Generative Modeling for Interpretable Anomaly Detection in Medical Imaging: Applications in Failure Detection and Data Curation
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
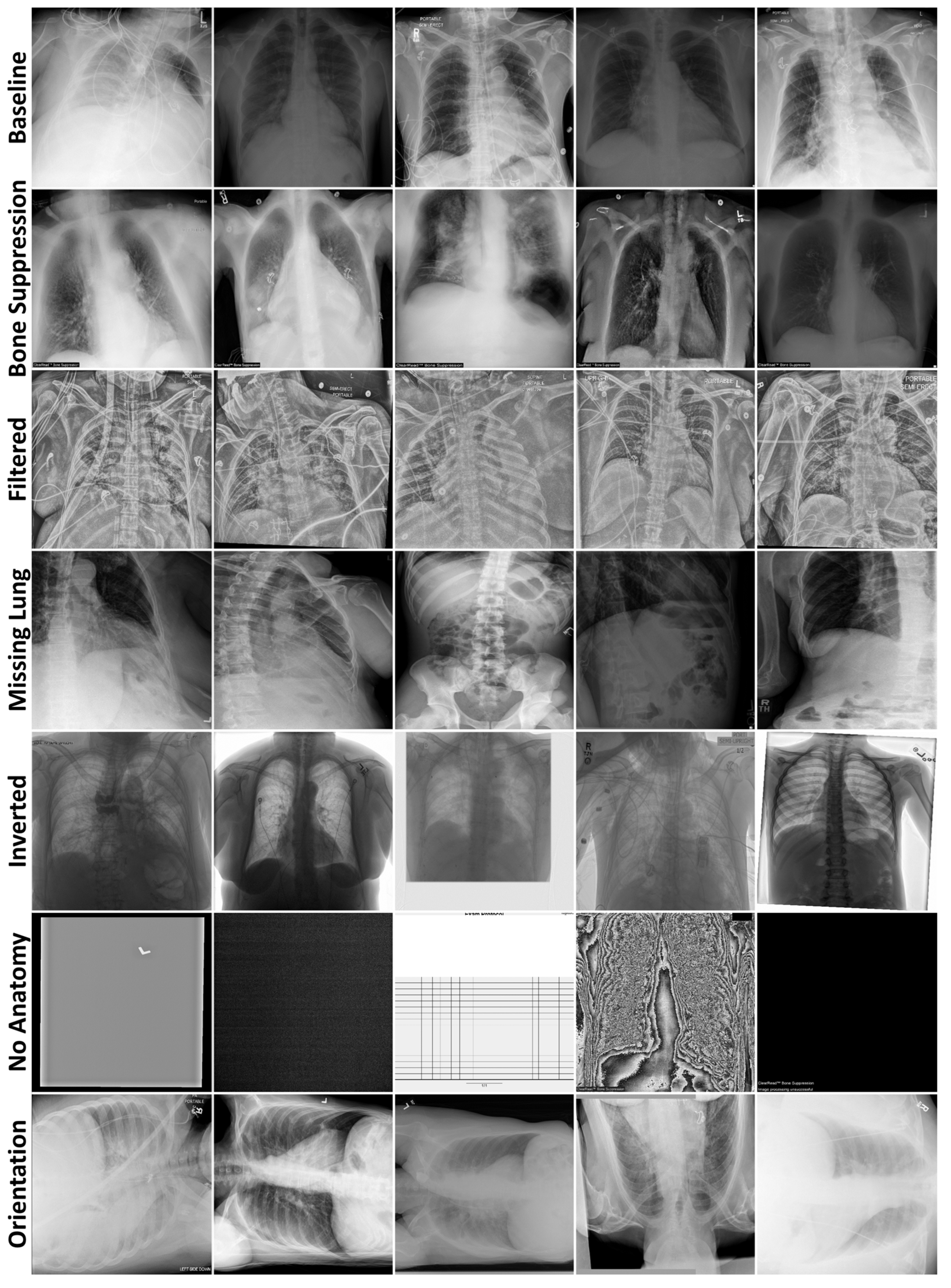
2.1.1. Failure Detection Datasets
2.1.2. Data Curation Datasets
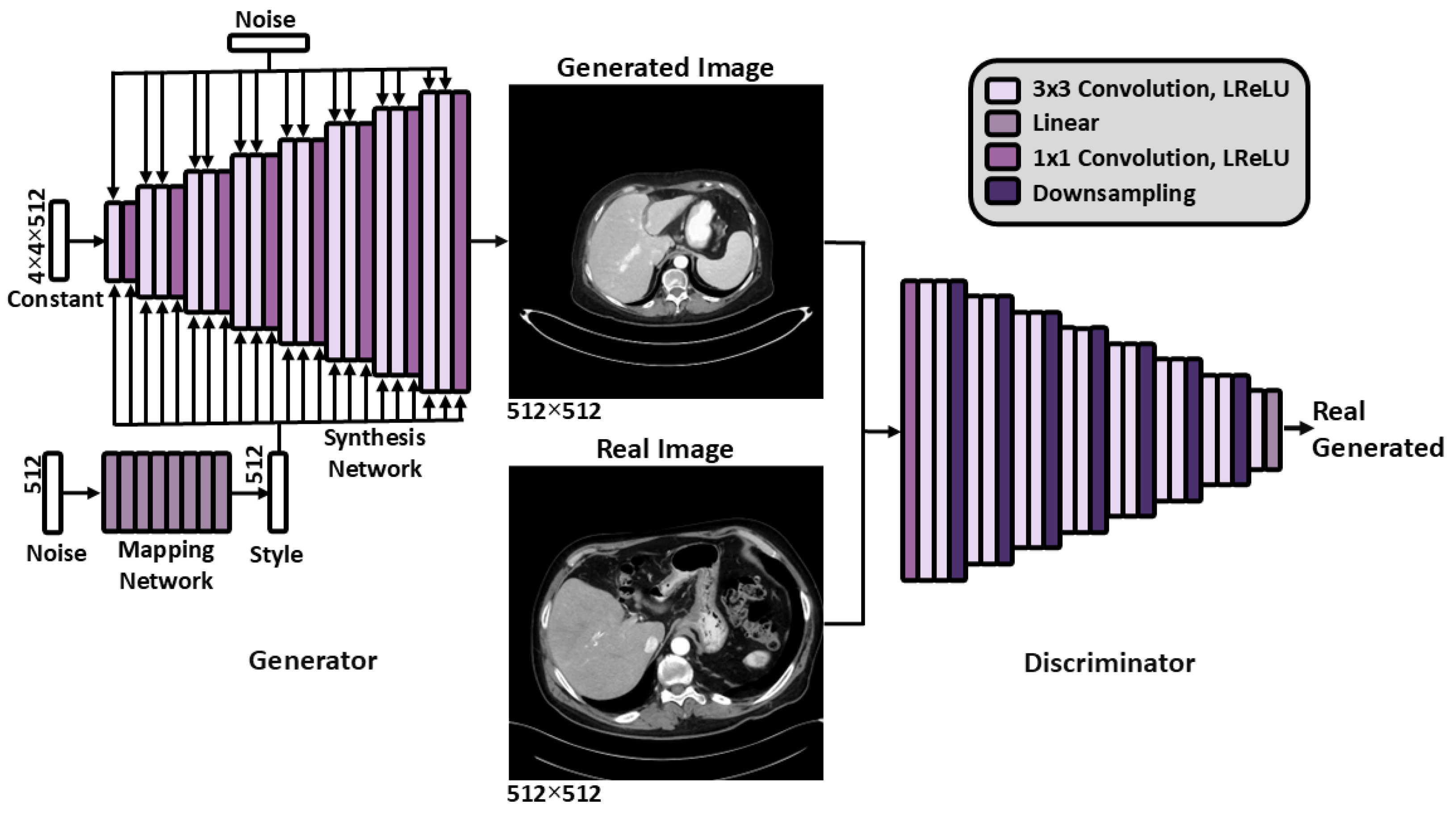
2.2. Generative Modeling for Anomaly Detection
2.2.1. Generative Model Training
2.2.2. Generative Modeling Evaluation
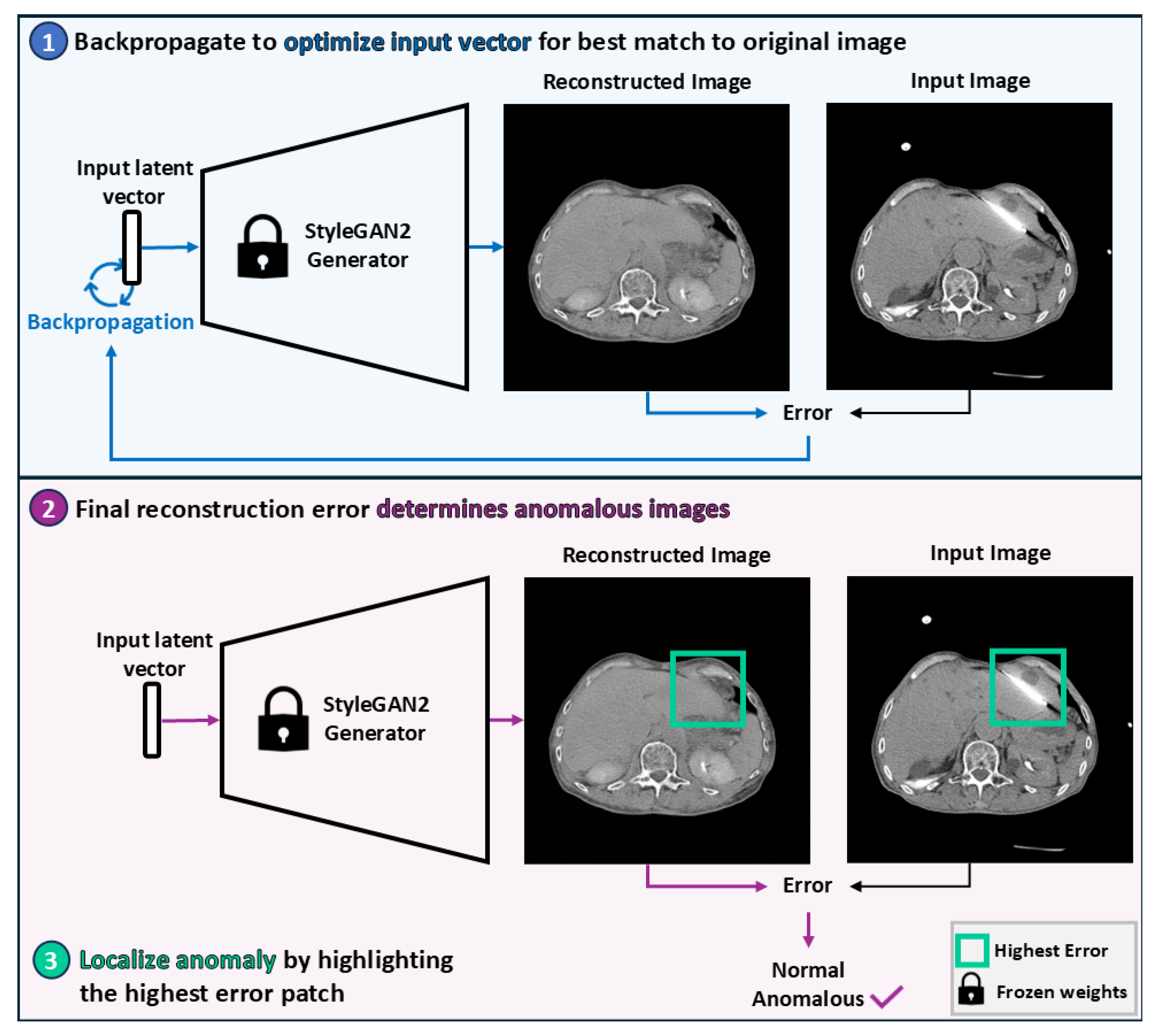
2.2.3. Image Reconstruction
2.2.4. Anomaly Detection
2.2.5. Anomaly Localization
2.3. Statistical Analysis
2.4. Code Availability
3. Results
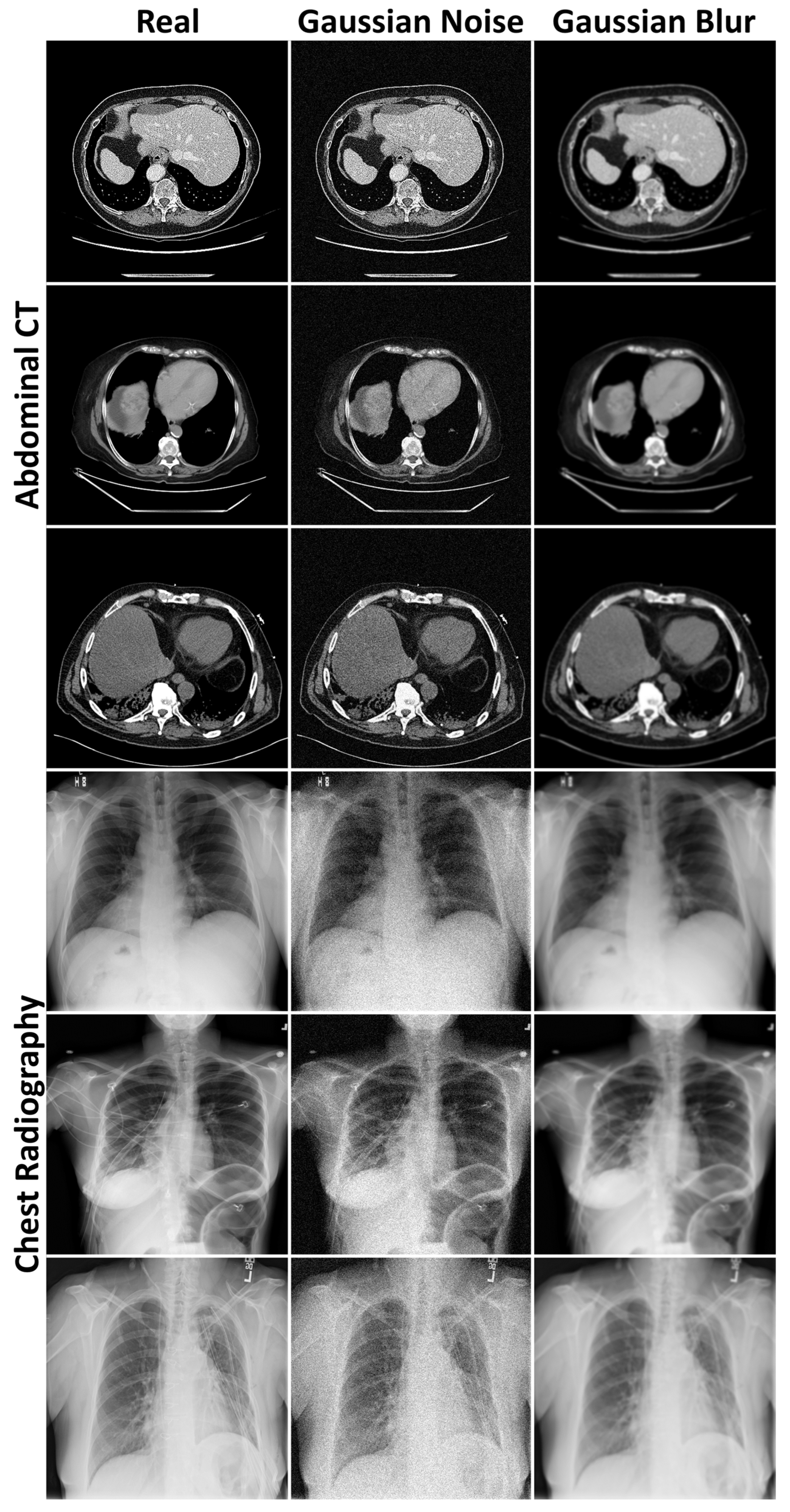
3.1. Evaluation of Generated Image Quality
3.2. Reconstruction Performance and Interpretation
3.3. Quantitative Anomaly Detection Performance
3.4. Localization of Anomalous Regions
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| AUROC | Area under the receiver operating characteristic curve |
| DICOM | Digital Imaging and Communication in Medicine |
| FD | Fréchet Distance |
| FID | Fréchet Inception Distance |
| FRD | Fréchet Radiomics Distance |
| FSD | Fréchet SwAV Distance |
| GAN | Generative Adversarial Network |
| HIPPA | Health Insurance Portability and Accountability Act |
| MIDRC | Medical Imaging and Data Resource Center |
| MSE | Mean squared error |
| PNG | Portable Network Graphic |
| SD | Standard deviation |
| WD | Wasserstein distance |
Appendix A
| Attribute | Baseline | Needles | Ascites | Brain, Head and Neck, Lung | Cervix |
|---|---|---|---|---|---|
| # Patients | 430 | 39 | 33 | 10 | 10 |
| Female | 194 (45) | 11 (28) | 14 (42) | 4 (40) | 10 (100) |
| Age | 63 (54–71) | 49 (48–58) | 66 (61–73) | 56 (54–62) | 38 (35–47) |
| # Images | 3235 | 48 | 33 | 10 | 10 |
| Contrast | 2134 (66) | 39 (66) | 31 (94) | 0 (0) | 1 (10) |
| Voxel Size | |||||
| X/Y | 0.8 (0.8–0.9) | 0.9 (0.8–0.9) | 0.8 (0.8–0.9) | 1.0 (0.9–1.0) | 1.2 (1.2–1.2) |
| Z | 3.0 (2.5–5.0) | 3.0 (3.0–3.0) | 2.5 (2.5–2.5) | 3.0 (3.0–3.0) | 3.0 (3.0–3.0) |
| Scanner | |||||
| GE BrightSpeed | 2 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| GE Discovery | 1116 (34) | 0 (0) | 8 (24) | 0 (0) | 1 (10) |
| GE LightSpeed | 123 (4) | 0 (0) | 3 (9) | 5 (50) | 0 (0) |
| GE Revolution | 665 (21) | 0 (0) | 4 (12) | 0 (0) | 0 (0) |
| Philips Big Bore | 0 (0) | 0 (0) | 2 (6) | 0 (0) | 9 (90) |
| Philips Brilliance 64 | 0 (0) | 0 (0) | 0 (0) | 2 (20) | 0 (0) |
| Philips Mx8000 IDT | 1 (0) | 0 (0) | 0 (0) | 3 (30) | 0 (0) |
| Siemens Sensation | 21 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Siemens SOMATOM | 1302 (40) | 48 (100) | 16 (48) | 0 (0) | 0 (0) |
| Toshiba Acquilion | 5 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Attribute | Baseline Train | Baseline Test | Bone Suppression | Filtering | Missing Lung | Inverted | No Anatomy | Orientation |
|---|---|---|---|---|---|---|---|---|
| # Images | 112,120 | 1,000 | 250 | 250 | 250 | 250 | 13 | 23 |
| Computed | - | 1000 (100) | 250 (100) | 250 (100) | 233 (93) | 250 (100) | 10 (77) | 1 (4) |
| Voxel Size X/Y | 0.1 (0.1–0.2) | 0.1 (0.1–0.1) | 0.1 (0.1–0.1) | 0.1 (0.1–0.1) | 0.1 (0.1–0.1) | 0.1 (0.1–0.1) | 0.1 (0.1–0.1) | 0.1 (0.1–0.1) |
| Unknown | 0 (0) | 42 (4) | 12 (5) | 14 (6) | 14 (6) | 8 (3) | 2 (15) | 8 (35) |
| Female | 48,780 (44) | 71 (43) | 16 (46) | 23 (55) | 18 (41) | 28 (47) | 1 (20) | 10 (48) |
| Unknown | 0 (0) | 833 (83) | 215 (86) | 208 (83) | 206 (82) | 191 (76) | 8 (62) | 2 (9) |
| Age | 49 (34–59) | 56 (44–65) | 55 (44–67) | 55 (43–65) | 55 (46–64) | 55 (40–64) | 50 (45–65) | 65 (55–70) |
| Unknown | 0 (0) | 367 (37) | 102 (41) | 89 (36) | 104 (42) | 78 (31) | 4 (31) | 0 (0) |
| Scanner | ||||||||
| AGFA CR 85 | 0 (0) | 9 (1) | 0 (0) | 2 (0) | 4 (2) | 3 (1) | 0 (0) | 0 (0) |
| Canon CXDI | 0 (0) | 6 (1) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 0 (0) |
| Carestream Classic CR | 0 (0) | 5 (1) | 1 (0) | 1 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) |
| Carestream DRX | 0 (0) | 27 (3) | 11 (4) | 3 (1) | 8 (3) | 8 (3) | 1 (8) | 0 (0) |
| GE Thunder | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 0 (0) |
| GE Revolution XRd | 0 (0) | 4 (0) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 1 (4) |
| GE WDR1Car | 0 (0) | 2 (0) | 2 (1) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) |
| Philips DigitalDiagnost | 0 (0) | 161 (16) | 38 (15) | 49 (20) | 44 (18) | 39 (16) | 4 (31) | 0 (0) |
| Philips Essenta | 0 (0) | 10 (1) | 4 (2) | 2 (1) | 0 (0) | 1 (0) | 0 (0) | 0 (0) |
| Philips MobileDiagnost | 0 (0) | 240 (24) | 61 (24) | 74 (30) | 66 (26) | 51 (20) | 0 (0) | 1 (4) |
| Siemens Fluorospot | 0 (0) | 6 (1) | 2 (1) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) |
| Unknown | 112,120 (100) | 534 (53) | 131 (52) | 118 (47) | 125 (50) | 145 (58) | 8 (62) | 21 (91) |
Appendix B

References
- Woodland, M.; Wood, J.; Anderson, B.M.; Kundu, S.; Lin, E.; Koay, E.; Odisio, B.; Chung, C.; Kang, H.C.; Venkatesan, A.M.; et al. Evaluating the performance of StyleGAN2-ADA on medical images. In Proceedings of the SASHIMI 2022, Singapore, 18 September 2022; Zhao, M., Svoboda, D., Wolterink, J.M., Escobar, M., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2022; Volume 13570, pp. 142–153. [Google Scholar]
- Yang, J.; Zhou, K.; Li, Y.; Liu, Z. Generalized out-of-distribution detection: A survey. Int. J. Comput. Vis. 2024, 132, 5635–5662. [Google Scholar] [CrossRef]
- Fernando, T.; Gammulle, H.; Denman, S.; Sridharan, S.; Fookes, C. Deep learning for medical anomaly detection—A survey. ACM Comput. Surv. 2021, 54, e141. [Google Scholar] [CrossRef]
- Xia, X.; Pan, X.; Li, N.; He, X.; Ma, L.; Zhang, X.; Ding, N. GAN-based anomaly detection: A review. Neurocomputing 2022, 493, 497–535. [Google Scholar] [CrossRef]
- Chen, X.; Konukoglu, E. Unsupervised abnormality detection in medical images with deep generative methods. In Biomedical Image Synthesis and Simulation: Methods and Applications; Burgos, N., Svoboda, D., Eds.; Academic Press: San Diego, CA, USA, 2022; pp. 303–324. [Google Scholar]
- Zech, J.R.; Badgeley, M.A.; Liu, M.; Costa, A.B.; Titano, J.J.; Oermann, E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med. 2018, 15, e1002683. [Google Scholar] [CrossRef]
- Willemink, M.J.; Koszek, W.A.; Hardell, C.; Wu, J.; Fleischmann, D.; Harvey, H.; Folio, L.R.; Summers, R.M.; Rubin, D.L.; Lungren, M.P. Preparing medical imaging data for machine learning. Radiology 2020, 295, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.M.; Lin, E.Y.; Cardenas, C.E.; Gress, D.A.; Erwin, W.D.; Odisio, B.C.; Koay, E.J.; Brock, K.K. Automated contouring of contrast and noncontrast computed tomography liver images with fully convolutional networks. Adv. Radiat. Oncol. 2021, 6, 100464. [Google Scholar] [CrossRef] [PubMed]
- Woodland, M.; Patel, N.; Castelo, A.; Al Taie, M.; Eltaher, M.; Yung, J.P.; Netherton, T.J.; Calderone, T.L.; Sanchez, J.I.; Cleere, D.W.; et al. Dimensionality reduction and nearest neighbors for improving out-of-distribution detection in medical image segmentation. J. Mach. Learn. Biomed. Imaging 2024, 2, 2006–2052. [Google Scholar] [CrossRef] [PubMed]
- Prior, F.; Almeida, J.; Kathiravelu, P.; Kurc, T.; Smith, K.; Fitzgerald, T.J.; Saltz, J. Open access image repositories: High-quality data to enable machine learning research. Clin. Radiol. 2020, 75, 7–12. [Google Scholar] [CrossRef]
- Johnston, L.R.; Curty, R.; Braxton, S.M.; Carlson, J.; Hadley, H.; Lafferty-Hess, S.; Luong, H.; Petters, J.L.; Kozlowski, W.A. Understanding the value of curation: A survey of US data repository curation practices and perceptions. PLoS ONE 2024, 19, e0301171. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the CVPR 2017, Honolulu, HI, USA, 21–26 June 2017; IEEE: New York, NY, USA, 2017; pp. 2097–2106. [Google Scholar]
- Odisio, B.C.; Albuquerque, J.; Lin, Y.-M.; Anderson, B.M.; O‘Connor, C.S.; Rigaud, B.; Briones-Dimayuga, M.; Jones, A.K.; Fellman, B.M.; Huang, S.Y.; et al. Software-based versus visual assessment of the minimal ablative margin in patients with liver tumours undergoing percutaneous thermal ablation (COVERALL): A randomized phase 2 trial. Lancet 2025, 10, 442–451. [Google Scholar]
- Baughan, N.; Whitney, H.M.; Drukker, K.; Sahiner, B.; Hu, T.; Kim, G.H.; McNitt-Gray, M.; Myers, K.J.; Giger, M.L. Sequestration of imaging studies in MIDRC: A multi-institutional data commons. J. Med. Imaging 2023, 10, e064501. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Advances in Neural Information Processing Systems 27, Proceedings of NIPS 2014, Montreal, QC, Canada, 8–13 December 2014; Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N.D., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2014; pp. 2672–2680. [Google Scholar]
- Karras, T.; Laine, S.; Aittala, M.; Hellsten, J.; Lehtinen, J.; Aila, T. Analyzing and improving the image quality of StyleGAN. In Proceedings of the CVPR 2020, Seattle, WA, USA, 13–19 June 2020; IEEE: New York, NY, USA, 2020; pp. 8107–8116. [Google Scholar]
- Karras, T.; Laine, S.; Aila, T. A style-based generator architecture for generative adversarial networks. In Proceedings of the CVPR 2019, Long Beach, CA, USA, 16–20 June 2019; IEEE: New York, NY, USA, 2019; pp. 4401–4410. [Google Scholar]
- Karras, T.; Aittala, M.; Hellsten, J.; Laine, S.; Lehtinen, J.; Aila, T. Training generative adversarial networks with limited data. In Advances in Neural Information Processing Systems 33, Proceedings of NeurIPS 2020, Online, 6–12 December 2020; Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M.F., Lin, H., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2020. [Google Scholar]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. GANs trained by a two time-scale update rule converge to a local Nash equilibrium. In Advances in Neural Information Processing Systems 30, Proceedings of NIPS 2017, Long Beach, CA, USA, 4–9 December 2017; Guyon, I., Von Luxburg, U., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017. [Google Scholar]
- Morozov, S.; Voynov, A.; Babenko, A. On self-supervised image representations for GAN evaluation. In Proceedings of the ICLR 2021, Online, 3–7 May 2021. [Google Scholar]
- Osuala, R.; Lang, D.M.; Verma, P.; Joshi, S.; Tsirikoglou, A.; Skorupko, G.; Kushibar, K.; Garrucho, L.; Pinaya, W.H.L.; Diaz, O.; et al. Towards learning contrast kinetics with multi-condition latent diffusion models. In LNCS, Vol. 15005, Proceedings of MICCAI 2024, Marrakesh, MA, Morocco, 6–10 October 2024; Linguraru, M.G., Dou, Q., Feragen, A., Giannarou, S., Glocker, B., Lekadir, K., Schnabel, J.A., Eds.; Springer: Cham, Switzerland, 2024; pp. 713–723. [Google Scholar]
- Borji, A. Pros and cons of GAN evaluation measures. Comput. Vis. Image Underst. 2018, 179, 41–65. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the CVPR 2009, Miami Beach, FL, USA, 20–25 June 2009; IEEE: New York, NY, USA, 2009; pp. 248–255. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the CVPR 2015, Boston, MA, USA, 7–12 June 2015; IEEE: New York, NY, USA, 2015. [Google Scholar]
- Caron, M.; Misra, I.; Mairal, J.; Goyal, P.; Bojanowski, P.; Joulin, A. Unsupervised learning of visual features by contrasting cluster assignments. In Advances in Neural Information Processing Systems 33, Proceedings of NeurIPS 2020, Online, 6–12 December 2020; Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M.F., Lin, H., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2020; pp. 9912–9924. [Google Scholar]
- Woodland, M.; Castelo, A.; Al Taie, M.; Albuquerque Marques Silva, J.; Eltaher, M.; Mohn, F.; Shieh, A.; Kundu, S.; Yung, J.P.; Patel, A.B.; et al. Feature extraction for generative medical imaging evaluation: New evidence against an evolving trend. In LNCS, Vol. 15012, Proceedings of MICCAI 2024, Marrakesh, Morocco, 6–10 October 2024; Linguraru, M.G., Dou, Q., Feragen, A., Giannarou, S., Glocker, B., Lekadir, K., Schnabel, J.A., Eds.; Springer: Cham, Switzerland, 2024; pp. 87–97. [Google Scholar]
- Schlegl, T.; Seeböck, P.; Waldstein, S.M.; Schmidt-Erfurth, U.; Langs, G. Unsupervised anomaly detection with generative adversarial networks to guide marker discovery. In LNCS, Vol. 10265, Proceedings of IPMI 2017, Boone, NC, USA, 25–30 June 2017; Niethammer, M., Styner, M., Aylward, S., Zhu, H., Oguz, I., Yap, P.-T., Shen, D., Eds.; Springer: Berlin, Germany, 2017; pp. 146–157. [Google Scholar]
- Gong, D.; Liu, L.; Le, V.; Saha, B.; Mansour, M.R.; Venkatesh, S.; van den Hengel, A. Memorizing normality to detect anomaly: Memory-augmented deep autoencoder for unsupervised anomaly detection. In Proceedings of the ICCV 2019, Seoul, Korea, 27 October–2 November 2019; IEEE: New York, NY, USA, 2019; pp. 1705–1714. [Google Scholar]
- Woodland, M.; Brock, K. Generative Modeling for Interpretable Failure Detection in Liver CT Segmentation and Scalable Data Curation of Chest Radiographs (Version v1). Zenodo 2025. Available online: https://zenodo.org/records/14901472 (accessed on 20 February 2025).
- Gal, Y.; Ghahramani, Z. Dropout as a Bayesian approximation: Representing model uncertainty in deep learning. In Proceedings of the 33rd International Conference on Machine Learning (ICML 2016), New York, NY, USA, 19–24 June 2016; Balcan, M.F., Weinberger, K.Q., Eds.; PMLR; 2016; Volume 48, pp. 1050–1059. [Google Scholar]
- Nguyen, A.; Yosinski, J.; Clune, J. Deep neural networks are easily fooled: High confidence predictions for unrecognizable images. In Proceedings of the CVPR 2015, Boston, MA, USA, 7–12 June 2015; IEEE: New York, NY, USA, 2015; pp. 427–436. [Google Scholar]
- Nakao, T.; Hanaoka, S.; Nomura, Y.; Murata, M.; Takenaga, T.; Miki, S.; Watadani, T.; Yoshikawa, T.; Hayashi, N.; Abe, O. Unsupervised deep anomaly detection in chest radiographs. J. Digit. Imaging 2021, 32, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, P.; Löwe, S.; Fauser, M.; Sattlegger, D.; Steger, C. Improving unsupervised defect segmentation by applying structural similarity to autoencoders. In Proceedings of the VISIGRAPP 2019, Prague, Czech Republic, 25–27 February 2019; Tremeau, A., Farinella, G.M., Braz, J., Eds.; SciTePress: Setúbal, Portugal, 2019; Volume 5, pp. 372–380. [Google Scholar]
- Yan, Z.; Fang, Q.; Lv, W.; Su, Q. AnomalySD: Few-shot multi-class anomaly detection with stable diffusion model. arXiv 2024, arXiv:2408.01960. [Google Scholar]
- Cheng, X.; Wen, M.; Gao, C.; Wang, Y. Hyperspectral anomaly detection based on Wasserstein distance and spatial filtering. Remote Sens 2022, 14, 2730. [Google Scholar] [CrossRef]

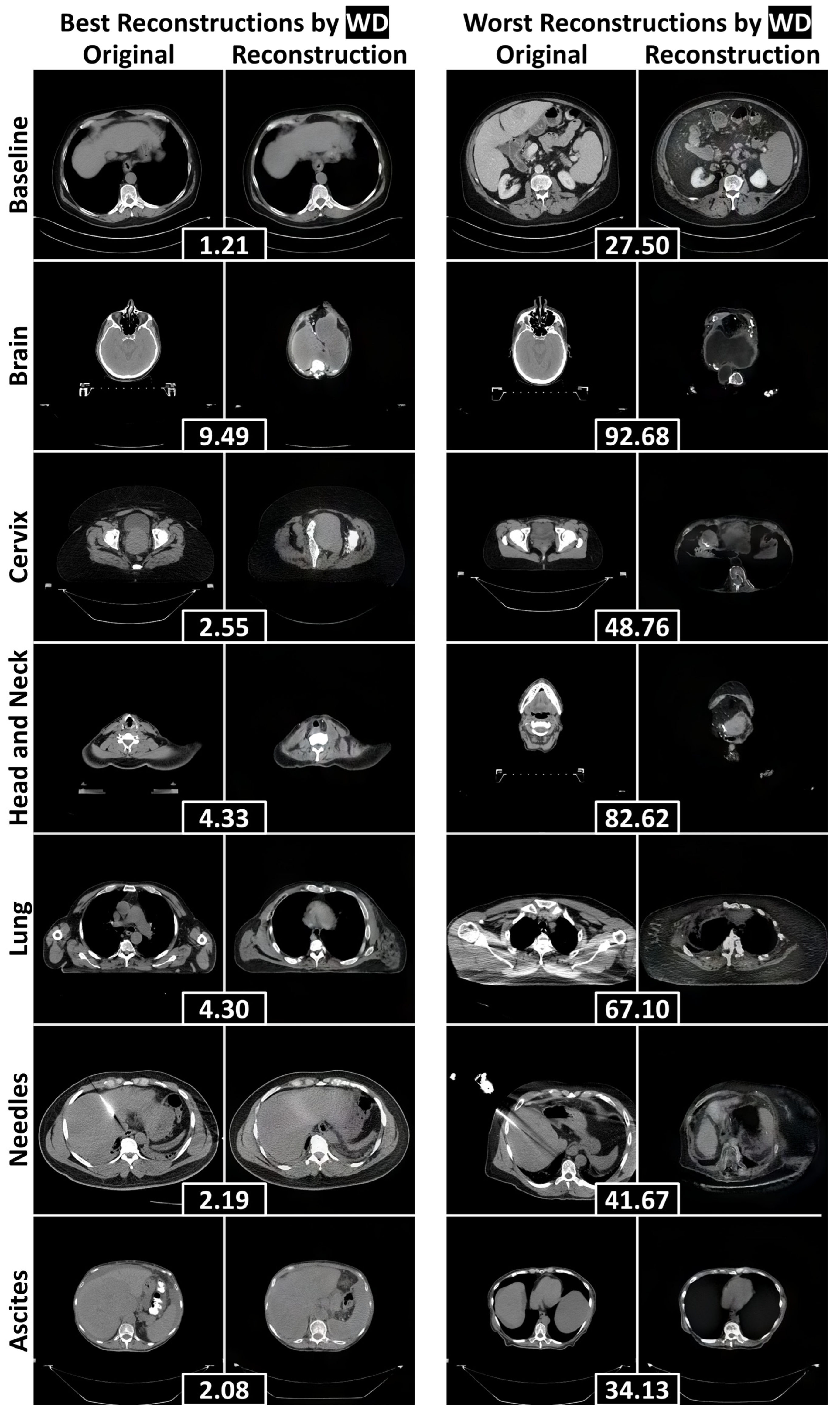
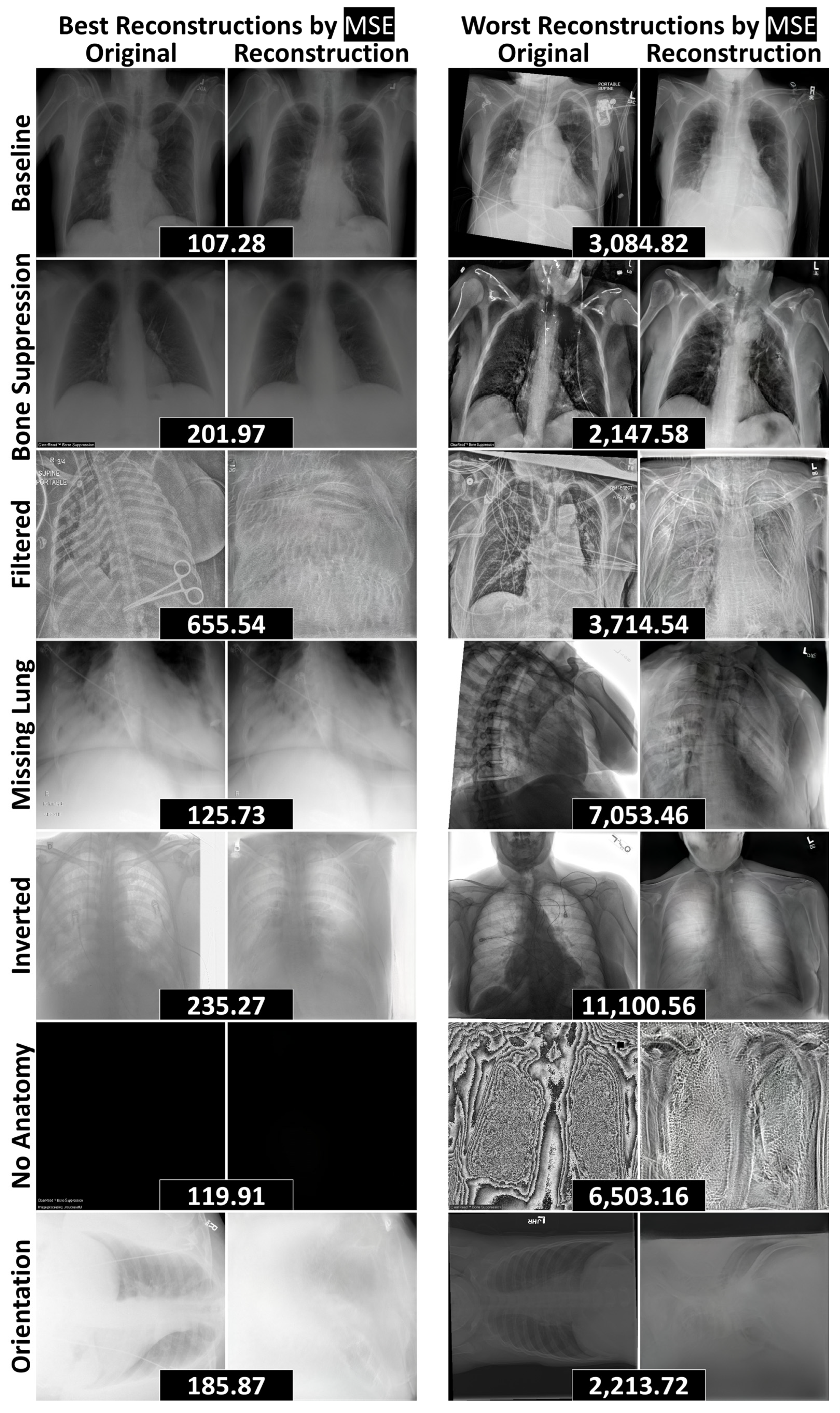
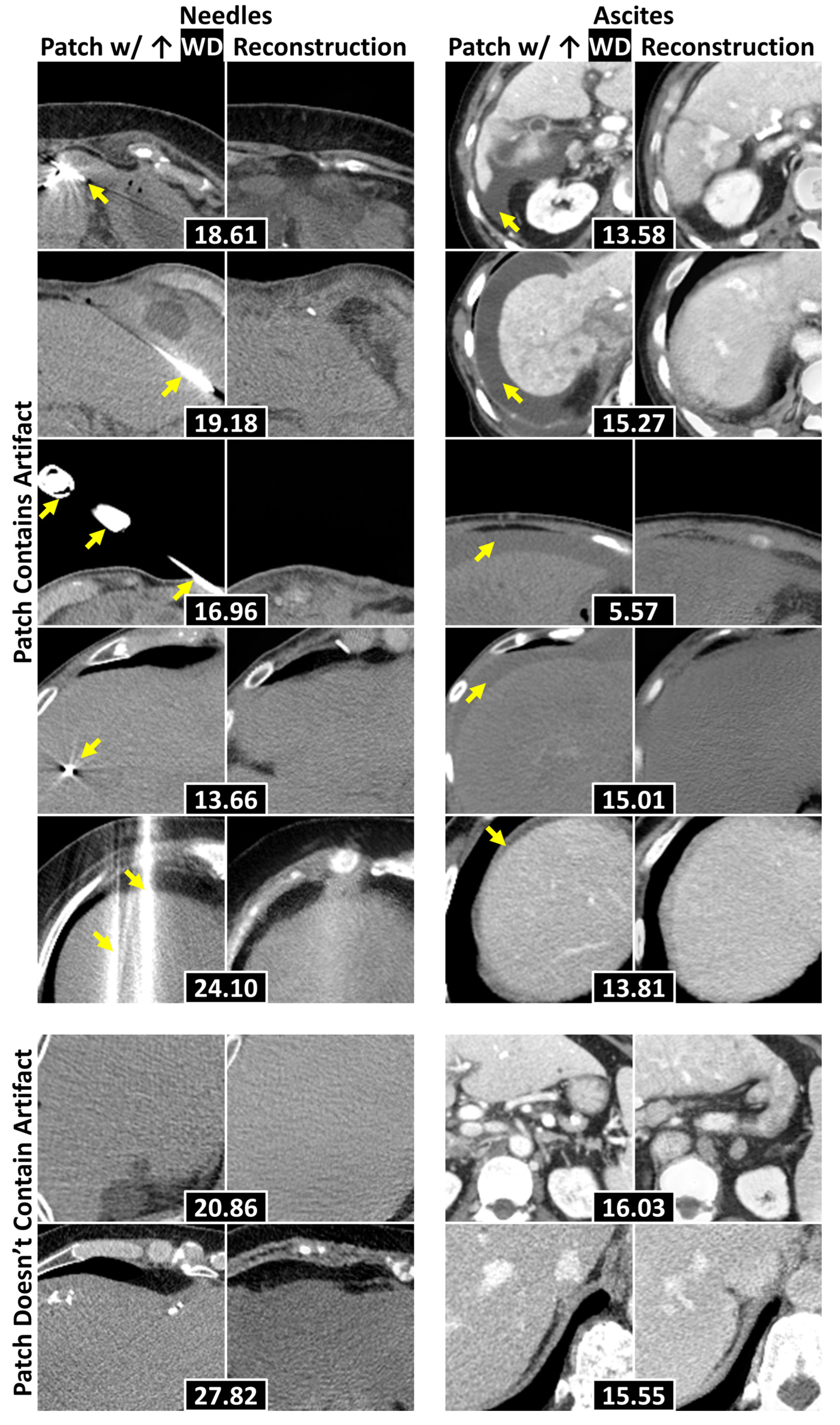
| Dataset | FID (±SD) ↓ | FSD (±SD) ↓ | FRD (±SD) ↓ | |
|---|---|---|---|---|
| Liver CT | Baseline | 0.26 (±0.00) | 0.01 (±0.00) | 0.01 (±0.00) |
| Generated | 3.37 (±0.05) - | 0.96 (±0.00) p < 0.001 * | 0.81 (±0.04) - | |
| Noise | 31.43 (±0.17) p < 0.001 * | 4.45 (±0.01) p < 0.001 * | 216.28 (±0.34) p < 0.001 * | |
| Blur | 47.65 (±0.13) p < 0.001 * | 0.44 (±0.00) - | 164.54 (±0.60) p < 0.001 * | |
| Chest Radiography | Baseline | 0.40 (±0.00) | 0.02 (±0.00) | 0.02 (±0.01) |
| Generated | 4.49 (±0.03) - | 0.92 (±0.00) - | 8.56 (±0.03) - | |
| Noise | 86.65 (±0.19) p < 0.001 * | 14.90 (±0.01) p < 0.001 * | 403.29 (±10.57) p < 0.001 * | |
| Blur | 36.98 (±0.12) p < 0.001 * | 1.41 (±0.01) p < 0.001 * | 49.64 (±5.30) p < 0.001 * |
| Dataset | WD-Based AUROC (±SD) ↑ | MSE-Based AUROC (±SD) ↑ | p | |
|---|---|---|---|---|
| Failure Detection | Brain | 0.66 (±0.03) | 0.20 (±0.02) | p < 0.001 * |
| Cervix | 0.71 (±0.02) | 0.48 (±0.02) | p < 0.001 * | |
| Head and Neck | 0.37 (±0.02) | 0.15 (±0.01) | p < 0.001 * | |
| Lung | 0.89 (±0.01) | 0.79 (±0.01) | p < 0.001 * | |
| Needles | 0.69 (±0.02) | 0.58 (±0.03) | p < 0.001 * | |
| Ascites | 0.60 (±0.02) | 0.43 (±0.03) | p < 0.001 * | |
| Data Curation | Bone Suppression | 0.68 (±0.02) | 0.79 (±0.01) | p < 0.001 * |
| Filtered | 0.57 (±0.02) | 0.98 (±0.00) | p < 0.001 * | |
| Missing Lung | 0.58 (±0.02) | 0.82 (±0.01) | p < 0.001 * | |
| Inverted | 0.84 (±0.01) | 0.90 (±0.01) | p < 0.001 * | |
| No Anatomy | 0.63 (±0.04) | 0.62 (±0.01) | p = 0.013 * | |
| Orientation | 0.74 (±0.05) | 0.78 (±0.03) | p < 0.001 * |
| Dataset | WD-Based AUROC (±SD) ↑ | MSE-Based AUROC (±SD) ↑ | p | |
|---|---|---|---|---|
| Non-liver | Brain | 1.00 (±0.00) † p < 0.001 ** | 0.90 (±0.01) † p < 0.001 ** | p < 0.001 * |
| Cervix | 0.90 (±0.01) † p < 0.001 ** | 0.70 (±0.02) † p < 0.001 ** | p < 0.001 * | |
| Head and Neck | 0.96 (±0.00) † p < 0.001 ** | 0.90 (±0.01) † p < 0.001 ** | p < 0.001 * | |
| Lung | 0.94 (±0.01) † p < 0.001 ** | 0.90 (±0.01) † p < 0.001 ** | p < 0.001 * | |
| Liver Anomaly | Needles | 0.69 (±0.02) p = 0.160 ** | 0.60 (±0.02) † p < 0.001 ** | p < 0.001 * |
| Ascites | 0.67 (±0.02) † p < 0.001 ** | 0.50 (±0.03) † p < 0.001 ** | p < 0.001 * |
| Dataset | WD ↑ | MSE ↑ | ||||
|---|---|---|---|---|---|---|
| 32 | 64 | 128 | 32 | 64 | 128 | |
| Needles | 0.43 (±0.01) p < 0.001 * | 0.41 (±0.02) p < 0.001 * | 0.70 (±0.03) | 0.44 (±0.01) p < 0.001 * | 0.38 (±0.03) p < 0.001 * | 0.35 (±0.03) p < 0.001 * |
| Ascites | 0.55 (±0.06) p < 0.001 * | 0.71 (±0.03) p < 0.001 * | 0.93 (±0.01) | 0.44 (±0.05) p < 0.001 * | 0.66 (±0.04) p < 0.001 * | 0.86 (±0.02) p < 0.001 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woodland, M.E.; Altaie, M.; O’Connor, C.S.; Castelo, A.H.; Lebimoyo, O.C.; Gupta, A.C.; Yung, J.P.; Kinahan, P.E.; Fuller, C.D.; Koay, E.J.; et al. Generative Modeling for Interpretable Anomaly Detection in Medical Imaging: Applications in Failure Detection and Data Curation. Bioengineering 2025, 12, 1106. https://doi.org/10.3390/bioengineering12101106
Woodland ME, Altaie M, O’Connor CS, Castelo AH, Lebimoyo OC, Gupta AC, Yung JP, Kinahan PE, Fuller CD, Koay EJ, et al. Generative Modeling for Interpretable Anomaly Detection in Medical Imaging: Applications in Failure Detection and Data Curation. Bioengineering. 2025; 12(10):1106. https://doi.org/10.3390/bioengineering12101106
Chicago/Turabian StyleWoodland, McKell E., Mais Altaie, Caleb S. O’Connor, Austin H. Castelo, Olubunmi C. Lebimoyo, Aashish C. Gupta, Joshua P. Yung, Paul E. Kinahan, Clifton D. Fuller, Eugene J. Koay, and et al. 2025. "Generative Modeling for Interpretable Anomaly Detection in Medical Imaging: Applications in Failure Detection and Data Curation" Bioengineering 12, no. 10: 1106. https://doi.org/10.3390/bioengineering12101106
APA StyleWoodland, M. E., Altaie, M., O’Connor, C. S., Castelo, A. H., Lebimoyo, O. C., Gupta, A. C., Yung, J. P., Kinahan, P. E., Fuller, C. D., Koay, E. J., Odisio, B. C., Patel, A. B., & Brock, K. K. (2025). Generative Modeling for Interpretable Anomaly Detection in Medical Imaging: Applications in Failure Detection and Data Curation. Bioengineering, 12(10), 1106. https://doi.org/10.3390/bioengineering12101106








