Visible Exocytosis of the Non-Photic Signal Neuropeptide Y to the Suprachiasmatic Nucleus in Fasted Transgenic Mice Throughout Their Circadian Rhythms
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of the Tg Mice
2.2. Genomic PCR Screening of the Tg Mice
2.3. Animals
2.4. Wheel-Running Activity for the Mice
2.5. Preparation of the Mouse Brain Slices and Immunostaining
2.6. Preparation of the Mouse Brain Slices and Transparency
2.7. Fluorescence Observation by Confocal Microscopy
2.8. Fluorescence Observation by Super-Resolution Microscopy
2.9. Statistical Analysis
3. Results
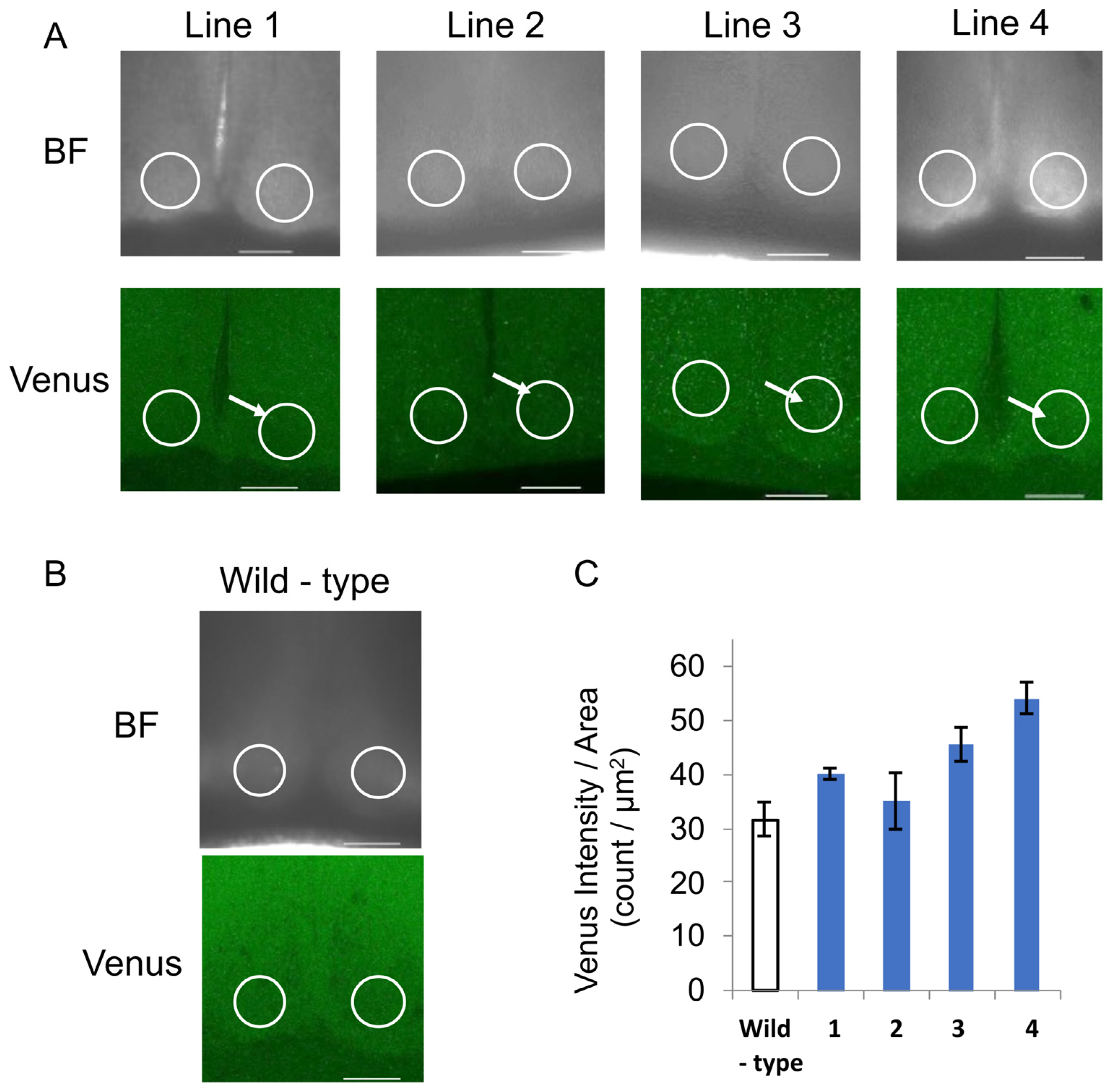
3.1. Generation of the Tg Animals
3.2. Selection of the NPY::Venus Tg Mice
3.3. Imaging of the SCN Slices of the NPY::Venus Tg Mice
3.4. Comparison of the NPY::Venus Expression Between the Feeding Condition Groups
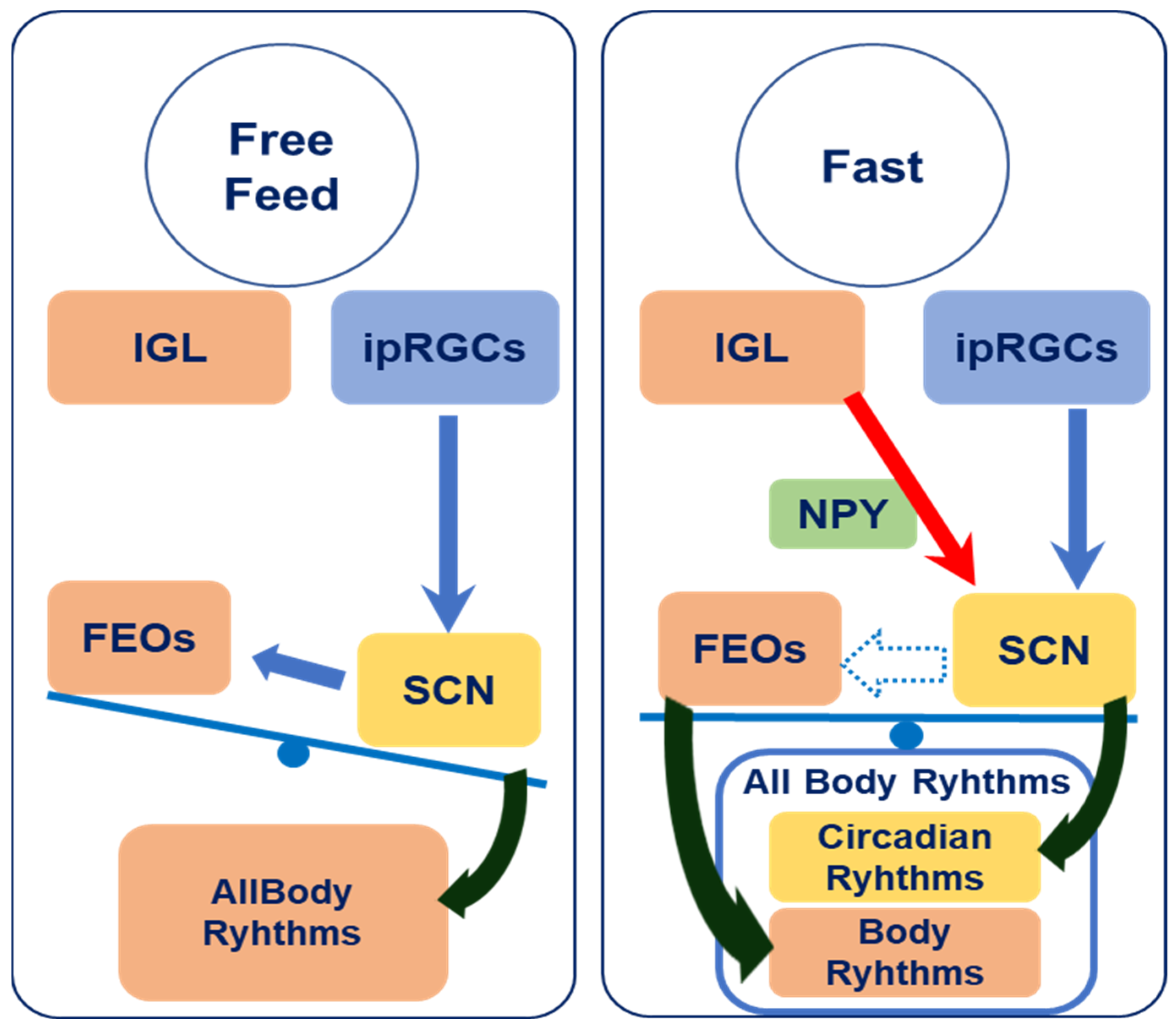
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SCN | the suprachiasmatic nucleus |
| FEO | the food-entrainable circadian oscillator |
| NPY | neuropeptide Y |
| IGL | intergeniculate leaflet |
| Tg | transgenic |
| Per | Periods |
| Per1::luc | Per1 promoter::luciferase |
| RF | restricted feeding |
| FF | free feeding |
| ZT | Zeitgeber time |
| FAA | food anticipatory activity |
| ORF | open reading frame |
| SV | simian virus |
| TUT | Toyohashi University of Technology |
| DD | constant dark |
| PBS | phosphate-buffered saline |
References
- Tei, H.; Okamura, H.; Shigeyoshi, Y.; Fukuhara, C.; Ozawa, R.; Hirose, M.; Sakaki, Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 1997, 389, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.S.; Albrecht, U.; Zhuchenko, O.; Bailey, J.; Eichele, G.; Lee, C.C. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell 1997, 90, 1003–1011. [Google Scholar] [CrossRef]
- Shearman, L.P.; Zylka, M.J.; Weaver, D.R.; Kolakowski, L.F., Jr.; Reppert, S.M. Two period Homologs: Circadian Expression and Photic Regulation in the Suprachiasmatic Nuclei. Neuron 1997, 19, 1261–1269. [Google Scholar] [CrossRef]
- Numano, R.; Yamazaki, S.; Umeda, N.; Samura, T.; Sujino, M.; Takahashi, R.-I.; Ueda, M.; Mori, A.; Yamada, K.; Sakaki, Y.; et al. Constitutive expression of the Period1 gene impairs behavioral and molecular circadian rhythms. Proc. Natl. Acad. Sci. USA 2006, 103, 3716–3721. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Numano, R.; Abe, M.; Hida, A.; Takahashi, R.-I.; Ueda, M.; Block, G.D.; Sakaki, Y.; Menaker, M.; Tei, H. Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000, 288, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Ohta, H.; Yamazaki, S.; McMahon, D.G. Constant light desynchronizes mammalian clock neurons. Nat. Neurosci. 2005, 8, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Pendergast, J.S.; Yamazaki, S. The Mysterious Food-Entrainable Oscillator: Insights from Mutant and Engineered Mouse Models. J. Biol. Rhythm. 2018, 33, 458–474. [Google Scholar] [CrossRef]
- Stokkan, K.-A.; Yamazaki, S.; Tei, H.; Sakaki, Y.; Menaker, M. Entrainment of the circadian clock in the liver by feeding. Science 2001, 291, 490–493. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.J.; Poole, A.S.; Yamazaki, S.; Menaker, M. Is the food-entrainable circadian oscillator in the digestive system? Genes Brain Behav. 2003, 2, 32–39. [Google Scholar] [CrossRef]
- Nakazawa, K.; Matsuo, M.; Kimura, N.; Numano, R. Restricted Feeding Resets the Peripheral Clocks of the Digestive System. Biomedicines 2023, 11, 1463. [Google Scholar] [CrossRef]
- Gribkoff, V.K.; Pieschl, R.L.; Wisialowski, T.A.; van den Pol, A.N.; Yocca, F.D. Phase shifting of circadian rhythms and depression of neuronal activity in the rat suprachiasmatic nucleus by neuropeptide Y: Mediation by different receptor subtypes. J. Neurosci. 1998, 18, 3014–3022. [Google Scholar] [CrossRef] [PubMed]
- Biello, S.M.; Golombek, D.A.; Schak, K.M.; Harrington, M.E. Circadian phase shifts to neuropeptide Y in vitro: Cellular communication and signal transduction. J. Neurosci. 1997, 17, 8468–8475. [Google Scholar] [CrossRef] [PubMed]
- Besing, R.C.; Hablitz, L.M.; Paul, J.R.; Johnson, R.L.; Prosser, R.A.; Gamble, K.L. Neuropeptide Y–Induced phase shifts of PER2::LUC rhythms are mediated by long-term suppression of neuronal excitability in a phase-specific manner. Chronobiol Int. 2012, 29, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Sindelar, D.K.; Palmiter, R.D.; Woods, S.C.; Schwartz, M.W. Attenuated feeding responses to circadian and palatability cues in mice lacking neuropeptide Y. Peptides 2005, 26, 2597–2602. [Google Scholar] [CrossRef]
- Saderi, N.; Cazarez-Márquez, F.; Buijs, F.; Salgado-Delgado, R.; Guzman-Ruiz, M.; del Carmen Basualdo, M.; Escobar, C.; Buijs, R. The NPY intergeniculate leaflet projections to the suprachiasmatic nucleus transmit metabolic conditions. Neuroscience 2013, 246, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.Y.; Card, J.P. Intergeniculate leaflet: An anatomically and functionally distinct subdivision of the lateral geniculate complex. J. Comp. Neurol. 1994, 344, 403–430. [Google Scholar] [CrossRef]
- Morin, L.P. Neuroanatomy of the extended circadian rhythm system. Exp. Neurol. 2013, 243, 4–20. [Google Scholar] [CrossRef]
- Wams, E.J.; Riede, S.J.; van der Laan, I.; ten Bulte, T.; Hut, R.A. Mechanisms of Non-photic Entrainment. In Biological Timekeeping: Clocks, Rhythms and Behaviour; Springer: New Delhi, India, 2017; pp. 395–404. [Google Scholar] [CrossRef]
- Fernandez, D.C.; Komal, R.; Langel, J.; Ma, J.; Duy, P.Q.; Penzo, M.A.; Zhao, H.; Hattar, S. Retinal innervation tunes circuits that drive nonphotic entrainment to food. Nature 2020, 581, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, T.; Ravier, M.A.; Parton, L.E.; Rutter, G.A. Sustained exposure to high glucose concentrations modifies glucose signaling and the mechanics of secretory vesicle fusion in primary rat pancreatic β-cells. Diabetes 2006, 55, 1057–1065. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Yannielli, P.C.; Harrington, M.E. Neuropeptide Y applied in vitro can block the phase shifts induced by light in vivo. NeuroReport 2000, 11, 1587–1591. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fukuhara, C.; Brewer, J.M.; Dirden, J.C.; Bittman, E.L.; Tosini, G.; Harrington, M.E. Neuropeptide Y rapidly reduces Period 1 and Period 2 mRNA levels in the hamster suprachiasmatic nucleus. Neurosci. Lett. 2001, 314, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Brewer, J.M.; Yannielli, P.C.; Harrington, M.E. Neuropeptide Y differentially suppresses per1 and per2 mRNA induced by light in the suprachiasmatic nuclei of the golden hamster. J. Biol. Rhythm. 2002, 17, 28–39. [Google Scholar] [CrossRef]
- Soscia, S.J.; Harrington, M.E. Neuropeptide Y attenuates NMDA-induced phase shifts in the SCN of NPY Y1 receptor knockout mice in vitro. Brain Res. 2004, 1023, 148–153. [Google Scholar] [CrossRef]
- Nakao, K.; Inoue, K. Cryopreservation for broader production of transgenic mice by DNA injection into zygotes. Exp. Anim. 2010, 59, 225–229. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Güler, A.D.; Ecker, J.L.; Lall, G.S.; Haq, S.; Altimus, C.M.; Liao, H.-W.; Barnard, A.R.; Cahill, H.; Badea, T.C.; Zhao, H.; et al. Melanopsin cells are the principal conduits for rod–cone input to non-image-forming vision. Nature 2010, 453, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lee, G.Y.; Biggers, J.D.; Toth, T.L.; Toner, M. Low cryoprotectant concentration rapid vitrification of mouse oocytes and embryos. Cryobiology 2021, 98, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Hattar, S.; Liao, H.-W.; Takao, M.; Berson, D.M.; Yau, K.-W. Melanopsin-Containing Retinal Ganglion Cells: Architecture, Projections, and Intrinsic Photosensitivity. Science 2002, 295, 1065–1070. [Google Scholar] [CrossRef]
- Fernandez, D.C.; Chang, Y.-T.; Hattar, S.; Chen, S.-K. Architecture of retinal projections to the central circadian pacemaker. Proc. Natl. Acad. Sci. USA 2016, 113, 6047–6052. [Google Scholar] [CrossRef]
- van Diepen, H.C.; Schoonderwoerd, R.A.; Ramkisoensing, A.; Janse, J.A.M.; Hattar, S.; Meijer, J.H. Distinct contribution of cone photoreceptor subtypes to the mammalian biological clock. Proc. Natl. Acad. Sci. USA 2021, 118, e2024500118. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, K.; Matsuo, M.; Kikuchi, Y.; Nakajima, Y.; Numano, R. Melanopsin DNA aptamers can regulate input signals of mammalian circadian rhythms by altering the phase of the molecular clock. Front. Neurosci. 2024, 18, 1186677. [Google Scholar] [CrossRef]
- Nakanishi-Kimura, A.; Takakura, A.; Hoshi-Numahata, M.; Watanabe, H.; Nishiura, M.; Sato, Y.; Takao-Kawabata, R.; Iimura, T. Dynamic morphometric changes in the mandibular osteocytic lacunae of ovariectomized rats in response to teriparatide, as revealed by three-dimensional fluorescence analyses: Possible involvement of osteocytic perilacunar remodeling. J. Oral Biosci. 2024, 66, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Stephan, F.K. Interaction between light- and feeding-entrainable circadian rhythms in the rat. Physiol. Behav. 1986, 38, 127–133. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakazawa, K.; Matsuo, M.; Nakao, K.; Nonaka, S.; Numano, R. Visible Exocytosis of the Non-Photic Signal Neuropeptide Y to the Suprachiasmatic Nucleus in Fasted Transgenic Mice Throughout Their Circadian Rhythms. Bioengineering 2025, 12, 192. https://doi.org/10.3390/bioengineering12020192
Nakazawa K, Matsuo M, Nakao K, Nonaka S, Numano R. Visible Exocytosis of the Non-Photic Signal Neuropeptide Y to the Suprachiasmatic Nucleus in Fasted Transgenic Mice Throughout Their Circadian Rhythms. Bioengineering. 2025; 12(2):192. https://doi.org/10.3390/bioengineering12020192
Chicago/Turabian StyleNakazawa, Kazuo, Minako Matsuo, Kazuki Nakao, Shigenori Nonaka, and Rika Numano. 2025. "Visible Exocytosis of the Non-Photic Signal Neuropeptide Y to the Suprachiasmatic Nucleus in Fasted Transgenic Mice Throughout Their Circadian Rhythms" Bioengineering 12, no. 2: 192. https://doi.org/10.3390/bioengineering12020192
APA StyleNakazawa, K., Matsuo, M., Nakao, K., Nonaka, S., & Numano, R. (2025). Visible Exocytosis of the Non-Photic Signal Neuropeptide Y to the Suprachiasmatic Nucleus in Fasted Transgenic Mice Throughout Their Circadian Rhythms. Bioengineering, 12(2), 192. https://doi.org/10.3390/bioengineering12020192



