Benchmarking Interpretability in Healthcare Using Pattern Discovery and Disentanglement
Abstract
1. Introduction
2. Related Work
2.1. AI in Healthcare
2.2. Model Interpretability and Its Importance
3. Proposed Model
3.1. Dataset
3.2. Dataset Preprocessing
3.3. Benchmarking Baseline Deep Learning Models
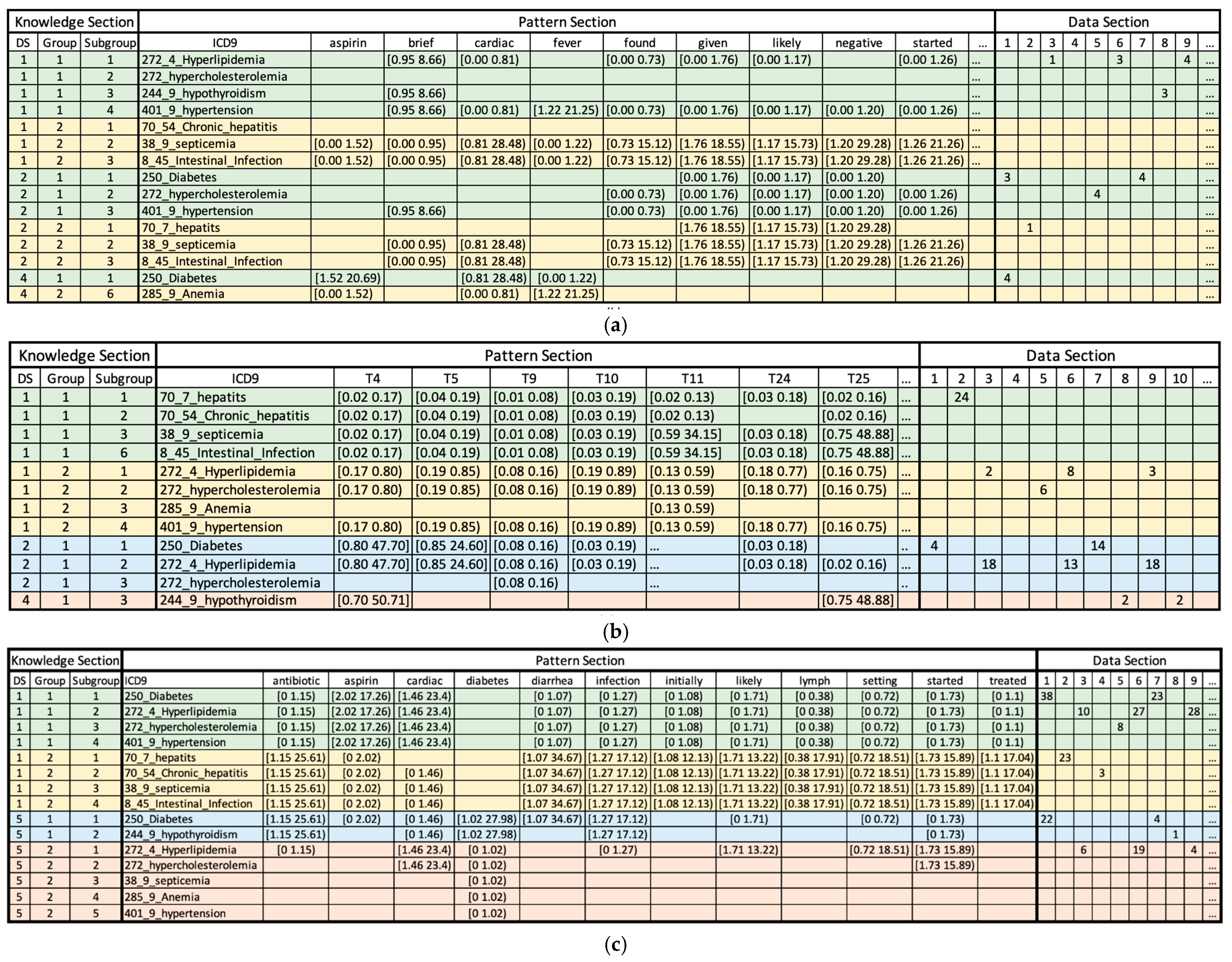
3.4. Clustering Using Pattern Discovery and Disentanglement
3.5. Post Hoc Interpretability Methods for Deep Learning Models
- Feature Permutation is a permutation-based algorithm that computes the importance of features at a global level [11]. Each feature’s value is randomly shuffled while keeping other features unchanged, and then, the algorithm computes the difference between the original model outputs and the shuffled model outputs.
- Integrated Gradients is a multi-step algorithm designed for feature importance estimation in deep learning models [12]. The process starts with establishing a baseline input, which serves as a point of comparison against the actual input. Following this, the algorithm constructs a linear path between the baseline and actual inputs. Crucially, Integrated Gradients compute the integral of the model’s output gradients with respect to each input feature along this linear path. Lastly, an importance score is assigned to each feature.
- Gradient SHAP estimates the SHapley Additive exPlanations (SHAP) value by calculating the expected value of gradients after conducting random sampling from baseline distribution [13]. This algorithm starts by adding white noise to every input observation by x times, where x is the total number of samples. From a particular baseline distribution, it picks a random point on the path between the baseline and the input. Then, the gradient of model outputs with respect to the randomly picked points is calculated. The described process is iterated a number of times so that the expected value of gradients can be approximated [13].
3.6. Evaluation of Interpretability Approaches
- m is the model.
- B is the set, [1, 5, 10, 20, 50] as stated in the original paper.
- is the input observation i.
- are the top k% rationales (important features) for input .
- are the top k% rationales (important features) applied to Gaussian noise for input .
- is the original predicted probability for class j.
- is the predicted probability for class j after removing rationales from .
4. Results and Discussions
4.1. Comparing the Performance of Baseline Classification Models
4.2. Discussions on the Interpretability of the PDD Model
- ICD-9 codes 70.7, 70.54, 38.9, and 8.45 (infectious hepatogastroenteric conditions) are associated with topic 4 (Orthostatic Hypotension in Elderly Patients), topic 5 (Neurological Examination: Focus on Cranial Nerves), topic 9 (The Role of Psychiatry in Managing Suicidal Ideation and Behavior) and topic 10 (Cerebral Infarcts and Aneurysms: A Focus on Vascular Neurology) in low probabilities.
- ICD-9 codes 272.4, 272, 285.9, and 401.9 (metabolic and cardiovascular conditions) exhibit different patterns and are associated with topic 4 (Orthostatic Hypotension in Elderly Patients), topic 5 (Neurological Examination: Focus on Cranial Nerves), topic 9 (The Role of Psychiatry in Managing Suicidal Ideation and Behavior), and high probability is associated with topic 10 (Cerebral Infarcts and Aneurysms: A Focus on Vascular Neurology) and topic 23 (Diagnostic Approach to Gastrointestinal Malignancies) in medium probabilities.
- ICD-9 codes 250, 272.4, and 272 (metabolic syndrome disorders) are associated with topic 4 (Orthostatic Hypotension in Elderly Patients), and topic 5 (Neurological Examination: Focus on Cranial Nerves) with higher probabilities.
- ICD-9 codes 250, 272.4, 272, and 401.9 (cardiometabolic disorders) are associated with low probabilities of tokens antibiotic, diarrhea, infection, initially, likely, lymph, setting, started, and treated. Meanwhile, they have a high probability for tokens such as aspirin and cardiac.
- ICD-9 codes 70.7, 70.54, 38.9, and 8.45 (infectious hepatointestinal conditions) demonstrate the opposite trend in relation to the first group (cardiometabolic disorders). These labels are associated with high probabilities of tokens antibiotics, diarrhea, infection, initially, likely, lymph, setting, started and treated but low probabilities of tokens aspirin and cardiac. Notably, this group of diseases is infectious, and ICD-9 code 8.45 (intestinal induction due to clostridium difficile) often accompanies symptoms of diarrhea.
- ICD-9 codes 250 and 244.9 (endocrine metabolic disorders) were associated with high probabilities of tokens antibiotics, diabetes, and infection, while they were also associated with low probabilities of tokens cardiac and started.
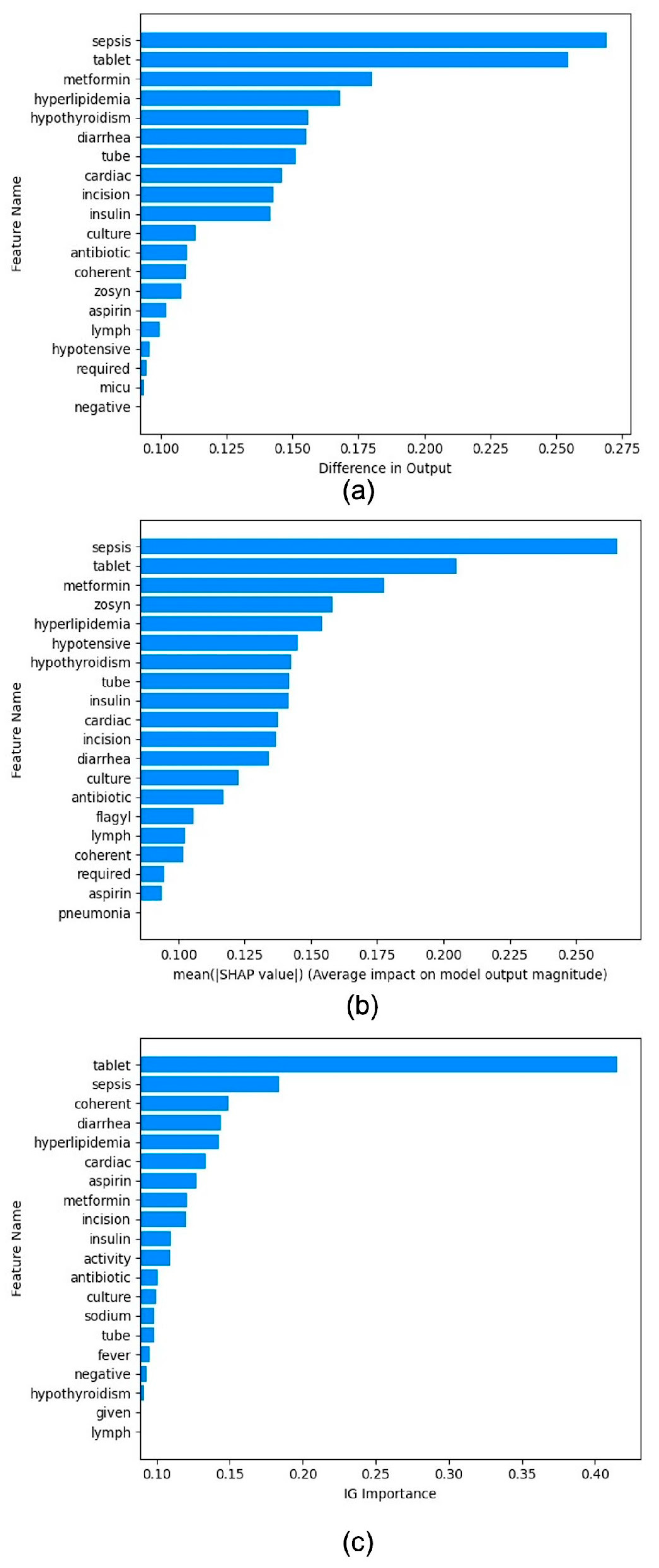
4.3. Benchmarking Post Hoc Interpretability Techniques
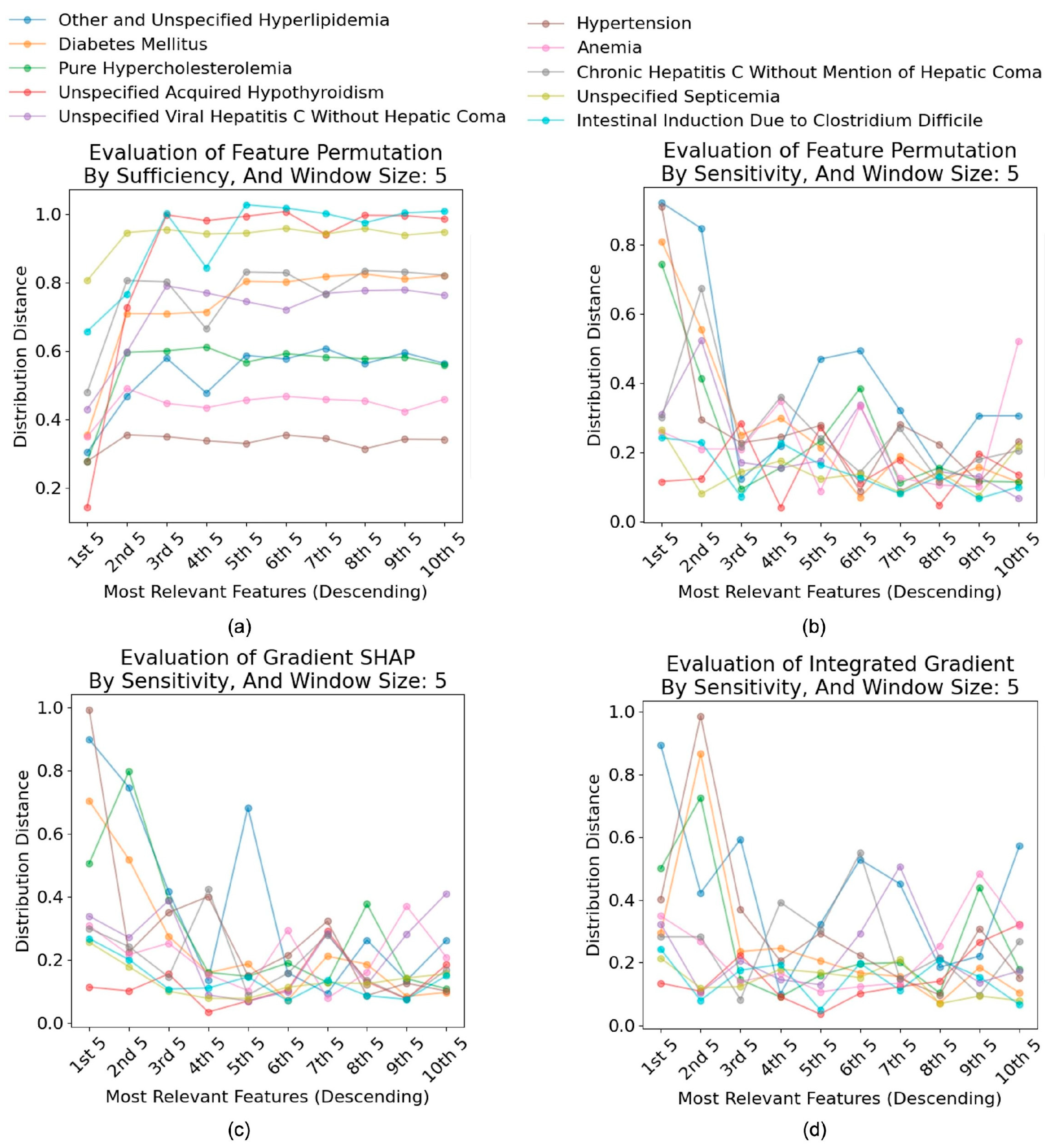
4.4. Evaluation of Interpretability Techniques
4.4.1. Faithfulness of Interpretability Tools
4.4.2. Effectiveness of Interpretability Tools
4.4.3. Discussions
4.5. Computational Cost Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ML | Machine Learning |
| MIMIC | Medical Information Mart for Intensive Care |
| PDD | Multidisciplinary Digital Publishing Institute |
| TF-IDF | Term Frequency-Inverse Document Frequency |
| TM | Topic Modeling |
| SHAP | SHapley Additive exPlanations |
| ICD-9 | International Classification of Disease |
| CNN | Convolutional Neural Network Model |
| GRU | Gate Recurrent Unit Model |
| LSTM | Long Short-Term Memory Model |
| RNN | Recurrent Neural Network Model |
| MultiResCNN | Multi-Filter Residual Convolutional Neural Network Model |
| PCA | Principal Component Analysis |
| SRM | Statistical Residual Matrix |
| AVs | Attributes Values |
| DSU | Disentangled Space Unit |
| AUC | Area Under the ROC curve |
| LLM | Large Language Model |
References
- Alowais, S.; Alghamdi, S.; Alsuhebany, N.; Alqahtani, T.; Alshaya, A.; Almohareb, S.N.; Aldairem, A.; Alrashed, M.; Saleh, K.B.; Badreldin, H.A.; et al. Revolutionizing healthcare: The role of artificial intelligence in clinical practice. BMC Med. Educ. 2023, 23, 689. [Google Scholar] [CrossRef] [PubMed]
- Alsentzer, E.; Murphy, J.R.; Boag, W.; Weng, W.-H.; Jin, D.; Naumann, T.; McDermott, M. Publicly available clinical BERT embeddings. arXiv 2019, arXiv:1904.03323. [Google Scholar]
- Hamid, S. The Opportunities and Risks of Artificial Intelligence in Medicine and Healthcare. 2016. Available online: https://www.repository.cam.ac.uk/items/4494a9ef-7256-4bd3-bd55-9d642ec9ab04 (accessed on 10 February 2025).
- Van Aken, B.; Papaioannou, J.-M.; Mayrdorfer, M.; Budde, K.; Gers, F.A.; Löser, A. Clinical outcome prediction from admission notes using self-supervised knowledge integration. arXiv 2021, arXiv:2102.04110. [Google Scholar]
- Hassija, V.; Chamola, V.; Mahapatra, A.; Singal, A.; Goel, D.; Huang, K.; Scardapane, S.; Spinelli, I.; Mahmud, M.; Hussain, A. Interpreting Black-Box Models: A Review on Explainable Artificial Intelligence. Cogn. Comput. 2024, 16, 45–74. [Google Scholar] [CrossRef]
- Ghannam, R.B.; Techtmann, S.M. Machine learning applications in microbial ecology, human microbiome studies, and environmental monitoring. Comput. Struct. Biotechnol. J. 2021, 19, 1092–1107. [Google Scholar] [CrossRef]
- Salahuddin, Z.; Woodruff, H.C.; Chatterjee, A.; Lambin, P. Transparency of deep neural networks for medical image analysis: A review of interpretability methods. Comput. Biol. Med. 2022, 140, 105111. [Google Scholar] [CrossRef]
- Ehghaghi, M.; Zhou, P.-Y.; Cheng, W.Y.; Rajabi, S.; Kuo, C.-H.; Lee, E.-S.A. Interpretable Disease Prediction from Clinical Text by Leveraging Pattern Disentanglement. In Proceedings of the 2023 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI), Pittsburgh, PA, USA, 15–18 October 2023. [Google Scholar]
- Wong, A.K.C.; Zhou, P.-Y.; Lee, A.E.-S. Theory and rationale of interpretable all-in-one pattern discovery and disentanglement system. npj Digit. Med. 2023, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.; Shen, L.B.L.; Gayles, A.; Shammout, A.; Horng, S.; Pollard, T.; Hao, S.; Moody, B.; Gow, B.; Lehman, L.-w.; et al. MIMIC-IV, a freely accessible electronic health record dataset. Sci. Data 2023, 10, 1. [Google Scholar] [CrossRef]
- Molnar, C. Interpretable Machine Learning, 3rd ed. 2022. Available online: https://christophm.github.io/interpretable-ml-book (accessed on 10 February 2025).
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic Attribution for Deep Networks. In International Conference on Machine Learning; PMLR: Birmingham, UK, 2017. [Google Scholar]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Yin, F.; Shi, Z.; Hsieh, C.-J.; Chang, K.-W. On the sensitivity and stability of model interpretations in NLP. arXiv 2021, arXiv:2104.08782. [Google Scholar]
- Ravı, D.; Wong, C.; Deligianni, F.; Berthelot, M.; Andreu-Perez, J.; Lo, B.; Yang, G.-Z. Deep learning for health informatics. IEEE J. Biomed. Health Inform. 2016, 21, 4–21. [Google Scholar] [CrossRef] [PubMed]
- Horng, S.; Sontag, D.A.; Halpern, Y.; Jernite, Y.; Shapiro, N.I.; Nathanson, L.A. Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PLoS ONE 2017, 12, e0174708. [Google Scholar] [CrossRef]
- DeYoung, J.; Jain, S.; Rajani, N.F.; Lehman, E.; Xiong, C.; Socher, R.; Wallace, B.C. ERASER: A Benchmark to Evaluate Rationalized NLP Models. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Johnson, S.L.J. AI, machine learning, and ethics in health care. J. Leg. Med. 2019, 39, 427–441. [Google Scholar] [CrossRef]
- Lyell, D.; Magrabi, F.; Raban, M.Z.; Pont, L.G.; Baysari, M.T.; Day, R.O.; Coiera, E. Automation bias in electronic prescribing. BMC Med. Inform. Decis. Mak. 2017, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Doshi-Velez, F.; Kim, B. Towards a rigorous science of interpretable machine learning. arXiv 2017, arXiv:1702.08608. [Google Scholar]
- Lipton, Z.C. The mythos of model interpretability: In machine learning, the concept of interpretability is both important and slippery. Queue 2018, 16, 31–57. [Google Scholar] [CrossRef]
- Johnson, A.; Bulgarelli, L.; Pollard, T.; Gow, B.; Moody, B.; Horng, S.; Celi, L.A.; Mark, R. MIMIC-IV (Version 3.1). PhysioNet 2024. Available online: https://physionet.org/content/mimiciv/3.1/ (accessed on 11 October 2024).
- Johnson, A.; Pollard, T.; Horng, S.; Celi, L.A.; Mark, R. MIMIC-IV-Note: Deidentified Free-Text Clinical Notes (Version 2.2). PhysioNet 2023. Available online: https://physionet.org/content/mimic-iv-note/2.2/ (accessed on 6 January 2023).
- Edin, J. Medical Coding Reproducibility. 2021. Available online: https://github.com/JoakimEdin/medical-coding-reproducibility (accessed on 11 October 2024).
- Sievert, C.; Shirley, K. LDAvis: A method for visualizing and interpreting topics. In Proceedings of the Workshop on Interactive Language Learning, Visualization, and Interfaces, Baltimore, MD, USA, 27 June 2014. [Google Scholar]
- Achiam, J.; Adler, S.; Agarwal, S.; Ahmad, L.; Akkaya, I.; Leoni Aleman, F.; Almeida, D.; Altenschmidt, J.; Altman, S.; Anadkat, S.; et al. Gpt-4 technical report. arXiv 2023, arXiv:2303.08774. [Google Scholar]
- Edin, J.; Junge, A.; Havtorn, J.D.; Borgholt, L.; Maistro, M.; Ruotsalo, T.; Maaløe, L. Automated Medical Coding on MIMIC-III and MIMIC-IV: A Critical Review and Replicability Study. In Proceedings of the 46th International ACM SIGIR Conference on Research and Development in Information Retrieval, Taipei, Taiwan, 23–27 July 2023. [Google Scholar]
- Huang, J.; Osorio, C.; Sy, L.W. An empirical evaluation of deep learning for ICD-9 code assignment using MIMIC-III clinical notes. Comput. Methods Programs Biomed. 2019, 177, 141–153. [Google Scholar] [CrossRef]
- Lipton, Z.C.; Kale, D.C.; Elkan, C.; Wetzel, R. Learning to diagnose with LSTM recurrent neural networks. arXiv 2015, arXiv:1511.03677. [Google Scholar]
- Li, F.; Yu, H. ICD coding from clinical text using multi-filter residual convolutional neural network. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020. [Google Scholar]
- Wong, A.K.C.; Zhou, P.-Y.; Butt, Z.A. Pattern discovery and disentanglement on relational datasets. Sci. Rep. 2021, 11, 5688. [Google Scholar] [CrossRef] [PubMed]
- Jacovi, A.; Goldberg, Y. Towards Faithfully Interpretable NLP Systems: How should we define and evaluate faithfulness? arXiv 2020, arXiv:2004.03685. [Google Scholar]
- Chan, C.S.; Kong, H.; Liang, G. A comparative study of faithfulness metrics for model interpretability methods. arXiv 2022, arXiv:2204.05514. [Google Scholar]

| ICD-9 Code | Disease Name | Frequency | Distribution |
|---|---|---|---|
| 250.0 | Diabetes mellitus without mention of complication, type II or unspecified type, not stated as uncontrolled | 12,539 | 24.1246% |
| 272.4 | Other and unspecified hyperlipidemia | 11,840 | 22.7797% |
| 244.9 | Unspecified acquired hypothyroidism | 10,224 | 19.6706% |
| 272.0 | Pure hypercholesterolemia | 3411 | 6.5626% |
| 38.9 | Unspecified septicemia | 2878 | 5.5372% |
| 401.9 | Unspecified essential hypertension | 2521 | 4.8503% |
| 70.54 | Chronic hepatitis C without mention of hepatic coma | 2466 | 4.7445% |
| 8.45 | Intestinal induction due to Clostridium difficile | 2327 | 4.4771% |
| 70.7 | Unspecified viral hepatitis C without hepatic coma | 2069 | 3.9807% |
| 285.9 | Anemia, unspecified | 1701 | 3.2727% |
| Data | Model | Macro ROC-AUC | Micro ROC-AUC | Macro F1 | Micro F1 | Balanced Acc. | Macro Prec. | Micro Prec. | Macro Recall | Micro Recall |
|---|---|---|---|---|---|---|---|---|---|---|
| TM_Top10 | CNN | 0.784 | 0.842 | 0.262 | 0.360 | 0.274 | 0.288 | 0.360 | 0.274 | 0.360 |
| GRU | 0.781 | 0.840 | 0.257 | 0.365 | 0.279 | 0.263 | 0.365 | 0.279 | 0.365 | |
| LSTM | 0.764 | 0.831 | 0.258 | 0.353 | 0.277 | 0.325 | 0.353 | 0.277 | 0.353 | |
| MultiResCNN | 0.793 | 0.846 | 0.269 | 0.373 | 0.289 | 0.267 | 0.373 | 0.289 | 0.373 | |
| SVM | 0.776 | 0.838 | 0.266 | 0.368 | 0.283 | 0.268 | 0.368 | 0.283 | 0.368 | |
| RF | 0.776 | 0.838 | 0.263 | 0.367 | 0.278 | 0.307 | 0.367 | 0.278 | 0.367 | |
| PDD | 0.689 | 0.643 | 0.370 | 0.358 | 0.447 | 0.440 | 0.358 | 0.450 | 0.358 | |
| TF-IDF_Top10 | CNN | 0.950 | 0.963 | 0.623 | 0.738 | 0.604 | 0.671 | 0.738 | 0.604 | 0.738 |
| GRU | 0.941 | 0.958 | 0.543 | 0.713 | 0.523 | 0.604 | 0.713 | 0.523 | 0.713 | |
| LSTM | 0.548 | 0.721 | 0.036 | 0.223 | 0.100 | 0.036 | 0.223 | 0.100 | 0.223 | |
| MultiResCNN | 0.956 | 0.968 | 0.611 | 0.743 | 0.596 | 0.689 | 0.743 | 0.596 | 0.743 | |
| SVM | 0.947 | 0.962 | 0.514 | 0.692 | 0.509 | 0.658 | 0.692 | 0.509 | 0.692 | |
| RF | 0.947 | 0.962 | 0.58 | 0.731 | 0.558 | 0.662 | 0.731 | 0.558 | 0.731 | |
| PDD | 0.825 | 0.855 | 0.640 | 0.738 | 0.679 | 0.650 | 0.738 | 0.680 | 0.738 | |
| TF-IDF_Top10 (Bigram included) | CNN | 0.943 | 0.960 | 0.616 | 0.736 | 0.603 | 0.647 | 0.736 | 0.603 | 0.736 |
| GRU | 0.933 | 0.955 | 0.507 | 0.702 | 0.526 | 0.558 | 0.702 | 0.526 | 0.702 | |
| LSTM | 0.937 | 0.958 | 0.570 | 0.713 | 0.575 | 0.598 | 0.713 | 0.575 | 0.713 | |
| MultiResCNN | 0.948 | 0.966 | 0.598 | 0.739 | 0.589 | 0.652 | 0.739 | 0.589 | 0.739 | |
| SVM | 0.942 | 0.962 | 0.591 | 0.719 | 0.586 | 0.614 | 0.719 | 0.586 | 0.719 | |
| RF | 0.942 | 0.962 | 0.604 | 0.738 | 0.589 | 0.646 | 0.738 | 0.589 | 0.738 | |
| PDD | 0.909 | 0.890 | 0.820 | 0.802 | 0.841 | 0.822 | 0.802 | 0.841 | 0.802 |
| Interpretability Method | Faithfulness Criteria | ICD_9 Code | Average | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 272.4 | 250.0 | 272 | 244.9 | 70.7 | 401.9 | 285.9 | 70.54 | 38.9 | 8.45 | |||
| Gradient SHAP | Comprehensiveness | 0.391 | 0.596 | 0.489 | 1.00 | 0.390 | 0.194 | 0.269 | 0.435 | 0.474 | 0.820 | 0.506 |
| Sufficiency | 0.1965 | 0.216 | 0.203 | 0.106 | 0.316 | 0.227 | 0.288 | 0.379 | 0.502 | 0.275 | 0.271 | |
| Sensitivity | 0.908 | 0.825 | 0.727 | 0.495 | 0.538 | 0.796 | 0.515 | 0.479 | 0.292 | 0.266 | 0.584 | |
| Feature Permutation | Comprehensiveness | 0.404 | 0.591 | 0.503 | 0.999 | 0.409 | 0.205 | 0.288 | 0.467 | 0.468 | 0.783 | 0.510 |
| Sufficiency | 0.192 | 0.212 | 0.206 | 0.102 | 0.310 | 0.220 | 0.281 | 0.344 | 0.518 | 0.290 | 0.267 | |
| Sensitivity | 0.928 | 0.842 | 0.748 | 0.475 | 0.514 | 0.761 | 0.559 | 0.528 | 0.287 | 0.265 | 0.591 | |
| TIIntegrated Gradient | Comprehensiveness | 0.410 | 0.573 | 0.491 | 0.981 | 0.356 | 0.220 | 0.277 | 0.424 | 0.426 | 0.679 | 0.484 |
| Sufficiency | 0.192 | 0.221 | 0.213 | 0.107 | 0.369 | 0.234 | 0.304 | 0.388 | 0.569 | 0.382 | 0.298 | |
| Sensitivity | 0.856 | 0.806 | 0.751 | 0.490 | 0.455 | 0.760 | 0.514 | 0.457 | 0.277 | 0.262 | 0.563 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, P.-Y.; Takeuchi, A.; Martinez-Lopez, F.; Ehghaghi, M.; Wong, A.K.C.; Lee, E.-S.A. Benchmarking Interpretability in Healthcare Using Pattern Discovery and Disentanglement. Bioengineering 2025, 12, 308. https://doi.org/10.3390/bioengineering12030308
Zhou P-Y, Takeuchi A, Martinez-Lopez F, Ehghaghi M, Wong AKC, Lee E-SA. Benchmarking Interpretability in Healthcare Using Pattern Discovery and Disentanglement. Bioengineering. 2025; 12(3):308. https://doi.org/10.3390/bioengineering12030308
Chicago/Turabian StyleZhou, Pei-Yuan, Amane Takeuchi, Fernando Martinez-Lopez, Malikeh Ehghaghi, Andrew K. C. Wong, and En-Shiun Annie Lee. 2025. "Benchmarking Interpretability in Healthcare Using Pattern Discovery and Disentanglement" Bioengineering 12, no. 3: 308. https://doi.org/10.3390/bioengineering12030308
APA StyleZhou, P.-Y., Takeuchi, A., Martinez-Lopez, F., Ehghaghi, M., Wong, A. K. C., & Lee, E.-S. A. (2025). Benchmarking Interpretability in Healthcare Using Pattern Discovery and Disentanglement. Bioengineering, 12(3), 308. https://doi.org/10.3390/bioengineering12030308









