machine learning models across five feature selection methods and five classifiers are performed to systematically validate the superiority and stability of the proposed 3D patch-based texture features relative to conventional texture features.
- (4)
- Extensive experiments are conducted on simulated data and two independent MRI datasets involving three MRI sequences and two clinical prediction tasks to validate the effectiveness and generalizability of the proposed method over the conventional method.
2. Related Work
3. Method
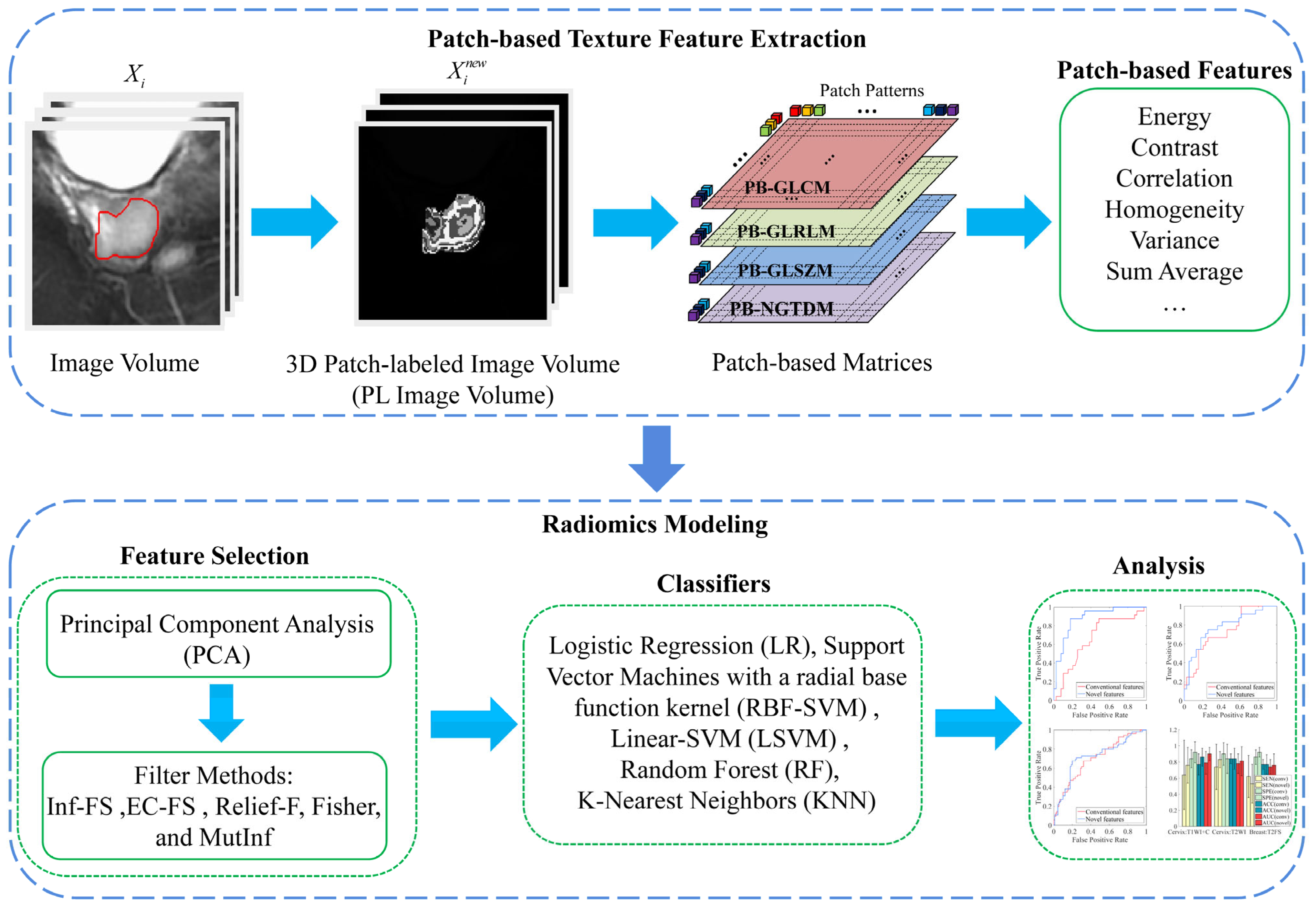
3.1. Patch-Based Texture Feature Construction
3.1.1. Patch-Labeled Image
3.1.2. Patch-Based Texture Features
3.2. Radiomics Analysis Framework
3.2.1. Segmentation of VOI
3.2.2. Image Preprocessing
3.2.3. Texture Feature Extraction
3.2.4. Feature Selection
3.2.5. Machine Learning Modeling
3.2.6. Clinical Utility
4. Materials and Experimental Configuration
4.1. Simulated Data
4.2. Clinical Data
4.3. Parameter Settings of Feature Extraction
4.4. Comparison of the Proposed Features with Conventional Features
4.5. Division of Training and Validation Sets
5. Results
5.1. Experiments on Simulated Data
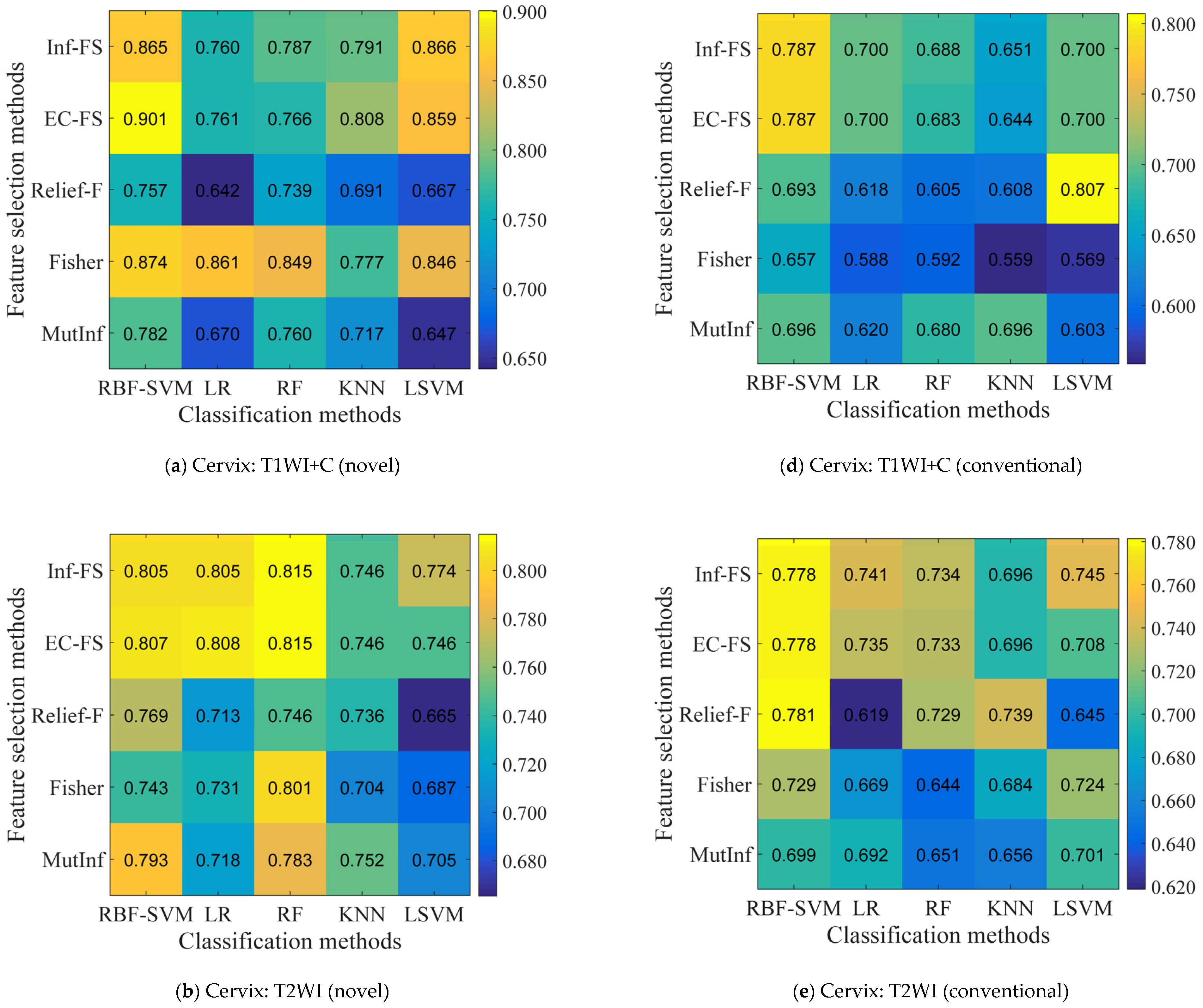
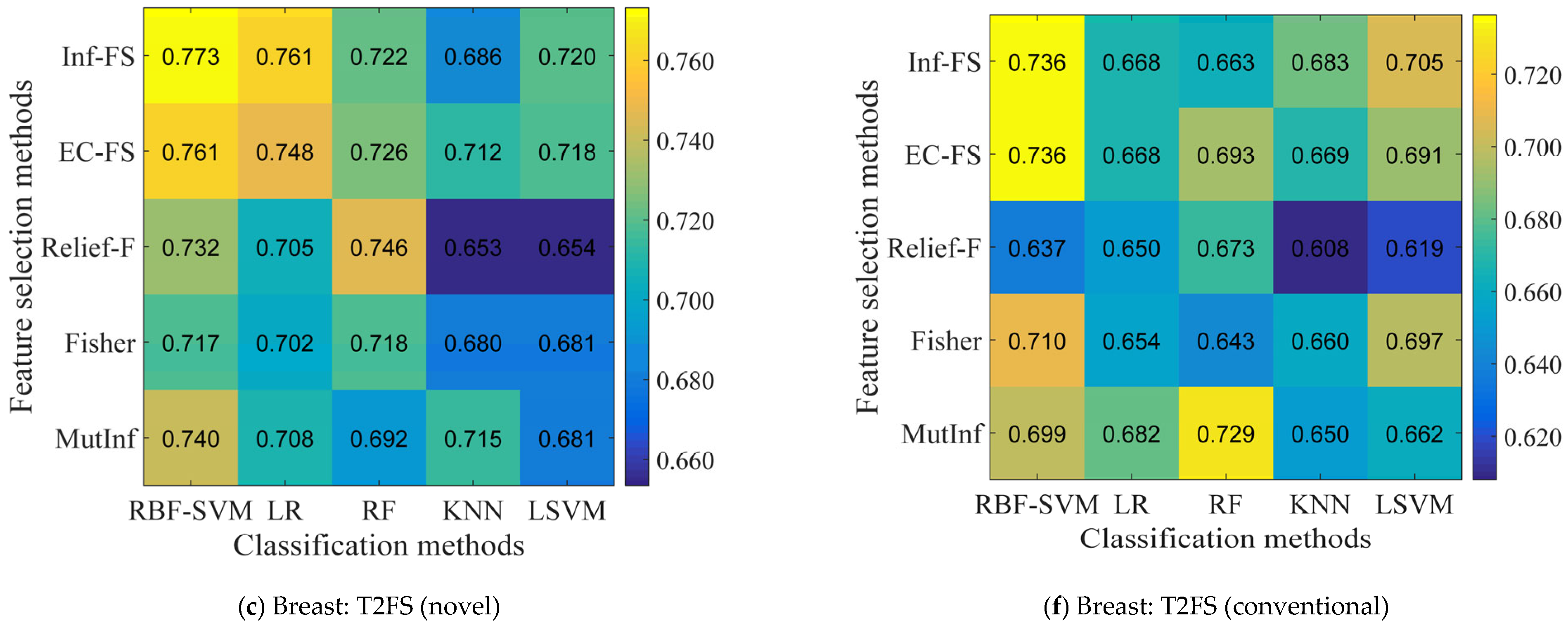
5.2. Identification of the Optimal Combination
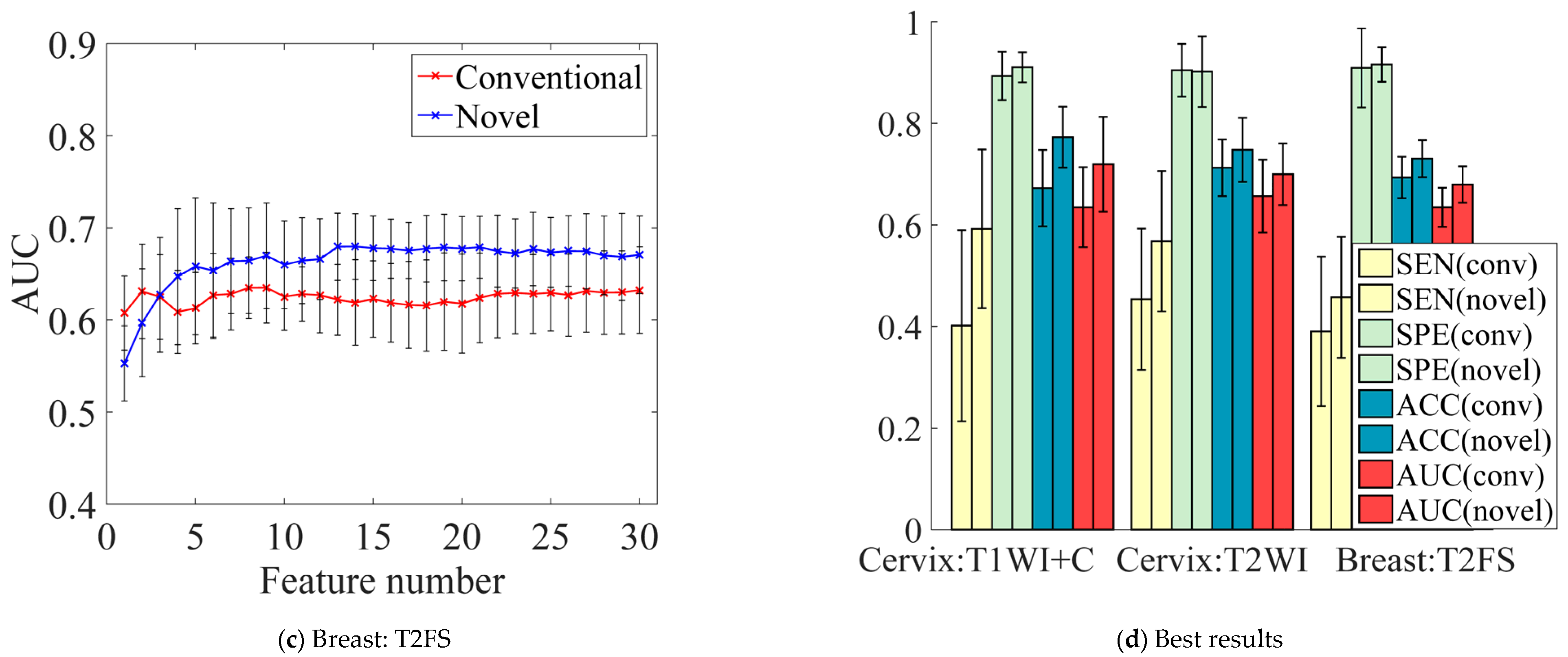
5.3. Influence of Feature Numbers on Prediction Performance
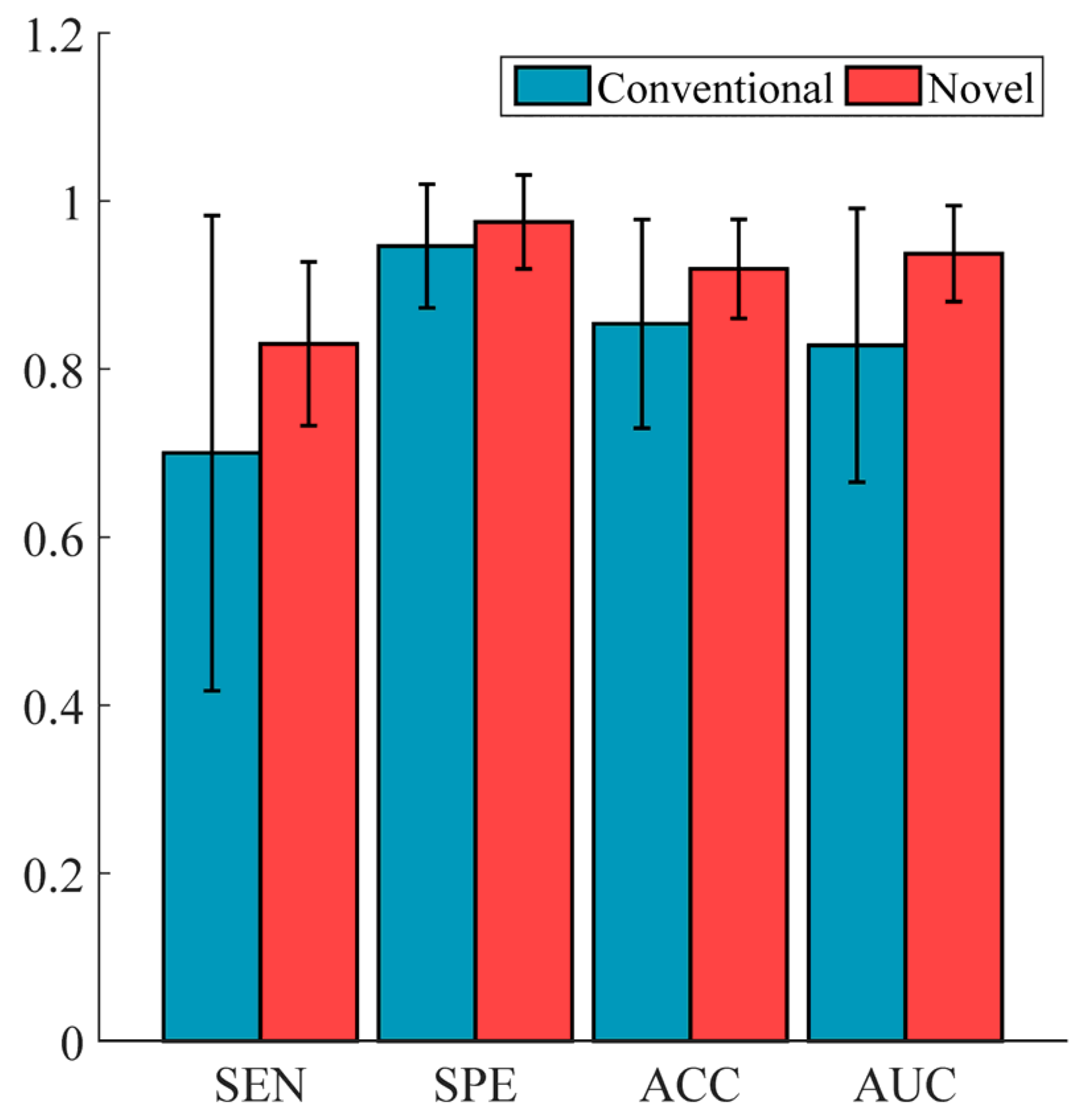
5.4. Performance Comparison of Conventional vs. Novel Texture Features in the “EC-FS + RBF-SVM” Combination
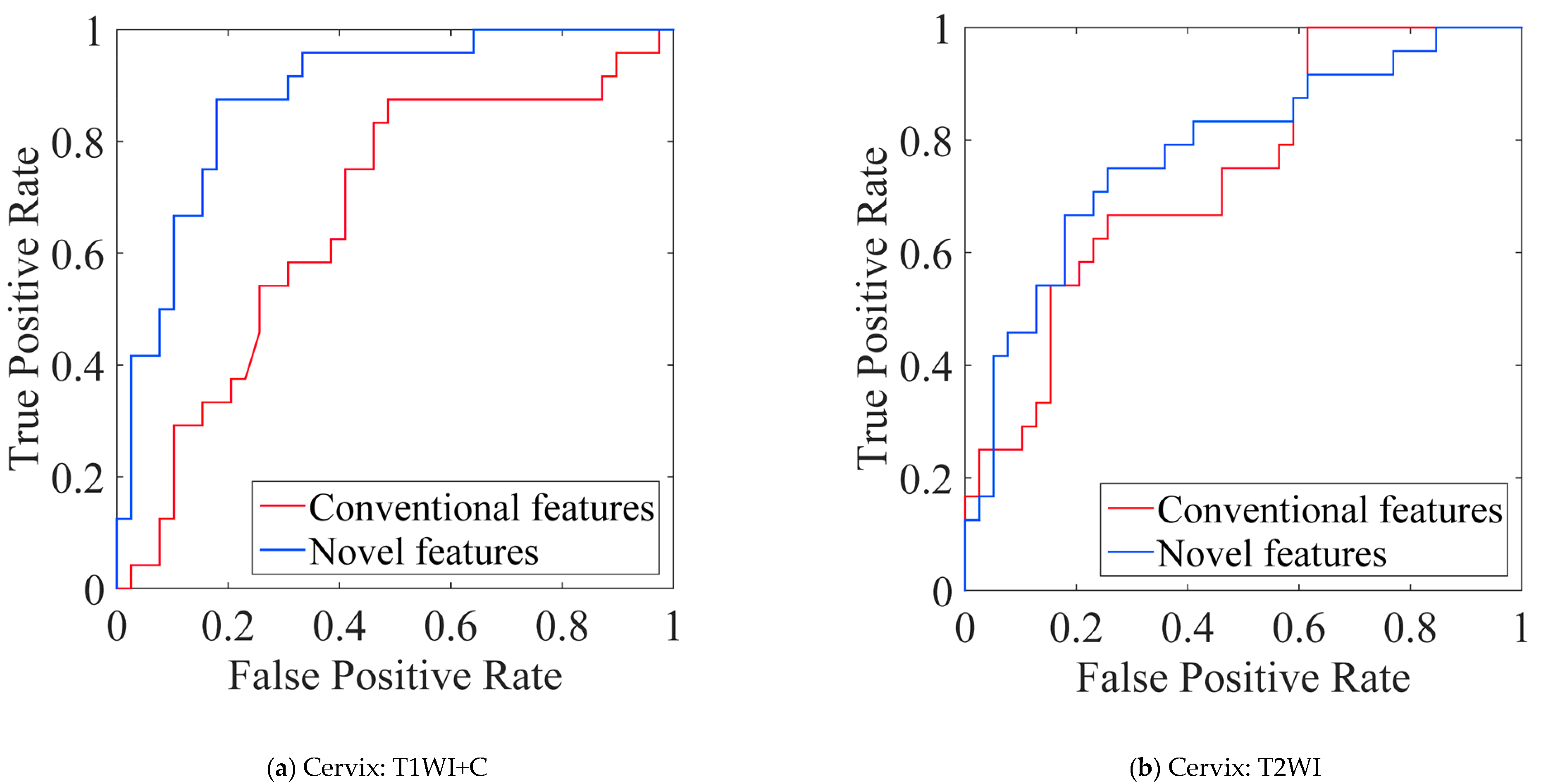
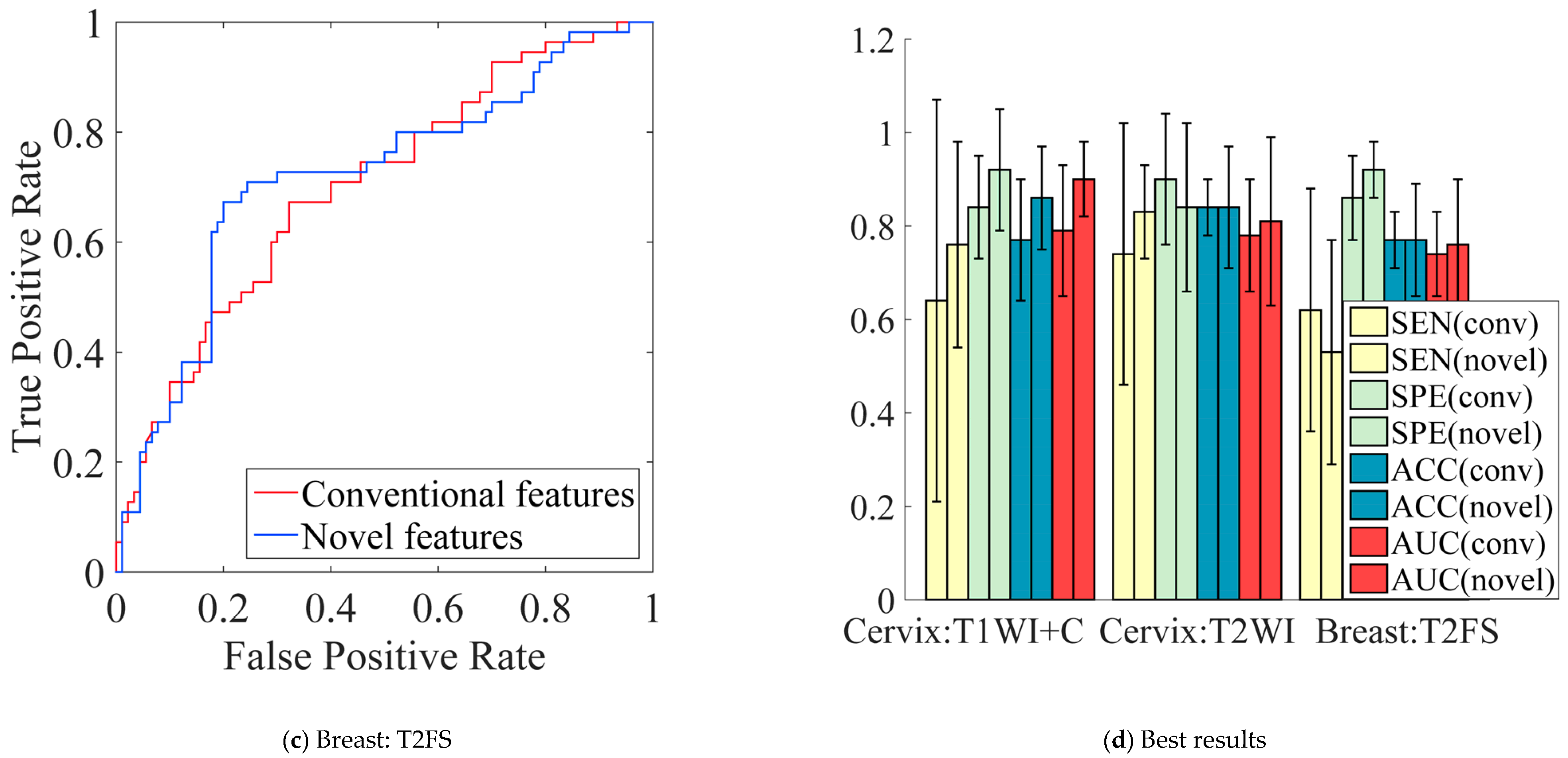
5.5. Performance Comparison of Conventional vs. Novel Texture Features in the Multi-Modality of Cervical Cancer
6. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Definition of Patch-Based Features
Appendix A.1. Patch-Based Gray-Level Co-Occurrence Matrix Features (Patch-Based GLCM Features)
- (1)
- Energy:
- (2)
- Contrast:
- (3)
- Correlation:
- (4)
- Homogeneity:
- (5)
- Variance:
- (6)
- Sum Average:
- (7)
- Entropy:
- (8)
- Auto Correlation:
Appendix A.2. Patch-Based Gray-Level Run-Length Matrix Features (Patch-Based GLRLM Features)
- (1)
- Short Run Emphasis (SRE):
- (2)
- Long Run Emphasis (LRE):
- (3)
- Gray-level Non-Uniformity (GLN):
- (4)
- Run Length Non-Uniformity (RLN):
- (5)
- Run Percentage (RP):
- (6)
- Low Gray-level Run Emphasis (LGRE):
- (7)
- High Gray-level Run Emphasis (HGRE):
- (8)
- Short Run Low Gray-level Emphasis (SRLGE):
- (9)
- Short Run High Gray-level Emphasis (SRHGE):
- (10)
- Long Run Low Gray-level Emphasis (LRLGE):
- (11)
- Long Run High Gray-level Emphasis (LRHGE):
- (12)
- Gray-level Variance (GLV):
- (13)
- Run length Variance (RLV):
Appendix A.3. Patch-Based Gray-Level Size Zone Matrix Features (Patch-Based GLSZM Features)
- (1)
- Small Zone Emphasis (SZE):
- (2)
- Large Zone Emphasis (LZE):
- (3)
- Gray-Level Non-uniformity (GLN):
- (4)
- Zone-Size Non-uniformity (ZSN):
- (5)
- Zone Percentage (ZP):
- (6)
- Low Gray-Level Zone Emphasis (LGZE):
- (7)
- High Gray-Level Zone Emphasis (HGZE):
- (8)
- Small Zone Low Gray-Level Emphasis (SZLGE):
- (9)
- Small Zone High Gray-Level Emphasis (SZHGE):
- (10)
- Large Zone Low Gray-Level Emphasis (LZLGE):
- (11)
- Large Zone High Gray-Level Emphasis (LZHGE):
- (12)
- Gray-Level Variance (GLV):
- (13)
- Zone-Size Variance (ZSV):
Appendix A.4. Patch-Based Neighborhood Gray Tone Difference Matrix Features (Patch-Based NGTDM Features)
- Then, the patch-based NGTDM texture features are defined as:
- (1)
- Coarseness:
- (2)
- Contrast:
- (3)
- Busyness:
- (4)
- Complexity:
- (5)
- Strength:
References
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Sala, E.; Mema, E.; Himoto, Y.; Veeraraghavan, H.; Brenton, J.D.; Snyder, A.; Weigelt, B.; Vargas, H.A. Unravelling Tumour Heterogeneity Using Next-Generation Imaging: Radiomics, Radiogenomics, and Habitat Imaging. Clin. Radiol. 2017, 72, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Wu, X.; Wang, B.; Lei, P.; Chen, Y.; Xu, X.; Lai, X.; Xu, J.; Wang, J. CT-Based Radiomics Analysis for Prediction of Pathological Subtypes of Lung Adenocarcinoma. J. Radiat. Res. Appl. Sci. 2024, 17, 101174. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, T.; Xu, F.; Cao, L.; Gu, W.; Zhu, W.; Fan, Y.; Wang, X.; Hu, C.; Yu, Y. MRI-Based Deep Learning Radiomics to Differentiate Dual-Phenotype Hepatocellular Carcinoma from HCC and Intrahepatic Cholangiocarcinoma: A Multicenter Study. Insights Imaging 2025, 16, 27. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Chen, Z.; Liang, Y.; Shen, W.; Yang, F.; Dai, R.; Wu, N.; Tian, J. Staging of Cervical Cancer Based on Tumor Heterogeneity Characterized by Texture Features on F-18-FDG PET Images. Phys. Med. Biol. 2015, 60, 5123–5139. [Google Scholar] [CrossRef]
- Zeng, S.; Wang, X.-L.; Yang, H. Radiomics and Radiogenomics: Extracting More Information from Medical Images for the Diagnosis and Prognostic Prediction of Ovarian Cancer. Mil. Med. Res. 2024, 11, 77. [Google Scholar] [CrossRef]
- Yip, S.S.F.; Coroller, T.; Sanford, N.N.; Mamon, H.J.; Aerts, H.J.W.L.; Berbeco, R. Relationship between the Temporal Changes in Positron-Emission-Tomography-Imaging-Based Textural Features and Pathologic Response and Survival in Esophageal Cancer Patients. Front. Oncol. 2016, 6, 72. [Google Scholar] [CrossRef]
- Coroller, T.; Agrawal, V.; Narayan, V.; Hou, Y.; Grossmann, P.; Lee, S.W.; Mak, R.H.; Aerts, H.J.W.L. Radiomic Phenotype Features Predict Pathological Response in Non-Small Cell Lung Cancer. Radiother. Oncol. 2016, 119, 480–486. [Google Scholar] [CrossRef]
- Dong, Y.; Feng, Q.; Yang, W.; Lu, Z.; Deng, C.; Zhang, L.; Lian, Z.; Liu, J.; Luo, X.; Pei, S.; et al. Preoperative Prediction of Sentinel Lymph Node Metastasis in Breast Cancer Based on Radiomics of T2-Weighted Fat-Suppression and Diffusion-Weighted MRI. Eur. Radiol. 2018, 28, 582–591. [Google Scholar] [CrossRef]
- Yan, M.; Zhang, Z.; Tian, J.; Yu, J.; Dekker, A.; Ruysscher, D.D.; Wee, L.; Zhao, L. Whole Lung Radiomic Features Are Associated with Overall Survival in Patients with Locally Advanced Non-Small Cell Lung Cancer Treated with Definitive Radiotherapy. Radiat Oncol. 2025, 20, 9. [Google Scholar] [CrossRef]
- Zhang, W.; Guo, Y.; Jin, Q. Radiomics and Its Feature Selection: A Review. Symmetry 2023, 15, 1834. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Galloway, M.M. Texture Analysis Using Gray Level Run Lengths. Comput. Graph. Image Process. 1975, 4, 172–179. [Google Scholar] [CrossRef]
- Thibault, G.; Angulo, J.; Meyer, F. Advanced Statistical Matrices for Texture Characterization: Application to DNA Chromatin and Microtubule Network Classification. In Proceedings of the 2011 18th IEEE International Conference on Image Processing, Brussels, Belgium, 11–14 September 2011; IEEE: Brussels, Belgium, 2011; pp. 53–56. [Google Scholar]
- Amadasun, M.; King, R.A. Textural Features Corresponding to Textural Properties. IEEE Trans. Syst. Man Cybern. 1989, 19, 1264–1274. [Google Scholar] [CrossRef]
- Depeursinge, A.; Van de Ville, D.; Platon, A.; Geissbuhler, A.; Poletti, P.A.; Muller, H. Near-Affine-Invariant Texture Learning for Lung Tissue Analysis Using Isotropic Wavelet Frames. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 665–675. [Google Scholar] [CrossRef]
- Sivic; Zisserman. Video Google: A Text Retrieval Approach to Object Matching in Videos. In Proceedings of the IEEE International Conference on Computer Vision, Nice, France, 13–16 October 2003; Volume 2, pp. 1470–1477. [Google Scholar]
- Wu, Y.; Liu, G.; Huang, M.; Guo, J.; Jiang, J.; Yang, W.; Chen, W.; Feng, Q. Prostate Segmentation Based on Variant Scale Patch and Local Independent Projection. IEEE Trans. Med. Imaging 2014, 33, 1290–1303. [Google Scholar] [CrossRef]
- Khalvati, F.; Zhang, J.; Wong, A.; Haider, M.A. Bag of Bags: Nested Multi Instance Classification for Prostate Cancer Detection. In Proceedings of the IEEE International Conference on Machine Learning and Applications, Anaheim, CA, USA, 18–20 December 2016; pp. 146–151. [Google Scholar]
- Cheng, J.; Yang, W.; Huang, M.; Huang, W.; Jiang, J.; Zhou, Y.; Yang, R.; Zhao, J.; Feng, Y.; Feng, Q.; et al. Retrieval of Brain Tumors by Adaptive Spatial Pooling and Fisher Vector Representation. PLoS ONE 2016, 11, e0157112. [Google Scholar] [CrossRef]
- Vallières, M.; Freeman, C.R.; Skamene, S.R.; El Naqa, I. A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities. Phys. Med. Biol. 2015, 60, 5471–5496. [Google Scholar] [CrossRef]
- Zhou, Y.; Ju, L.; Wang, S. Multiscale Superpixels and Supervoxels Based on Hierarchical Edge-Weighted Centroidal Voronoi Tessellation. IEEE Trans. Image Process. 2015, 24, 3834–3845. [Google Scholar] [CrossRef]
- Yang, W.; Feng, Q.; Yu, M.; Lu, Z.; Gao, Y.; Xu, Y.; Chen, W. Content-Based Retrieval of Brain Tumor in Contrast-Enhanced MRI Images Using Tumor Margin Information and Learned Distance Metric. Med. Phys. 2012, 39, 6929. [Google Scholar] [CrossRef]
- Huang, M.; Yang, W.; Wu, Y.; Jiang, J.; Gao, Y.; Chen, Y.; Feng, Q.; Chen, W.; Lu, Z. Content-Based Image Retrieval Using Spatial Layout Information in Brain Tumor T1-Weighted Contrast-Enhanced MR Images. PLoS ONE 2014, 9, e102754. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xu, L.; Yang, P.; Lin, N.; Huang, X.; Pan, W.; Li, H.; Lin, P.; Li, B.; Bunpetch, V.; et al. Survival Prediction in High-Grade Osteosarcoma Using Radiomics of Diagnostic Computed Tomography. EBioMedicine 2018, 34, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, J.; Meng, X.; Yue, J.; Feng, R.; Ma, L. Prognostic Value of Serial [18F]Fluorodeoxyglucose PET-CT Uptake in Stage III Patients with Non-Small Cell Lung Cancer Treated by Concurrent Chemoradiotherapy. Eur. J. Radiol. 2011, 77, 92–96. [Google Scholar] [CrossRef]
- Higgins, K.A.; Hoang, J.K.; Roach, M.C.; Chino, J.; Yoo, D.S.; Turkington, T.G.; Brizel, D.M. Analysis of Pretreatment FDG-PET SUV Parameters in Head-and-Neck Cancer: Tumor SUVmean Has Superior Prognostic Value. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, 548–553. [Google Scholar] [CrossRef]
- Cheng, N.-M.; Fang, Y.-H.D.; Yen, T.-C. The Promise and Limits of PET Texture Analysis. Ann. Nucl. Med. 2013, 27, 867–869. [Google Scholar] [CrossRef]
- Kido, S.; Kuriyama, K.; Higashiyama, M.; Kasugai, T.; Kuroda, C. Fractal Analysis of Small Peripheral Pulmonary Nodules in Thin-Section CT: Evaluation of the Lung-Nodule Interfaces. J. Comput. Assist. Tomogr. 2002, 26, 573–578. [Google Scholar] [CrossRef]
- Johansen, R.; Jensen, L.R.; Rydland, J.; Goa, P.E.; Kvistad, K.A.; Bathen, T.F.; Axelson, D.E.; Lundgren, S.; Gribbestad, I.S. Predicting Survival and Early Clinical Response to Primary Chemotherapy for Patients with Locally Advanced Breast Cancer Using DCE-MRI. J. Magn. Reson. Imaging 2009, 29, 1300–1307. [Google Scholar] [CrossRef]
- Peng, S.-L.; Chen, C.-F.; Liu, H.-L.; Lui, C.-C.; Huang, Y.-J.; Lee, T.-H.; Chang, C.-C.; Wang, F.-N. Analysis of Parametric Histogram from Dynamic Contrast-Enhanced MRI: Application in Evaluating Brain Tumor Response to Radiotherapy. NMR Biomed. 2013, 26, 443–450. [Google Scholar] [CrossRef]
- Coroller, T.P.; Grossmann, P.; Hou, Y.; Rios Velazquez, E.; Leijenaar, R.T.H.; Hermann, G.; Lambin, P.; Haibe-Kains, B.; Mak, R.H.; Aerts, H.J.W.L. CT-Based Radiomic Signature Predicts Distant Metastasis in Lung Adenocarcinoma. Radiother. Oncol. 2015, 114, 345–350. [Google Scholar] [CrossRef]
- Petkovska, I.; Shah, S.K.; McNitt-Gray, M.F.; Goldin, J.G.; Brown, M.S.; Kim, H.J.; Brown, K.; Aberle, D.R. Pulmonary Nodule Characterization: A Comparison of Conventional with Quantitative and Visual Semi-Quantitative Analyses Using Contrast Enhancement Maps. Eur. J. Radiol. 2006, 59, 244–252. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cook, G.J.R.; Yip, C.; Siddique, M.; Goh, V.; Chicklore, S.; Roy, A.; Marsden, P.; Ahmad, S.; Landau, D. Are Pretreatment 18F-FDG PET Tumor Textural Features in Non-Small Cell Lung Cancer Associated with Response and Survival After Chemoradiotherapy? J. Nucl. Med. 2013, 54, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Ganeshan, B.; Abaleke, S.; Young, R.C.D.; Chatwin, C.R.; Miles, K.A. Texture Analysis of Non-Small Cell Lung Cancer on Unenhanced Computed Tomography: Initial Evidence for a Relationship with Tumour Glucose Metabolism and Stage. Cancer Imaging 2010, 10, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Keys, R. Cubic Convolution Interpolation for Digital Image Processing. IEEE Trans. Acoust. Speech Signal Process. 1981, 29, 1153–1160. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, X.; Tian, Q.; Zhang, G.; Liu, Y.; Cui, G.; Meng, J.; Wu, Y.; Liu, T.; Yang, Z.; et al. Three-Dimensional Texture Features from Intensity and High-Order Derivative Maps for the Discrimination between Bladder Tumors and Wall Tissues via MRI. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 645–656. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, L.; Laguna, A.; Wu, J.; Zou, B.; Mohseni, A.; Chandra, R.S.; Mehta, T.I.; Zaki, H.A.; Zhang, P.; et al. Defining a Radiomics Feature Selection Method for Predicting Response to Transarterial Chemoembolization in Hepatocellular Carcinoma Patients. Meta-Radiol. 2024, 2, 100067. [Google Scholar] [CrossRef]
- Chen, W.; Lin, G.; Kong, C.; Wu, X.; Hu, Y.; Chen, M.; Xia, S.; Lu, C.; Xu, M.; Ji, J. Non-Invasive Prediction Model of Axillary Lymph Node Status in Patients with Early-Stage Breast Cancer: A Feasibility Study Based on Dynamic Contrast-Enhanced-MRI Radiomics. Br. J. Radiol. 2024, 97, 439–450. [Google Scholar] [CrossRef]
- Jiang, M.; Li, C.-L.; Luo, X.-M.; Chuan, Z.-R.; Chen, R.-X.; Tang, S.-C.; Lv, W.-Z.; Cui, X.-W.; Dietrich, C.F. Radiomics Model Based on Shear-Wave Elastography in the Assessment of Axillary Lymph Node Status in Early-Stage Breast Cancer. Eur. Radiol. 2022, 32, 2313–2325. [Google Scholar] [CrossRef]
- Hu, K.; Wang, W.; Liu, X.; Meng, Q.; Zhang, F. Comparison of Treatment Outcomes between Squamous Cell Carcinoma and Adenocarcinoma of Cervix after Definitive Radiotherapy or Concurrent Chemoradiotherapy. Radiat Oncol. 2018, 13, 249. [Google Scholar] [CrossRef]
- Abrol, S.; Kotrotsou, A.; Salem, A.; Zinn, P.O.; Colen, R.R. Radiomic Phenotyping in Brain Cancer to Unravel Hidden Information in Medical Images. Top. Magn. Reson. Imaging 2017, 26, 43–53. [Google Scholar] [CrossRef]
- Max, J. Quantizing for Minimum Distortion. IEEE Trans. Inf. Theory 1960, 6, 7–12. [Google Scholar] [CrossRef]
- Lloyd, S. Least Squares Quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Limkin, E.J.; Sun, R.; Dercle, L.; Zacharaki, E.I.; Robert, C.; Reuzé, S.; Schernberg, A.; Paragios, N.; Deutsch, E.; Ferté, C. Promises and Challenges for the Implementation of Computational Medical Imaging (Radiomics) in Oncology. Ann. Oncol. 2017, 28, 1191–1206. [Google Scholar] [CrossRef]
- Lee, G.; Lee, H.Y.; Park, H.; Schiebler, M.L.; van Beek, E.J.R.; Ohno, Y.; Seo, J.B.; Leung, A. Radiomics and Its Emerging Role in Lung Cancer Research, Imaging Biomarkers and Clinical Management: State of the Art. Eur. J. Radiol. 2017, 86, 297–307. [Google Scholar] [CrossRef]
- Khalvati, F.; Wong, A.; Haider, M.A. Automated Prostate Cancer Detection via Comprehensive Multi-Parametric Magnetic Resonance Imaging Texture Feature Models. BMC Med. Imaging 2015, 15, 27. [Google Scholar] [CrossRef]
- Zhou, Z.; Folkert, M.; Iyengar, P.; Westover, K.; Zhang, Y.; Choy, H.; Timmerman, R.; Jiang, S.; Wang, J. Multi-Objective Radiomics Model for Predicting Distant Failure in Lung SBRT. Phys. Med. Biol. 2017, 62, 4460–4478. [Google Scholar] [CrossRef]
- Bomhals, B.; Cossement, L.; Maes, A.; Sathekge, M.; Mokoala, K.M.G.; Sathekge, C.; Ghysen, K.; Van De Wiele, C. Principal Component Analysis Applied to Radiomics Data: Added Value for Separating Benign from Malignant Solitary Pulmonary Nodules. J. Clin. Med. 2023, 12, 7731. [Google Scholar] [CrossRef]
- Du, D.; Feng, H.; Lv, W.; Ashrafinia, S.; Yuan, Q.; Wang, Q.; Yang, W.; Feng, Q.; Chen, W.; Rahmim, A.; et al. Machine Learning Methods for Optimal Radiomics-Based Differentiation Between Recurrence and Inflammation: Application to Nasopharyngeal Carcinoma Post-Therapy PET/CT Images. Mol. Imaging Biol. 2020, 22, 730–738. [Google Scholar] [CrossRef]
- Laajili, R.; Said, M.; Tagina, M. Application of Radiomics Features Selection and Classification Algorithms for Medical Imaging Decision: MRI Radiomics Breast Cancer Cases Study. Inform. Med. Unlocked 2021, 27, 100801. [Google Scholar] [CrossRef]
- Salmanpour, M.R.; Hosseinzadeh, M.; Bakhtiyari, M.; Maghsudi, M.; Rahmim, A. Prediction of Drug Amount in Parkinson’s Disease Using Hybrid Machine Learning Systems and Radiomics Features. Int. J. Imaging Syst. Tech. 2023, 33, 1437–1449. [Google Scholar] [CrossRef]
- Liu, H.; Lao, M.; Zhang, Y.; Chang, C.; Yin, Y.; Wang, R. Radiomics-Based Machine Learning Models for Differentiating Pathological Subtypes in Cervical Cancer: A Multicenter Study. Front. Oncol. 2024, 14, 1346336. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Jiao, Y.; Zhang, L.; Fu, C.; Zhu, X.; Wang, Q.; Gu, Y. Multiparametric MRI-Based Radiomics Analysis: Differentiation of Subtypes of Cervical Cancer in the Early Stage. Acta Radiol. 2022, 63, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, L.; Dong, X.; Luo, R.; Ge, Y.; Liu, H.; Zhang, Y.; Wang, D. Deep Learning Radiomics of Preoperative Breast MRI for Prediction of Axillary Lymph Node Metastasis in Breast Cancer. J. Digit Imaging 2023, 36, 1323–1331. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lin, Y.; Ding, C.; Guan, W.; Zhang, X.; Jia, J.; Zhou, W.; Liu, Z.; Bai, G. Multi-Modality Radiomics Model Predicts Axillary Lymph Node Metastasis of Breast Cancer Using MRI and Mammography. Eur. Radiol. 2024, 34, 6121–6131. [Google Scholar] [CrossRef]
- Liu, H.; Zou, L.; Xu, N.; Shen, H.; Zhang, Y.; Wan, P.; Wen, B.; Zhang, X.; He, Y.; Gui, L.; et al. Deep Learning Radiomics Based Prediction of Axillary Lymph Node Metastasis in Breast Cancer. NPJ Breast Cancer 2024, 10, 22. [Google Scholar] [CrossRef]


| Texture Type | Texture Feature Description |
|---|---|
| Patch-based GLCM | Energy Contrast Correlation Homogeneity Variance Sum Average Entropy Auto Correlation |
| Patch-based GLRLM | Short Run Emphasis (SRE) Long Run Emphasis (LRE) Gray-Level Non-uniformity (GLN) Run-Length Non-uniformity (RLN) Run Percentage (RP) Low Gray-Level Run Emphasis (LGRE) High Gray-Level Run Emphasis (HGRE) Short Run Low Gray-Level Emphasis (SRLGE) Short Run High Gray-Level Emphasis (SRHGE) Long Run Low Gray-Level Emphasis (LRLGE) Long Run High Gray-Level Emphasis (LRHGE) Gray-Level Variance (GLV) Run-Length Variance (RLV) |
| Patch-based GLSZM | Small Zone Emphasis (SZE) Large Zone Emphasis (LZE) Gray-Level Non-uniformity (GLN) Zone-Size Non-uniformity (ZSN) Zone Percentage (ZP) Low Gray-Level Zone Emphasis (LGZE) High Gray-Level Zone Emphasis (HGZE) Small Zone Low Gray-Level Emphasis (SZLGE) Small Zone High Gray-Level Emphasis (SZHGE) Large Zone Low Gray-Level Emphasis (LZLGE) Large Zone High Gray-Level Emphasis (LZHGE) Gray-Level Variance (GLV) Zone-Size Variance (ZSV) |
| Patch-based NGTDM | Coarseness Contrast Busyness Complexity Strength |
| Feature Source | Modality | Parameter | |||
|---|---|---|---|---|---|
| R | S | K | P | ||
| Novel features | Breast-T2FS | 0.5, 1, 1.5 | pixelCS, 1, 1.3, 1.5, 1.7, 2 | 32, 128, 160, 256 | 3, 5 |
| Cervical-T1WI+C | 0.5, 1, 1.5 | PixelCS, 1, 2, 3, 4, 5 | 64, 128, 192, 256 | 3, 5 | |
| Cervical-T2WI | 0.5, 1, 1.5 | PixelCS, 1, 2, 3, 4, 5 | 64, 128, 192, 256 | 3, 5 | |
| R | S | Quan.algo | Ng | ||
| Conventional features | Breast-T2FS | 0.5, 1, 1.5 | PixelCS, 1, 2, 3, 4, 5 | Equal, Lloyd | 8, 16, 32, 64 |
| Cervical-T1WI+C | 0.5, 1, 1.5 | PixelCS, 1, 2, 3, 4, 5 | Equal, Lloyd | 8, 16, 32, 64 | |
| Cervical-T2WI | 0.5, 1, 1.5 | PixelCS, 1, 2, 3, 4, 5 | Equal, Lloyd | 8, 16, 32, 64 | |
| Clinical Task | Study | Modality | Method | Sample Size (Train/Validation) | Validation Performance |
|---|---|---|---|---|---|
| Breast cancer axillary lymph node metastasis prediction | This study | T2FS | Three-dimensional patch-based texture features | 145 (5-fold CV) | AUC = 0.76; ACC = 0.77 |
| Chen et al. [56] | DWI-ADC + DCE-MRI | Deep learning features and clinicopathological factors | 479 (366/122) | AUC = 0.71; ACC = 0.75 | |
| Wang et al. [57] | MRI | Conventional radiomics features | 379 (247/132) | AUC = 0.810; ACC = 0.765 | |
| MRI + Mammography | Multi-modality radiomics features and clinical predictors | 379 (247/132) | AUC = 0.892; ACC = 0.818 | ||
| Liu et al. [58] | Ultrasound | Deep learning features and conventional radiomics features | 883 (621/262) | AUC = 0.914–0.952; ACC = 0.87–0.89 | |
| Cervical cancer histological subtype prediction | This study | T1WI+C + T2WI | Three-dimensional patch-based texture features | 63 (5-fold CV) | AUC = 0.937; ACC = 0.919 |
| Liu et al. [54] | PET | Conventional radiomics features | 168 (136/59) | AUC = 0.851; ACC = 0.915 | |
| CT | Conventional radiomics features | AUC = 0.513; ACC = 0.661 | |||
| Wang et al. [55] | T2SAG + T2TRA + CESAG + CETRA + ADC | Conventional radiomics features | 96 | AUC = 0.89; ACC = 0.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, T.; Deng, C.; Feng, Q. Patch-Based Texture Feature Extraction Towards Improved Clinical Task Performance. Bioengineering 2025, 12, 404. https://doi.org/10.3390/bioengineering12040404
Lian T, Deng C, Feng Q. Patch-Based Texture Feature Extraction Towards Improved Clinical Task Performance. Bioengineering. 2025; 12(4):404. https://doi.org/10.3390/bioengineering12040404
Chicago/Turabian StyleLian, Tao, Chunyan Deng, and Qianjin Feng. 2025. "Patch-Based Texture Feature Extraction Towards Improved Clinical Task Performance" Bioengineering 12, no. 4: 404. https://doi.org/10.3390/bioengineering12040404
APA StyleLian, T., Deng, C., & Feng, Q. (2025). Patch-Based Texture Feature Extraction Towards Improved Clinical Task Performance. Bioengineering, 12(4), 404. https://doi.org/10.3390/bioengineering12040404






