Fibroblast Growth Factor-Derived Peptides: Sources, Functions, and Applications
Abstract
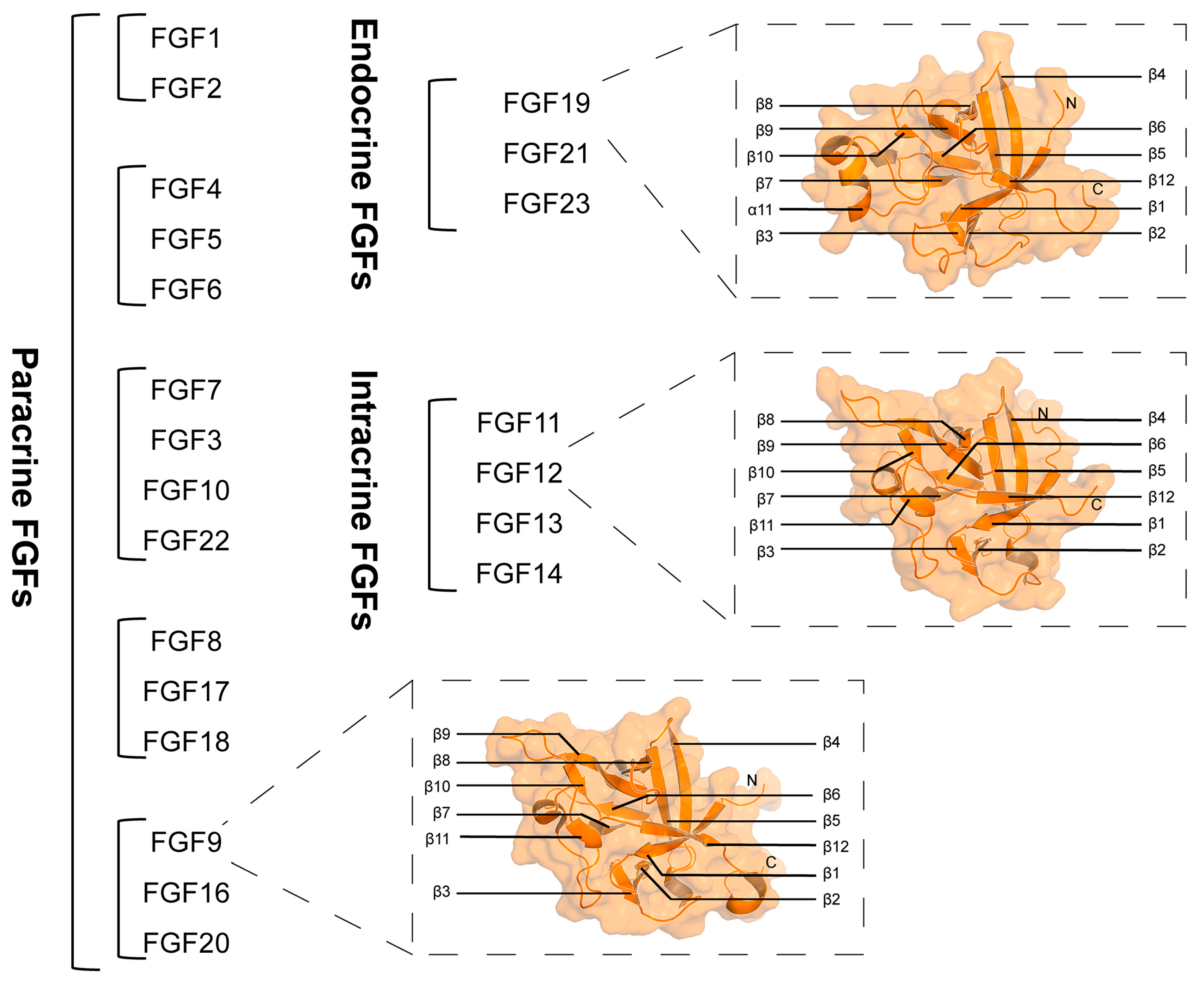
1. Introduction
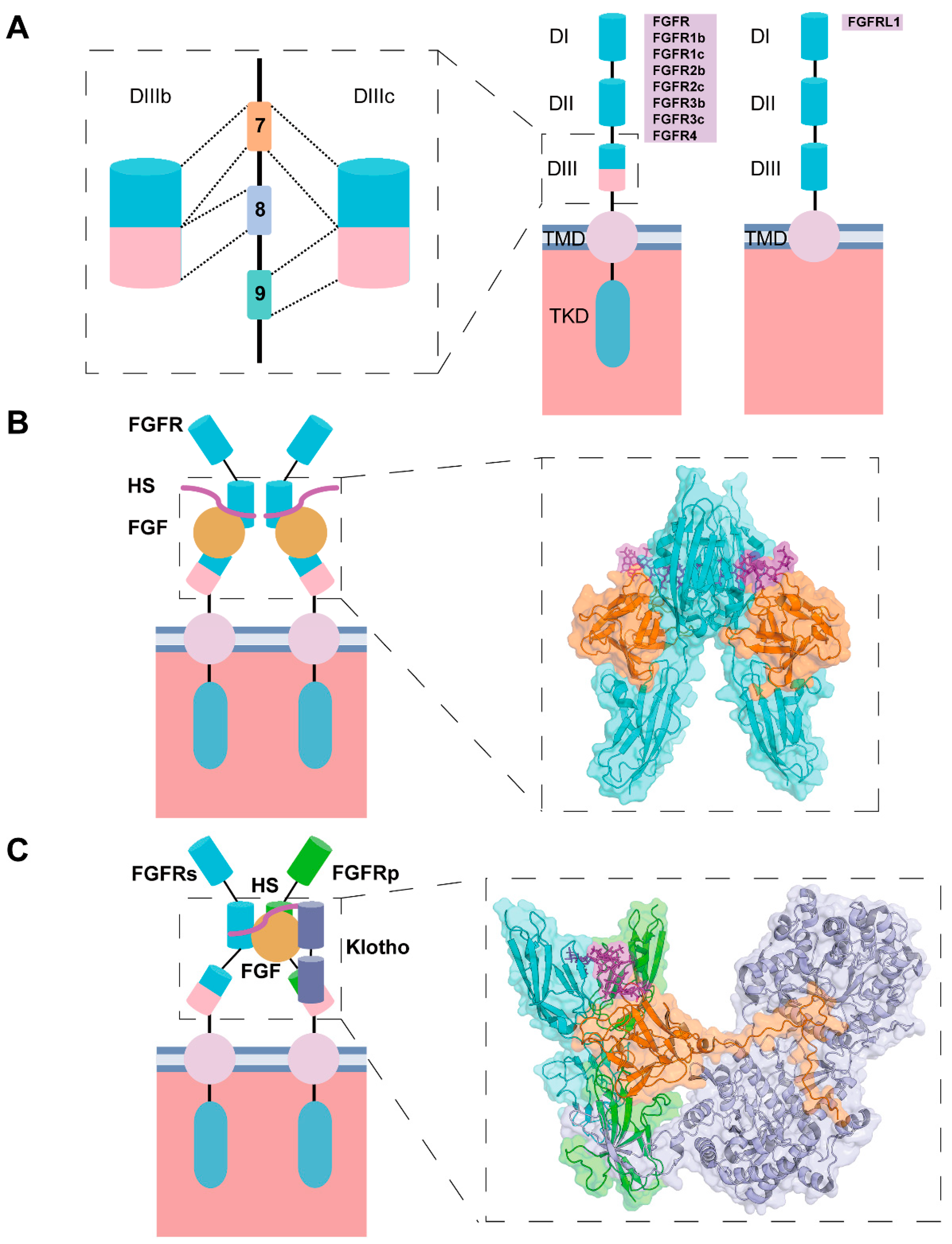
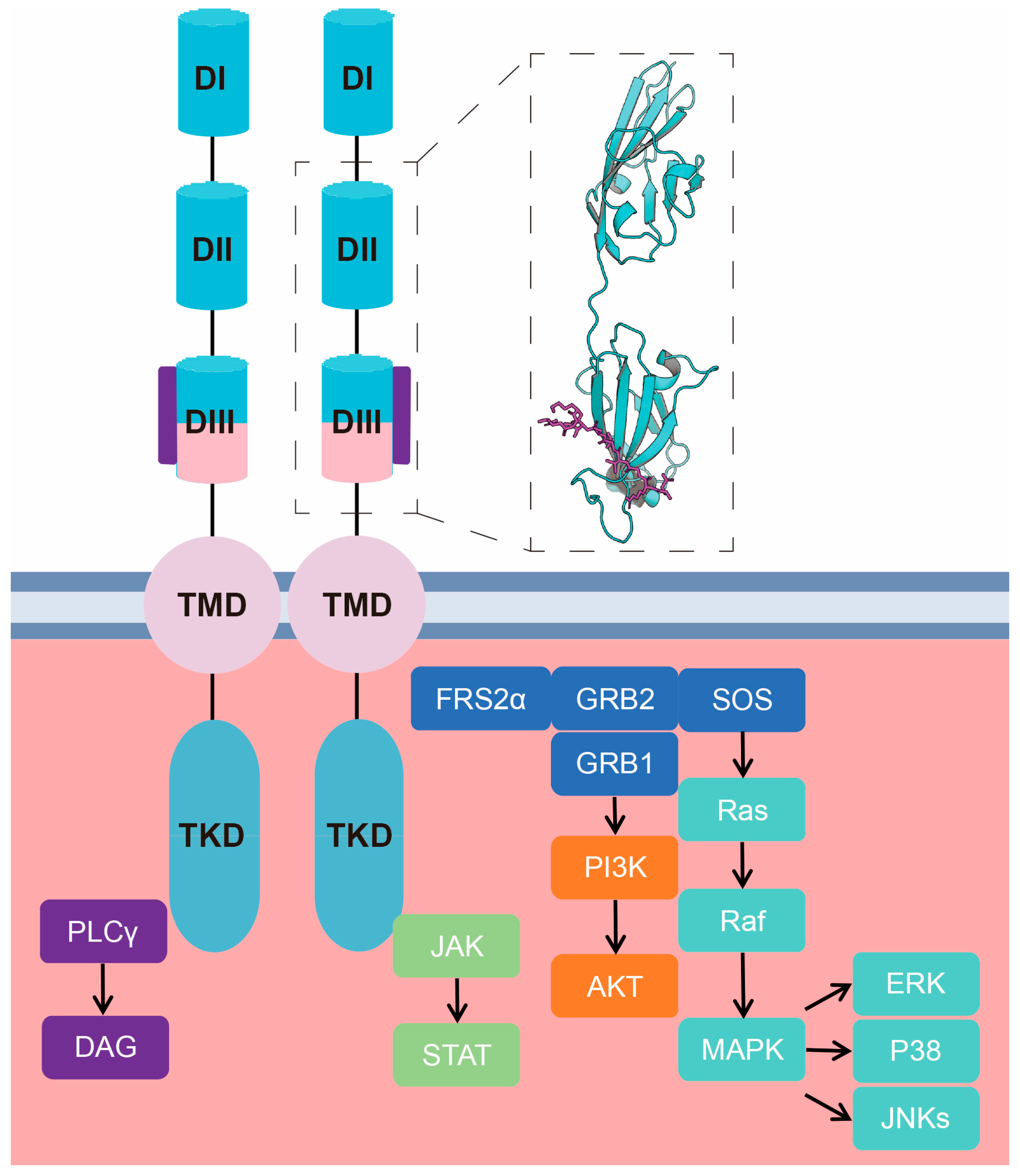
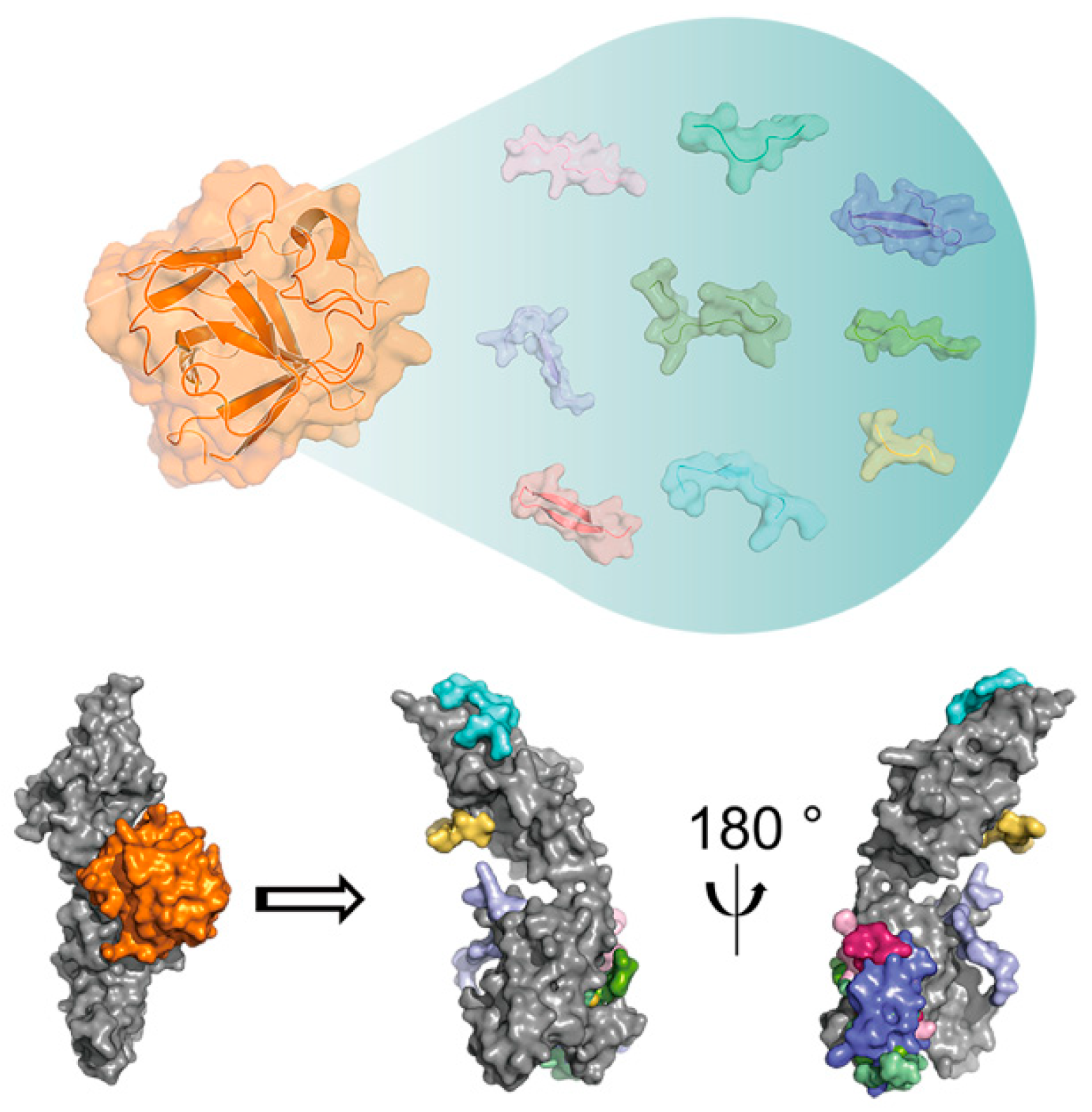
2. Molecular Basis of the Functional Activity of FGF-Derived Peptides
3. Activation Functions of FGF-Derived Peptides
3.1. FGF-Derived Peptides from Natural Sequences
3.2. Artificially Designed FGF-Derived Peptides
4. Antagonistic Effects of FGF-Derived Peptides
4.1. FGF-Derived Peptides from Natural Sequences
4.2. Artificially Designed FGF-Derived Peptides
5. Drug Delivery
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | artificial intelligence |
| ARS | acute radiation syndrome |
| BSA | bovine serum albumin |
| CAM | chorioallantoic membrane |
| DSPE | distearoyl phosphatidylethanolamine |
| DSS | dextran sulfate sodium |
| ECD | extracellular domain |
| FGF | fibroblast growth factor |
| FGFR | fibroblast growth factor receptor |
| HBcAg | hepatitis B core antigen |
| hBM-MSCs | human bone marrow mesenchymal stem cells |
| HS | heparan sulfate |
| HUVECs | human umbilical vein endothelial cells |
| hWJ-MSCs | human Wharton’s jelly mesenchymal stem cells |
| I/R | ischemia–reperfusion |
| IFN-γ | interferon-γ |
| LPS | lipopolysaccharide |
| NCAM | neural cell adhesion molecule |
| NLS | nuclear localization signal |
| OGD | oxygen-glucose deprivation |
| ONs | oligonucleotides |
| PAMAM | polyamidoamine |
| PAs | peptide amphiphiles |
| PCNA | proliferating cell nuclear antigen |
| PDGFR-α | platelet-derived growth factor receptor-α |
| PE | phosphatidylethanolamine |
| PEG | polyethylene glycol |
| PEI | polyethylenimine |
| SCs | skeletal muscle satellite cells |
| SP | signal peptide |
| SPPS | solid-phase peptide synthesis |
| TGF-βRII | transforming growth factor-β receptor type II |
| TKD | tyrosine kinase domain |
| TMD | transmembrane domain |
| VEGF | vascular endothelial growth factor |
| VLPs | virus-like particles |
References
- Liu, Y.; Liu, Y.; Deng, J.; Li, W.; Nie, X. Fibroblast Growth Factor in Diabetic Foot Ulcer: Progress and Therapeutic Prospects. Front. Endocrinol. 2021, 12, 744868. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Ornitz, D.M. Fibroblast Growth Factors: From Molecular Evolution to Roles in Development, Metabolism and Disease. J. Biochem. 2011, 149, 121–130. [Google Scholar] [CrossRef]
- Jin, L.; Yang, R.; Geng, L.; Xu, A. Fibroblast Growth Factor–Based Pharmacotherapies for the Treatment of Obesity-Related Metabolic Complications. Annu. Rev. Pharmacol. Toxicol. 2023, 63, 359–382. [Google Scholar] [CrossRef]
- Phan, P.; Saikia, B.B.; Sonnaila, S.; Agrawal, S.; Alraawi, Z.; Kumar, T.K.S.; Iyer, S. The Saga of Endocrine FGFs. Cells 2021, 10, 2418. [Google Scholar] [CrossRef] [PubMed]
- Armelin, H.A. Pituitary Extracts and Steroid Hormones in the Control of 3T3 Cell Growth. Proc. Natl. Acad. Sci. USA 1973, 70, 2702–2706. [Google Scholar] [CrossRef]
- Li, X.; Lu, W.; Kharitonenkov, A.; Luo, Y. Targeting the FGF19–FGFR4 Pathway for Cholestatic, Metabolic, and Cancerous Diseases. J. Intern. Med. 2024, 295, 292–312. [Google Scholar] [CrossRef]
- Biadun, M.; Sidor, S.; Kalka, M.; Karelus, R.; Sochacka, M.; Krowarsch, D.; Opalinski, L.; Zakrzewska, M. Production and Purification of Recombinant Long Protein Isoforms of FGF11 Subfamily. J. Biotechnol. 2025, 403, 9–16. [Google Scholar] [CrossRef]
- Ornitz, D.M.; Itoh, N. New Developments in the Biology of Fibroblast Growth Factors. WIREs Mech. Dis. 2022, 14, e1549. [Google Scholar] [CrossRef]
- Ornitz, D.M.; Itoh, N. Fibroblast Growth Factors. Genome Biol. 2001, 2, reviews3005.1. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, J.; Beenken, A.; Srinivasan, L.; Eliseenkova, A.V.; Mohammadi, M. Regulation of Receptor Binding Specificity of FGF9 by an Autoinhibitory Homodimerization. Structure 2017, 25, 1325–1336.e3. [Google Scholar] [CrossRef] [PubMed]
- Olsen, S.K.; Garbi, M.; Zampieri, N.; Eliseenkova, A.V.; Ornitz, D.M.; Goldfarb, M.; Mohammadi, M. Fibroblast Growth Factor (FGF) Homologous Factors Share Structural but Not Functional Homology with FGFs. J. Biol. Chem. 2003, 278, 34226–34236. [Google Scholar] [CrossRef]
- Goetz, R.; Beenken, A.; Ibrahimi, O.A.; Kalinina, J.; Olsen, S.K.; Eliseenkova, A.V.; Xu, C.; Neubert, T.A.; Zhang, F.; Linhardt, R.J.; et al. Molecular Insights into the Klotho-Dependent, Endocrine Mode of Action of Fibroblast Growth Factor 19 Subfamily Members. Mol. Cell. Biol. 2007, 27, 3417–3428. [Google Scholar] [CrossRef]
- Asada, M.; Shinomiya, M.; Suzuki, M.; Honda, E.; Sugimoto, R.; Ikekita, M.; Imamura, T. Glycosaminoglycan Affinity of the Complete Fibroblast Growth Factor Family. Biochim. Biophys. Acta (BBA) Gen. Subj. 2009, 1790, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Goetz, R.; Mohammadi, M. Exploring Mechanisms of FGF Signalling through the Lens of Structural Biology. Nat. Rev. Mol. Cell Biol. 2013, 14, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Jiang, W.; Chen, Z.; Huang, Y.; Mao, J.; Zheng, W.; Hu, Y.; Shi, J. Advance in Peptide-Based Drug Development: Delivery Platforms, Therapeutics and Vaccines. Signal Transduct. Target. Ther. 2025, 10, 74. [Google Scholar] [CrossRef]
- Lubell, W.D. Peptide-Based Drug Development. Biomedicines 2022, 10, 2037. [Google Scholar] [CrossRef]
- Apostolopoulos, V.; Bojarska, J.; Chai, T.-T.; Elnagdy, S.; Kaczmarek, K.; Matsoukas, J.; New, R.; Parang, K.; Lopez, O.P.; Parhiz, H.; et al. A Global Review on Short Peptides: Frontiers and Perspectives. Molecules 2021, 26, 430. [Google Scholar] [CrossRef]
- Ternier, G.; Shahzad, K.; Edirisinghe, O.; Okoto, P.; Alraawi, Z.; Sonnaila, S.; Phan, P.; Adams, P.D.; Thallapuranam, S.K. Fibroblast Growth Factors: Roles and Emerging Therapeutic Applications. Curr. Drug Targets 2025, 26, 551–570. [Google Scholar] [CrossRef] [PubMed]
- Komi, A.; Ishisaki, A.; Suzuki, M.; Imamura, T. A Permeable FGF-1 Nuclear Localization Sequence Peptide Induces DNA Synthesis Independently of Ras Activation. Exp. Cell Res. 2003, 283, 91–100. [Google Scholar] [CrossRef]
- Li, S.; Christensen, C.; Køhler, L.B.; Kiselyov, V.V.; Berezin, V.; Bock, E. Agonists of Fibroblast Growth Factor Receptor Induce Neurite Outgrowth and Survival of Cerebellar Granule Neurons. Dev. Neurobiol. 2009, 69, 837–854. [Google Scholar] [CrossRef]
- Kiyota, S.; Franzoni, L.; Nicastro, G.; Benedetti, A.; Oyama, S.; Viviani, W.; Gambarini, A.G.; Spisni, A.; Miranda, M.T.M. Introduction of a Chemical Constraint in a Short Peptide Derived from Human Acidic Fibroblast Growth Factor Elicits Mitogenic Structural Determinants. J. Med. Chem. 2003, 46, 2325–2333. [Google Scholar] [CrossRef]
- Manfè, V.; Kochoyan, A.; Bock, E.; Berezin, V. Peptides Derived from Specific Interaction Sites of the Fibroblast Growth Factor 2—FGF Receptor Complexes Induce Receptor Activation and Signaling. J. Neurochem. 2010, 114, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Fujii, I.; Kinoshita, R.; Akiyama, H.; Nakamura, A.; Iwamori, K.; Fukada, S.; Honda, H.; Shimizu, K. Discovery of Fibroblast Growth Factor 2-Derived Peptides for Enhancing Mice Skeletal Muscle Satellite Cell Proliferation. Biotechnol. J. 2024, 19, 2400278. [Google Scholar] [CrossRef]
- Lee, J.; Choo, J.; Choi, Y.; Shim, I.; Lee, S.; Seol, Y.; Chung, C.; Park, Y. Effect of Immobilized Cell-binding Peptides on Chitosan Membranes for Osteoblastic Differentiation of Mesenchymal Stem Cells. Biotech. Appl. Biochem. 2009, 52, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Facchiano, A.; Russo, K.; Facchiano, A.M.; De Marchis, F.; Facchiano, F.; Ribatti, D.; Aguzzi, M.S.; Capogrossi, M.C. Identification of a Novel Domain of Fibroblast Growth Factor 2 Controlling Its Angiogenic Properties. J. Biol. Chem. 2003, 278, 8751–8760. [Google Scholar] [CrossRef] [PubMed]
- Aguzzi, M.S.; Faraone, D.; D’Arcangelo, D.; De Marchis, F.; Toietta, G.; Ribatti, D.; Parazzoli, A.; Colombo, P.; Capogrossi, M.C.; Facchiano, A. The FGF-2-Derived Peptide FREG Inhibits Melanoma Growth in Vitro and in Vivo. Mol. Ther. 2011, 19, 266–273. [Google Scholar] [CrossRef]
- Zhang, Y.; Ouyang, M.; Wang, H.; Zhang, B.; Guang, W.; Liu, R.; Li, X.; Shih, T.; Li, Z.; Cao, J.; et al. A Cyclic Peptide Retards the Proliferation of DU145 Prostate Cancer Cells in Vitro and in Vivo through Inhibition of FGFR2. MedComm 2020, 1, 362–375. [Google Scholar] [CrossRef]
- Su, Z.; Zhang, Y.; Cao, J.; Sun, Y.; Cai, Y.; Zhang, B.; He, L.; Zhang, Z.; Xie, J.; Meng, Q.; et al. Hyaluronic Acid-FGF2-Derived Peptide Bioconjugates for Suppression of FGFR2 and AR Simultaneously as an Acne Antagonist. J. Nanobiotechnol. 2023, 21, 55. [Google Scholar] [CrossRef]
- Lee, S.B.; Abdal Dayem, A.; Kmiecik, S.; Lim, K.M.; Seo, D.S.; Kim, H.-T.; Kumar Biswas, P.; Do, M.; Kim, D.-H.; Cho, S.-G. Efficient Improvement of the Proliferation, Differentiation, and Anti-Arthritic Capacity of Mesenchymal Stem Cells by Simply Culturing on the Immobilized FGF2 Derived Peptide, 44-ERGVVSIKGV-53. J. Adv. Res. 2024, 62, 119–141. [Google Scholar] [CrossRef]
- Xiong, S.; Xu, Y.; Ma, M.; Wang, H.; Wei, F.; Gu, Q.; Xu, X. Neuroprotective Effects of a Novel Peptide, FK18, under Oxygen-Glucose Deprivation in SH-SY5Y Cells and Retinal Ischemia in Rats via the Akt Pathway. Neurochem. Int. 2017, 108, 78–90. [Google Scholar] [CrossRef]
- Shu, C.; Sun, P.; Xie, H.; Huang, W.; Qi, J.; Ma, Y. Virus-Like Particles Presenting the FGF-2 Protein or Identified Antigenic Peptides Promoted Antitumor Immune Responses in Mice. Int. J. Nanomed. 2020, 15, 1983–1996. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, W.; Wang, J.; Zhang, M.; Yang, S.; Tian, Y.; Vidyasagar, S.; Peña, L.A.; Zhang, K.; Cao, Y.; et al. Mitigation Effect of an FGF-2 Peptide on Acute Gastrointestinal Syndrome after High-Dose Ionizing Radiation. Int. J. Radiat. Oncol. Biol. Phys. 2010, 77, 261–268. [Google Scholar] [CrossRef]
- Casey-Sawicki, K.; Zhang, M.; Kim, S.; Zhang, A.; Zhang, S.B.; Zhang, Z.; Singh, R.; Yang, S.; Swarts, S.; Vidyasagar, S.; et al. A Basic Fibroblast Growth Factor Analog for Protection and Mitigation against Acute Radiation Syndromes. Health Phys. 2014, 106, 704–712. [Google Scholar] [CrossRef]
- Zhang, K.; Tian, Y.; Yin, L.; Zhang, M.; Beck, L.A.; Zhang, B.; Okunieff, P.; Zhang, L.; Vidyasagar, S. Fibroblast Growth Factor-Peptide Improves Barrier Function and Proliferation in Human Keratinocytes After Radiation. Int. J. Radiat. Oncol. Biol. Phys. 2011, 81, 248–254. [Google Scholar] [CrossRef][Green Version]
- Lin, X.; Takahashi, K.; Campion, S.; Liu, Y.; Gustavsen, G.; Peña, L.; Zamora, P. Synthetic Peptide F2A4-K-NS Mimics Fibroblast Growth Factor-2 in Vitro and Is Angiogenic in Vivo. Int. J. Mol. Med. 2006, 17, 833–839. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, X.; Zamora, P.O.; Takahashi, K.; Lui, Y. Alleviation of Experimental Ulcerative Colitis with the Synthetic Peptide, F2A4-K-NS (Fibratide). Dig. Dis. Sci. 2007, 52, 2054–2062. [Google Scholar] [CrossRef] [PubMed]
- Rubert Pérez, C.M.; Álvarez, Z.; Chen, F.; Aytun, T.; Stupp, S.I. Mimicking the Bioactivity of Fibroblast Growth Factor-2 Using Supramolecular Nanoribbons. ACS Biomater. Sci. Eng. 2017, 3, 2166–2175. [Google Scholar] [CrossRef]
- Álvarez, Z.; Kolberg-Edelbrock, A.N.; Sasselli, I.R.; Ortega, J.A.; Qiu, R.; Syrgiannis, Z.; Mirau, P.A.; Chen, F.; Chin, S.M.; Weigand, S.; et al. Bioactive Scaffolds with Enhanced Supramolecular Motion Promote Recovery from Spinal Cord Injury. Science 2021, 374, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Choo, J.; Choi, Y.; Lee, K.; Min, D.; Pi, S.; Seol, Y.; Lee, S.; Jo, I.; Chung, C.; et al. Characterization of the Surface Immobilized Synthetic Heparin Binding Domain Derived from Human Fibroblast Growth Factor-2 and Its Effect on Osteoblast Differentiation. J. Biomed. Mater. Res. 2007, 83, 970–979. [Google Scholar] [CrossRef]
- Allahmoradi, H.; Asghari, S.M.; Ahmadi, A.; Assareh, E.; Nazari, M. Anti-Tumor and Anti-Metastatic Activity of the FGF2 118–126 Fragment Dependent on the Loop Structure. Biochem. J. 2022, 479, 1285–1302. [Google Scholar] [CrossRef]
- Terada, T.; Mizobata, M.; Kawakami, S.; Yabe, Y.; Yamashita, F.; Hashida, M. Basic Fibroblast Growth Factor-Binding Peptide as a Novel Targeting Ligand of Drug Carrier to Tumor Cells. J. Drug Target. 2006, 14, 536–545. [Google Scholar] [CrossRef]
- Lee, J.; Jung, J.; Kim, Y.-J.; Lee, E.; Choi, J.S. Gene Delivery of PAMAM Dendrimer Conjugated with the Nuclear Localization Signal Peptide Originated from Fibroblast Growth Factor 3. Int. J. Pharm. 2014, 459, 10–18. [Google Scholar] [CrossRef]
- Mahlum, E.; Mandal, D.; Halder, C.; Maran, A.; Yaszemski, M.J.; Jenkins, R.B.; Bolander, M.E.; Sarkar, G. Engineering a Noncarrier to a Highly Efficient Carrier Peptide for Noncovalently Delivering Biologically Active Proteins into Human Cells. Anal. Biochem. 2007, 365, 215–221. [Google Scholar] [CrossRef]
- Dokka, S.; Toledo-Velasquez, D.; Shi, X.; Wang, L.; Rojanasakul, Y. Cellular Delivery of Oligonucleotides by Synthetic Import Peptide Carrier. Pharm. Res. 1997, 14, 1759–1764. [Google Scholar] [CrossRef]
- Ito, C.; Saitoh, Y.; Fujita, Y.; Yamazaki, Y.; Imamura, T.; Oka, S.; Suzuki, S. Decapeptide with Fibroblast Growth Factor (FGF)-5 Partial Sequence Inhibits Hair Growth Suppressing Activity of FGF-5. J. Cell. Physiol. 2003, 197, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.; Madhu Krishna, M.; Sen, D.; Ghosh, S.; Basak, P.; Das, A. 3D Porous Polymer Scaffold-Conjugated KGF-Mimetic Peptide Promotes Functional Skin Regeneration in Chronic Diabetic Wounds. ACS Appl. Mater. Interfaces 2024, 16, 37418–37434. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Luo, W.; He, D.; Wang, R.; Huang, Y.; Zeng, X.; Wang, W.; Chen, X.; Gao, S.; Yu, Y.; et al. A Short Peptide Derived from the gN Helix Domain of FGF8b Suppresses the Growth of Human Prostate Cancer Cells. Cancer Lett. 2013, 339, 226–236. [Google Scholar] [CrossRef]
- Lin, X.; Song, L.; He, D.; Zeng, X.; Wu, J.; Luo, W.; Yang, Q.; Wang, J.; Wang, T.; Cai, J.; et al. An FGF8b-Mimicking Peptide with Potent Antiangiogenic Activity. Mol. Med. Rep. 2017, 16, 894–900. [Google Scholar] [CrossRef]
- Liu, H.; Lin, X.; Huang, T.; Song, L.; Zhu, C.; Ma, H.; Long, T.; Zeng, H.; Li, R.; Wang, H.; et al. A Short Peptide Reverses the Aggressive Phenotype of Prostate Cancer Cells. Eur. J. Pharmacol. 2018, 838, 129–137. [Google Scholar] [CrossRef]
- Berndt, T.J.; Craig, T.A.; McCormick, D.J.; Lanske, B.; Sitara, D.; Razzaque, M.S.; Pragnell, M.; Bowe, A.E.; O’Brien, S.P.; Schiavi, S.C.; et al. Biological Activity of FGF-23 Fragments. Pflug. Arch. Eur. J. Physiol. 2007, 454, 615–623. [Google Scholar] [CrossRef]
- Farooq, M.; Hwang, M.; Khan, A.W.; Batool, M.; Ahmad, B.; Kim, W.; Kim, M.S.; Choi, S. Identification of a Novel Fibroblast Growth Factor Receptor-Agonistic Peptide and Its Effect on Diabetic Wound Healing. Life Sci. 2025, 364, 123432. [Google Scholar] [CrossRef]
- Ballinger, M.D.; Shyamala, V.; Forrest, L.D.; Deuter-Reinhard, M.; Doyle, L.V.; Wang, J.; Panganiban-Lustan, L.; Stratton, J.R.; Apell, G.; Winter, J.A.; et al. Semirational Design of a Potent, Artificial Agonist of Fibroblast Growth Factor Receptors. Nat. Biotechnol. 1999, 17, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- Lipok, M.; Szlachcic, A.; Kindela, K.; Czyrek, A.; Otlewski, J. Identification of a Peptide Antagonist of the FGF 1– FGFR 1 Signaling Axis by Phage Display Selection. FEBS Open Bio 2019, 9, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Kawata, Y.; Masuda, Y.; Umemoto, T.; Ito, T.; Asami, T.; Takekawa, S.; Ohtaki, T.; Inooka, H. Discovery of an Artificial Peptide Agonist to the Fibroblast Growth Factor Receptor 1c/βKlotho Complex from Random Peptide T7 Phage Display. Biochem. Biophys. Res. Commun. 2016, 480, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Q.; Xie, C.; Cai, Y.; Chen, X.; Hou, Y.; He, L.; Li, J.; Yao, M.; Chen, S.; et al. Peptide Ligands Targeting FGF Receptors Promote Recovery from Dorsal Root Crush Injury via AKT/mTOR Signaling. Theranostics 2021, 11, 10125–10147. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Q.; Jin, Y.; Li, Y.; Nie, C.; Huang, P.; Li, Z.; Zhang, B.; Su, Z.; Hong, A.; et al. Discovery and Characterization of a High-Affinity Small Peptide Ligand, H1, Targeting FGFR2IIIc for Skin Wound Healing. Cell Physiol. Biochem. 2018, 49, 1074–1089. [Google Scholar] [CrossRef]
- Jin, M.; Yu, Y.; Qi, H.; Xie, Y.; Su, N.; Wang, X.; Tan, Q.; Luo, F.; Zhu, Y.; Wang, Q.; et al. A Novel FGFR3-Binding Peptide Inhibits FGFR3 Signaling and Reverses the Lethal Phenotype of Mice Mimicking Human Thanatophoric Dysplasia. Hum. Mol. Genet. 2012, 21, 5443–5455. [Google Scholar] [CrossRef]
- Rao, G.A.; Tsai, R.; Roura, D.; Hughes, J.A. Evaluation of the Transfection Property of a Peptide Ligand for the Fibroblast Growth Factor Receptor as Part of PEGylated Polyethylenimine Polyplex. J. Drug Target. 2008, 16, 79–89. [Google Scholar] [CrossRef]
- Huang, T.; Lin, X.; Li, Q.; Luo, W.; Song, L.; Tan, X.; Wang, W.; Li, X.; Wu, X. Selection of a Novel FGF23-Binding Peptide Antagonizing the Inhibitory Effect of FGF23 on Phosphate Uptake. Appl. Microbiol. Biotechnol. 2015, 99, 3169–3177. [Google Scholar] [CrossRef]
- Yamada, R.; Fukumoto, R.; Noyama, C.; Fujisawa, A.; Oka, S.; Imamura, T. An Epidermis-permeable Dipeptide Is a Potential Cosmetic Ingredient with Partial Agonist/Antagonist Activity toward Fibroblast Growth Factor Receptors. J. Cosmet. Dermatol. 2020, 19, 477–484. [Google Scholar] [CrossRef]
- Dai, X.; Cai, C.; Xiao, F.; Xiong, Y.; Huang, Y.; Zhang, Q.; Xiang, Q.; Lou, G.; Lian, M.; Su, Z.; et al. Identification of a Novel aFGF-Binding Peptide with Anti-Tumor Effect on Breast Cancer from Phage Display Library. Biochem. Biophys. Res. Commun. 2014, 445, 795–801. [Google Scholar] [CrossRef]
- Wu, X. A Novel bFGF Antagonist Peptide Inhibits Breast Cancer Cell Growth. Mol. Med. Report. 2012, 6, 210–214. [Google Scholar] [CrossRef][Green Version]
- Huang, Y.; Wang, T.; Lin, X.; Wang, H.; Li, R.; Zeng, X.; Zhu, C.; Chen, L.; Guo, Q.; Liu, H.; et al. Antitumor Effect of a Short Peptide on P53-Null SKOV3 Ovarian Cancer Cells. Anti Cancer Drugs 2019, 30, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Yu, Y.; Wang, R.; He, D.; Wang, C.; Zeng, X.; Chen, X.; Tan, X.; Huang, T.; Wu, X. P7 Peptides Targeting bFGF Sensitize Colorectal Cancer Cells to CPT-11. Int. J. Mol. Med. 2014, 33, 194–200. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, X.; Yan, Q.; Huang, Y.; Huang, H.; Su, Z.; Xiao, J.; Zeng, Y.; Wang, Y.; Nie, C.; Yang, Y.; et al. Isolation of a Novel Basic FGF-Binding Peptide with Potent Antiangiogenetic Activity. J. Cell Mol. Med. 2010, 14, 351–356. [Google Scholar] [CrossRef]
- Wang, W.; Chen, T.; Li, H.; Chen, Y.; Wu, Z.; Feng, T.; Zhang, X.; Zhong, Q.; Zhong, Q.; Li, G.; et al. Screening a Novel FGF3 Antagonist Peptide with Anti-Tumor Effects on Breast Cancer from a Phage Display Library. Mol. Med. Rep. 2015, 12, 7051–7058. [Google Scholar] [CrossRef]
- Wang, W.; Chen, X.; Li, T.; Li, Y.; Wang, R.; He, D.; Luo, W.; Li, X.; Wu, X. Screening a Phage Display Library for a Novel FGF8b-Binding Peptide with Anti-Tumor Effect on Prostate Cancer. Exp. Cell Res. 2013, 319, 1156–1164. [Google Scholar] [CrossRef]
- Wang, J.; Tan, X.; Guo, Q.; Lin, X.; Huang, Y.; Chen, L.; Zeng, X.; Li, R.; Wang, H.; Wu, X. FGF9 Inhibition by a Novel Binding Peptide Has Efficacy in Gastric and Bladder Cancer per Se and Reverses Resistance to Cisplatin. Pharmacol. Res. 2020, 152, 104575. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Su, N.; Yang, J.; Tan, Q.; Huang, S.; Jin, M.; Ni, Z.; Zhang, B.; Zhang, D.; Luo, F.; et al. FGF/FGFR Signaling in Health and Disease. Signal Transduct. Target. Ther. 2020, 5, 181. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.; Grose, R. Fibroblast Growth Factor Signalling: From Development to Cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef]
- Plotnikov, A.N.; Schlessinger, J.; Hubbard, S.R.; Mohammadi, M. Structural Basis for FGF Receptor Dimerization and Activation. Cell 1999, 98, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ibrahimi, O.A.; Olsen, S.K.; Umemori, H.; Mohammadi, M.; Ornitz, D.M. Receptor Specificity of the Fibroblast Growth Factor Family. J. Biol. Chem. 2006, 281, 15694–15700. [Google Scholar] [CrossRef]
- Park, J.S.; Choi, J.; Cao, L.; Mohanty, J.; Suzuki, Y.; Park, A.; Baker, D.; Schlessinger, J.; Lee, S. Isoform-Specific Inhibition of FGFR Signaling Achieved by a de-Novo-Designed Mini-Protein. Cell Rep. 2022, 41, 111545. [Google Scholar] [CrossRef]
- Chen, G.; Chen, L.; Li, X.; Mohammadi, M. FGF-Based Drug Discovery: Advances and Challenges. Nat. Rev. Drug Discov. 2025, 24, 335–357. [Google Scholar] [CrossRef]
- Schlessinger, J.; Plotnikov, A.N.; Ibrahimi, O.A.; Eliseenkova, A.V.; Yeh, B.K.; Yayon, A.; Linhardt, R.J.; Mohammadi, M. Crystal Structure of a Ternary FGF-FGFR-Heparin Complex Reveals a Dual Role for Heparin in FGFR Binding and Dimerization. Mol. Cell 2000, 6, 743–750. [Google Scholar] [CrossRef]
- Kuro-o, M. The Klotho Proteins in Health and Disease. Nat. Rev. Nephrol. 2019, 15, 27–44. [Google Scholar] [CrossRef]
- Chen, L.; Fu, L.; Sun, J.; Huang, Z.; Fang, M.; Zinkle, A.; Liu, X.; Lu, J.; Pan, Z.; Wang, Y.; et al. Structural Basis for FGF Hormone Signalling. Nature 2023, 618, 862–870. [Google Scholar] [CrossRef]
- Wen, X.; Jiao, L.; Tan, H. MAPK/ERK Pathway as a Central Regulator in Vertebrate Organ Regeneration. Int. J. Mol. Sci. 2022, 23, 1464. [Google Scholar] [CrossRef] [PubMed]
- Glaviano, A.; Foo, A.S.C.; Lam, H.Y.; Yap, K.C.H.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B.; et al. PI3K/AKT/mTOR Signaling Transduction Pathway and Targeted Therapies in Cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef]
- Mandal, S.; Bandyopadhyay, S.; Tyagi, K.; Roy, A. Recent Advances in Understanding the Molecular Role of Phosphoinositide-Specific Phospholipase C Gamma 1 as an Emerging Onco-Driver and Novel Therapeutic Target in Human Carcinogenesis. Biochim. Biophys. Acta (BBA) Rev. Cancer 2021, 1876, 188619. [Google Scholar] [CrossRef] [PubMed]
- Philips, R.L.; Wang, Y.; Cheon, H.; Kanno, Y.; Gadina, M.; Sartorelli, V.; Horvath, C.M.; Darnell, J.E.; Stark, G.R.; O’Shea, J.J. The JAK-STAT Pathway at 30: Much Learned, Much More to Do. Cell 2022, 185, 3857–3876. [Google Scholar] [CrossRef] [PubMed]
- Ornitz, D.M.; Itoh, N. The Fibroblast Growth Factor Signaling Pathway. WIREs Dev. Biol. 2015, 4, 215–266. [Google Scholar] [CrossRef] [PubMed]
- Santhana Kumar, K.; Brunner, C.; Schuster, M.; Kopp, L.L.; Gries, A.; Yan, S.; Jurt, S.; Moehle, K.; Bruns, D.; Grotzer, M.; et al. Discovery of a Small Molecule Ligand of FRS2 That Inhibits Invasion and Tumor Growth. Cell Oncol. 2023, 46, 331–356. [Google Scholar] [CrossRef]
- Peng, M.; Deng, J.; Li, X. Clinical Advances and Challenges in Targeting FGF/FGFR Signaling in Lung Cancer. Mol. Cancer 2024, 23, 256. [Google Scholar] [CrossRef]
- Katoh, M.; Loriot, Y.; Brandi, G.; Tavolari, S.; Wainberg, Z.A.; Katoh, M. FGFR-Targeted Therapeutics: Clinical Activity, Mechanisms of Resistance and New Directions. Nat. Rev. Clin. Oncol. 2024, 21, 312–329. [Google Scholar] [CrossRef]
- Edirisinghe, O.; Ternier, G.; Alraawi, Z.; Suresh Kumar, T.K. Decoding FGF/FGFR Signaling: Insights into Biological Functions and Disease Relevance. Biomolecules 2024, 14, 1622. [Google Scholar] [CrossRef]
- Jafari, B.; Hamzeh-Mivehroud, M.; Morris, M.B.; Dastmalchi, S. Exploitation of Phage Display for the Development of Anti-Cancer Agents Targeting Fibroblast Growth Factor Signaling Pathways: New Strategies to Tackle an Old Challenge. Cytokine Growth Factor. Rev. 2019, 46, 54–65. [Google Scholar] [CrossRef]
- Istomina, P.V.; Gorchakov, A.A.; Paoin, C.; Yamabhai, M. Phage Display for Discovery of Anticancer Antibodies. New Biotechnol. 2024, 83, 205–218. [Google Scholar] [CrossRef]
- Sousa-Victor, P.; García-Prat, L.; Muñoz-Cánoves, P. Control of Satellite Cell Function in Muscle Regeneration and Its Disruption in Ageing. Nat. Rev. Mol. Cell Biol. 2022, 23, 204–226. [Google Scholar] [CrossRef]
- Lee, Y.; Lim, K.-M.; Bong, H.; Lee, S.-B.; Jeon, T.-I.; Lee, S.-Y.; Park, H.-S.; Kim, J.-Y.; Song, K.; Kang, G.-H.; et al. The Immobilization of an FGF2-Derived Peptide on Culture Plates Improves the Production and Therapeutic Potential of Extracellular Vesicles from Wharton’s Jelly Mesenchymal Stem Cells. Int. J. Mol. Sci. 2024, 25, 10709. [Google Scholar] [CrossRef]
- Verrecchio, A.; Germann, M.W.; Schick, B.P.; Kung, B.; Twardowski, T.; San Antonio, J.D. Design of Peptides with High Affinities for Heparin and Endothelial Cell Proteoglycans. J. Biol. Chem. 2000, 275, 7701–7707. [Google Scholar] [CrossRef]
- Kiselyov, V.V.; Skladchikova, G.; Hinsby, A.M.; Jensen, P.H.; Kulahin, N.; Soroka, V.; Pedersen, N.; Tsetlin, V.; Poulsen, F.M.; Berezin, V.; et al. Structural Basis for a Direct Interaction between FGFR1 and NCAM and Evidence for a Regulatory Role of ATP. Structure 2003, 11, 691–701. [Google Scholar] [CrossRef]
- Cherian, K.E.; Paul, T.V. Inherited Fibroblast Growth Factor 23 Excess. Best Pract. Res. Clin. Endocrinol. Metab. 2024, 38, 101844. [Google Scholar] [CrossRef]
- Lin, Y.-Z.; Yao, S.; Veach, R.A.; Torgerson, T.R.; Hawiger, J. Inhibition of Nuclear Translocation of Transcription Factor NF-κB by a Synthetic Peptide Containing a Cell Membrane-Permeable Motif and Nuclear Localization Sequence. J. Biol. Chem. 1995, 270, 14255–14258. [Google Scholar] [CrossRef]
- Komi, A.; Suzuki, M.; Imamura, T. Permeable FGF-1 Nuclear Localization Signal Peptide Stimulates DNA Synthesis in Various Cell Types but Is Cell-Density Sensitive and Unable to Support Cell Proliferation. Exp. Cell Res. 1998, 243, 408–414. [Google Scholar] [CrossRef]
- Li, A.-J.; Tsuboyama, H.; Komi, A.; Ikekita, M.; Imamura, T. Strong Suppression of Feeding by a Peptide Containing Both the Nuclear Localization Sequence of Fibroblast Growth Factor-1 and a Cell Membrane-Permeable Sequence. Neurosci. Lett. 1998, 255, 41–44. [Google Scholar] [CrossRef] [PubMed]
- She, Q.-Y.; Bao, J.-F.; Wang, H.-Z.; Liang, H.; Huang, W.; Wu, J.; Zhong, Y.; Ling, H.; Li, A.; Qin, S.-L. Fibroblast Growth Factor 21: A “Rheostat” for Metabolic Regulation? Metabolism 2022, 130, 155166. [Google Scholar] [CrossRef]
- Adams, A.C.; Cheng, C.C.; Coskun, T.; Kharitonenkov, A. FGF21 Requires Βklotho to Act In Vivo. PLoS ONE 2012, 7, e49977. [Google Scholar] [CrossRef]
- Ruan, R.; Li, L.; Li, X.; Huang, C.; Zhang, Z.; Zhong, H.; Zeng, S.; Shi, Q.; Xia, Y.; Zeng, Q.; et al. Unleashing the Potential of Combining FGFR Inhibitor and Immune Checkpoint Blockade for FGF/FGFR Signaling in Tumor Microenvironment. Mol. Cancer 2023, 22, 60. [Google Scholar] [CrossRef]
- Terada, T.; Mizobata, M.; Kawakami, S.; Yamashita, F.; Hashida, M. Optimization of Tumor-Selective Targeting by Basic Fibroblast Growth Factor-Binding Peptide Grafted PEGylated Liposomes. J. Control. Release 2007, 119, 262–270. [Google Scholar] [CrossRef]
- Kimura-Ueki, M.; Oda, Y.; Oki, J.; Komi-Kuramochi, A.; Honda, E.; Asada, M.; Suzuki, M.; Imamura, T. Hair Cycle Resting Phase Is Regulated by Cyclic Epithelial FGF18 Signaling. J. Investig. Dermatol. 2012, 132, 1338–1345. [Google Scholar] [CrossRef]
- Higgins, C.A.; Petukhova, L.; Harel, S.; Ho, Y.Y.; Drill, E.; Shapiro, L.; Wajid, M.; Christiano, A.M. FGF5 Is a Crucial Regulator of Hair Length in Humans. Proc. Natl. Acad. Sci. USA 2014, 111, 10648–10653. [Google Scholar] [CrossRef]
- Saw, P.E.; Song, E.-W. Phage Display Screening of Therapeutic Peptide for Cancer Targeting and Therapy. Protein Cell 2019, 10, 787–807. [Google Scholar] [CrossRef]
- Suh, J.; Kim, D.; Lee, Y.; Jang, J.; Surh, Y. Fibroblast Growth Factor-2, Derived from Cancer-associated Fibroblasts, Stimulates Growth and Progression of Human Breast Cancer Cells via FGFR1 Signaling. Mol. Carcinog. 2020, 59, 1028–1040. [Google Scholar] [CrossRef]
- Dienstmann, R.; Rodon, J.; Prat, A.; Perez-Garcia, J.; Adamo, B.; Felip, E.; Cortes, J.; Iafrate, A.J.; Nuciforo, P.; Tabernero, J. Genomic Aberrations in the FGFR Pathway: Opportunities for Targeted Therapies in Solid Tumors. Ann. Oncol. 2014, 25, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Mattila, M.; Harkonen, P. Role of Fibroblast Growth Factor 8 in Growth and Progression of Hormonal Cancer. Cytokine Growth Factor Rev. 2007, 18, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ren, C.; Han, C.; Wang, D.; Chen, Y.; Fu, D. Expression and Prognostic Value of miR-486-5p in Patients with Gastric Adenocarcinoma. PLoS ONE 2015, 10, e0119384. [Google Scholar] [CrossRef]
- Knowles, M.A. Novel Therapeutic Targets In Bladder Cancer: Mutation and Expression of Fgf Receptors. Future Oncol. 2008, 4, 71–83. [Google Scholar] [CrossRef]
- Wrobel, W.; Pach, E.; Ben-Skowronek, I. Advantages and Disadvantages of Different Treatment Methods in Achondroplasia: A Review. Int. J. Mol. Sci. 2021, 22, 5573. [Google Scholar] [CrossRef]
- Zhao, J.; Lin, H.; Wang, L.; Guo, K.; Jing, R.; Li, X.; Chen, Y.; Hu, Z.; Gao, S.; Xu, N. Suppression of FGF5 and FGF18 Expression by Cholesterol-Modified siRNAs Promotes Hair Growth in Mice. Front. Pharmacol. 2021, 12, 666860. [Google Scholar] [CrossRef]
- Tsuruda, A.; Kawano, Y.; Maekawa, T.; Oka, S. A Short Peptide GPIGS Promotes Proliferation of Hair Bulb Keratinocytes and Accelerates Hair Regrowth in Mice. Biol. Pharm. Bull. 2005, 28, 485–489. [Google Scholar] [CrossRef][Green Version]
- Cai, J.; Dou, G.; Zheng, L.; Yang, T.; Jia, X.; Tang, L.; Huang, Y.; Wu, W.; Li, X.; Wang, X. Pharmacokinetics of Topically Applied Recombinant Human Keratinocyte Growth Factor-2 in Alkali-Burned and Intact Rabbit Eye. Exp. Eye Res. 2015, 136, 93–99. [Google Scholar] [CrossRef]
- Sun, J.; Wu, J.; Jin, H.; Ying, T.; Jin, W.; Fan, M.; Zhou, J.; Chen, H.; Jin, L.; Zhou, J. Structure-Guided Design, Generation, and Biofunction of PEGylated Fibroblast Growth Factor 2 Variants for Wound Healing. Nanoscale 2020, 12, 18200–18213. [Google Scholar] [CrossRef]
- Xia, X.; Babcock, J.P.; Blaber, S.I.; Harper, K.M.; Blaber, M. Pharmacokinetic Properties of 2nd-Generation Fibroblast Growth Factor-1 Mutants for Therapeutic Application. PLoS ONE 2012, 7, e48210. [Google Scholar] [CrossRef]
- Subbiah, V.; Verstovsek, S. Clinical Development and Management of Adverse Events Associated with FGFR Inhibitors. Cell Rep. Med. 2023, 4, 101204. [Google Scholar] [CrossRef]
- Shinbara, K.; Liu, W.; Van Neer, R.H.P.; Katoh, T.; Suga, H. Methodologies for Backbone Macrocyclic Peptide Synthesis Compatible With Screening Technologies. Front. Chem. 2020, 8, 447. [Google Scholar] [CrossRef]
- Li, X.; Wang, N.; Liu, Y.; Li, W.; Bai, X.; Liu, P.; He, C.-Y. Backbone N-Methylation of Peptides: Advances in Synthesis and Applications in Pharmaceutical Drug Development. Bioorg. Chem. 2023, 141, 106892. [Google Scholar] [CrossRef]
- Fischer, N.H.; Nielsen, D.S.; Palmer, D.; Meldal, M.; Diness, F. C-Terminal Lactamization of Peptides. Chem. Commun. 2021, 57, 895–898. [Google Scholar] [CrossRef]
- Andrianov, A.K. Noncovalent PEGylation of Protein and Peptide Therapeutics. WIREs Nanomed. Nanobiotechnol. 2023, 15, e1897. [Google Scholar] [CrossRef]
- Martin, V.; Egelund, P.H.G.; Johansson, H.; Thordal Le Quement, S.; Wojcik, F.; Sejer Pedersen, D. Greening the Synthesis of Peptide Therapeutics: An Industrial Perspective. RSC Adv. 2020, 10, 42457–42492. [Google Scholar] [CrossRef]
- Noki, S.; De La Torre, B.G.; Albericio, F. Safety-Catch Linkers for Solid-Phase Peptide Synthesis. Molecules 2024, 29, 1429. [Google Scholar] [CrossRef]
- Zhang, Z.-X.; Nong, F.-T.; Wang, Y.-Z.; Yan, C.-X.; Gu, Y.; Song, P.; Sun, X.-M. Strategies for Efficient Production of Recombinant Proteins in Escherichia Coli: Alleviating the Host Burden and Enhancing Protein Activity. Microb. Cell Fact. 2022, 21, 191. [Google Scholar] [CrossRef]
- Eskandari, A.; Nezhad, N.G.; Leow, T.C.; Rahman, M.B.A.; Oslan, S.N. Current Achievements, Strategies, Obstacles, and Overcoming the Challenges of the Protein Engineering in Pichia Pastoris Expression System. World J. Microbiol. Biotechnol. 2024, 40, 39. [Google Scholar] [CrossRef]
- Stevens, T.A.; Tomaleri, G.P.; Hazu, M.; Wei, S.; Nguyen, V.N.; DeKalb, C.; Voorhees, R.M.; Pleiner, T. A Nanobody-Based Strategy for Rapid and Scalable Purification of Human Protein Complexes. Nat. Protoc. 2024, 19, 127–158. [Google Scholar] [CrossRef]
- Silverman, A.D.; Karim, A.S.; Jewett, M.C. Cell-Free Gene Expression: An Expanded Repertoire of Applications. Nat. Rev. Genet. 2020, 21, 151–170. [Google Scholar] [CrossRef]
- Hartrampf, N.; Saebi, A.; Poskus, M.; Gates, Z.P.; Callahan, A.J.; Cowfer, A.E.; Hanna, S.; Antilla, S.; Schissel, C.K.; Quartararo, A.J.; et al. Synthesis of Proteins by Automated Flow Chemistry. Science 2020, 368, 980–987. [Google Scholar] [CrossRef]
- Haddadzadegan, S.; Dorkoosh, F.; Bernkop-Schnürch, A. Oral Delivery of Therapeutic Peptides and Proteins: Technology Landscape of Lipid-Based Nanocarriers. Adv. Drug Deliv. Rev. 2022, 182, 114097. [Google Scholar] [CrossRef]
- Shan, B.; Wu, F. Hydrogel-Based Growth Factor Delivery Platforms: Strategies and Recent Advances. Adv. Mater. 2024, 36, 2210707. [Google Scholar] [CrossRef]
- Mohammadi, Z.; Eini, M.; Rastegari, A.; Tehrani, M.R. Chitosan as a Machine for Biomolecule Delivery: A Review. Carbohydr. Polym. 2021, 256, 117414. [Google Scholar] [CrossRef]
- Li, C.; Li, T.; Tian, X.; An, W.; Wang, Z.; Han, B.; Tao, H.; Wang, J.; Wang, X. Research Progress on the PEGylation of Therapeutic Proteins and Peptides (TPPs). Front. Pharmacol. 2024, 15, 1353626. [Google Scholar] [CrossRef]
- Holz, E.; Darwish, M.; Tesar, D.B.; Shatz-Binder, W. A Review of Protein- and Peptide-Based Chemical Conjugates: Past, Present, and Future. Pharmaceutics 2023, 15, 600. [Google Scholar] [CrossRef]
- Zhao, J.; Qiu, P.; Wang, Y.; Wang, Y.; Zhou, J.; Zhang, B.; Zhang, L.; Gou, D. Chitosan-Based Hydrogel Wound Dressing: From Mechanism to Applications, a Review. Int. J. Biol. Macromol. 2023, 244, 125250. [Google Scholar] [CrossRef]
- Cao, H.; Duan, L.; Zhang, Y.; Cao, J.; Zhang, K. Current Hydrogel Advances in Physicochemical and Biological Response-Driven Biomedical Application Diversity. Signal Transduct. Target. Ther. 2021, 6, 426. [Google Scholar] [CrossRef]
- Abdel-Mageed, H.M.; Abd El Aziz, A.E.; Mohamed, S.A.; AbuelEzz, N.Z. The Tiny Big World of Solid Lipid Nanoparticles and Nanostructured Lipid Carriers: An Updated Review. J. Microencapsul. 2022, 39, 72–94. [Google Scholar] [CrossRef]
- Li, X.; Naeem, A.; Xiao, S.; Hu, L.; Zhang, J.; Zheng, Q. Safety Challenges and Application Strategies for the Use of Dendrimers in Medicine. Pharmaceutics 2022, 14, 1292. [Google Scholar] [CrossRef]
- Lauko, A.; Pellock, S.J.; Sumida, K.H.; Anishchenko, I.; Juergens, D.; Ahern, W.; Jeung, J.; Shida, A.; Hunt, A.; Kalvet, I.; et al. Computational Design of Serine Hydrolases. Science 2025, 388, eadu2454. [Google Scholar] [CrossRef]
- Liu, H.; Song, Z.; Zhang, Y.; Wu, B.; Chen, D.; Zhou, Z.; Zhang, H.; Li, S.; Feng, X.; Huang, J.; et al. De Novo Design of Self-Assembling Peptides with Antimicrobial Activity Guided by Deep Learning. Nat. Mater. 2025, 24, 1295–1306. [Google Scholar] [CrossRef]
- Watson, J.L.; Juergens, D.; Bennett, N.R.; Trippe, B.L.; Yim, J.; Eisenach, H.E.; Ahern, W.; Borst, A.J.; Ragotte, R.J.; Milles, L.F.; et al. De Novo Design of Protein Structure and Function with RFdiffusion. Nature 2023, 620, 1089–1100. [Google Scholar] [CrossRef]
- Yang, W.; Hicks, D.R.; Ghosh, A.; Schwartze, T.A.; Conventry, B.; Goreshnik, I.; Allen, A.; Halabiya, S.F.; Kim, C.J.; Hinck, C.S.; et al. Design of High-Affinity Binders to Immune Modulating Receptors for Cancer Immunotherapy. Nat. Commun. 2025, 16, 2001. [Google Scholar] [CrossRef]
| Peptide Name [Refs.] | Peptide Sequence | Origin | Corresponding Residues | Working Concentration | Mode of Action | Model | Application |
|---|---|---|---|---|---|---|---|
| FGF1 NLS [19] | NYKKPKL | Human FGF1 | 21~27 | 20~100 μg/mL | fusion with a cell-penetrating peptide | In Vitro Model: NIH3T3 Cell Line | Metabolic Regulation |
| Hexafin 1 [20] | TGQYLAMDTDGLLYGS | Human FGF1 | 76~91 | 0~100 μmol/L | Tetramer | In Vitro Model: Neurite Outgrowth Assay Using Cerebellar Granule Neurons | Neural Functional Recovery |
| peptide 1 [21] | SKKHAEKNWF | human FGF1 | 114~123 | 10~200 μmol/L | Cyclic peptide | In Vitro Model: 3T3 Cell Proliferation Assay | Rational Design of Peptides |
| Canofin 1 [22] | HFKDPKRLYCK | human FGF2 | 25~35 | 0~100 μmol/L | Tetramer | In Vitro Model: Neurite Outgrowth Assay of Cerebellar Granule Neurons | Neural Functional Recovery |
| Peptide 33 [23] | CKNGGFF | human FGF2 | 34~40 | 10~400 μmol/L | Linear monomer | In Vitro Model: Skeletal Muscle Satellite Cell Proliferation Assay | Muscle Regeneration |
| Peptide 33-13 [23] | CKNGGFFLRIHPD | human FGF2 | 34~46 | 10~400 μmol/L | Linear monomer | In Vitro Model: Skeletal Muscle Satellite Cell Proliferation Assay | Muscle Regeneration |
| F36 [24] | PDGRVD | human FGF2 | 45~50 | 100 μmol/L (conjugation dose) | immobilized on the surface of a chitosan membrane | In Vitro Model: Human Mesenchymal Stem Cell Adhesion and Osteogenic Differentiation Assay | Bone Regeneration |
| FREG [25,26] | DPHIKLQLQAE | human FGF2 | 57~67 | 0~100 ng/mL(cells) 3~6 mg·kg−1·d−1 (animals) | Linear monomer | In Vitro Model: Human Melanoma Cell Proliferation and Invasion Assay In Vivo Model: Melanoma Mouse Model | Tumor Suppression |
| P5 and DcP5 [27,28] | LQLQAEER | human FGF2 | 62~69 | 5~15 μmol/L (cells) 10 mg/kg (animals) 20~200 μmol/L (conjugation dose) | Linear monomer Cyclic peptide conjugated with the polysaccharide hyaluronic acid | In Vitro Model: DU145 Prostate Cancer Cell Proliferation Model In Vivo Models: Tumor Model and Acne Model | Tumor Suppression Alleviation of Acne |
| FP2 [29] | ERGVVSIKGV | human FGF2 | 68~77 | 0.05 μg/mL (conjugation dose) | fused with mussel adhesive protein immobilized on the surface of the culture plate | In Vitro Model: Proliferation and Differentiation of Human Wharton’s Jelly-Derived Mesenchymal Stem Cells In Vivo Model: Osteoarthritis Model | Bone Regeneration |
| Hexafin 2 [20] | ANRYLAMKEDGRLLAS | human FGF2 | 79~94 | 0~100 μmol/L | Tetramer | In Vitro Model: Neurite Outgrowth Assay of Cerebellar Granule Neurons | Neural Functional Recovery |
| F77 [24] | KEDGRLL | human FGF2 | 86~92 | 100 μmol/L (conjugation dose) | immobilized on the surface of a chitosan membrane | In Vitro Model: Adhesion and Osteogenic Differentiation of Human Mesenchymal Stem Cells | Bone Regeneration |
| FK18 [30] | FFFERLESNNYNTYRSRK | human FGF2 | 102~119 | 0~100 μg/mL | Linear monomer | In Vitro Model: Oxygen-Glucose Deprivation (OGD) Model in SH-SY5Y Cells In Vivo Model: Retinal Ischemia Model | Neural Functional Recovery |
| Peptide 12 [31] | FFFERLESNNYNTYRSRKYSSWYVA | human FGF2 | 102~126 | 50 μg/animal | conjugated with VLPs | In Vivo Model: Breast Tumor Model | Tumor Suppression |
| FGF-P [32,33,34,35,36,37,38] | YRSRKYSSWYVALKR | human FGF2 | 115~129 | 200 ng/mL (cells) 0~20 mg/kg (animals) 3~6 mmol/L (conjugation dose) 40~400 ng/sample (fusion protein) | Linear monomer conjugated with PA fused with a heparin-binding sequence | In Vitro Model: Proliferation and Migration Assays Using Hs-27 Fibroblasts and Keratinocytes In Vivo Model: Total Body Irradiation (TBI) Model, Bone Marrow Syndrome Model, Skin Burn Injury Model, Spinal Cord Injury (SCI) Model | Multi-Organ Repair Spinal Cord Injury Repair Tissue Regeneration |
| F105 [39] | YKRSRYT | human FGF2 | 120~114 | 100 μmol/L (conjugation dose) | immobilized on the surface of the culture plate | In Vitro Model: Adhesion and Osteogenic Differentiation of Human Mesenchymal Stem Cells | Bone Regeneration |
| BGF1 [40] | CLKRTGQYKLC | human FGF2 | 127~135 | 0~1.8 mmol/L (cells) 2~10 mg/kg (animals) | Cyclic peptide | In Vitro Model: Proliferation models of human umbilical vein endothelial cells (HUVECs), 4T1 breast cancer cells, U87 glioblastoma cells, and SKOV3 ovarian cancer cells In Vivo Model: 4T1 Breast Cancer Model | Tumor Suppression |
| bFGFp [41] | KRTGQYKLC | human FGF2 | 128~135 | 100 mg/mL (conjugation dose) | conjugated with bovine serum albumin or liposomes | In Vitro Model: 3T3 Cell Proliferation Assay | Tumor Suppression |
| F119 [39] | KRTGQYKLGSKTGPGQK | human FGF2 | 128~144 | 100 μmol/L (conjugation dose) | immobilized on the surface of the culture plate | In Vitro Model: Adhesion and Osteogenic Differentiation of Human Mesenchymal Stem Cells | Bone Regeneration |
| Canofin 3 [22] | KTGPGQKAIL | human FGF2 | 138~147 | 0~100 μmol/L | Tetramer | In Vitro Model: Neurite Outgrowth Assay of Cerebellar Granule Neurons | Neural Functional Recovery |
| Canofin 2 [22] | FLPMSAKS | human FGF2 | 147~155 | 0~100 μmol/L | Tetramer | In Vitro Model: Neurite Outgrowth Assay of Cerebellar Granule Neurons | Neural Functional Recovery |
| FGF3 NLS [42] | RRRK | human FGF3 | 44~47 | 0~100 μg/mL | conjugated with PAMAM | In Vitro Model: Transfection of HEK293 and HeLa Cells | Cell Transfection |
| Hexafin 3 [20] | SGRYLAMNKRGRLYKS | human FGF3 | 93~108 | 0~10 μmol/L | Tetramer | In Vitro Model: Neurite Outgrowth Assay of Cerebellar Granule Neurons | Neural Functional Recovery |
| SP and IP [43,44] | AAVALLPAVLLALLAP | human FGF4 | 7~22 | 0~80 μmol/L (conjugation dose) 0~500 μmol/L (conjugation dose) | conjugated with lysine conjugated with PL | In Vitro Model: Protein Delivery into 143B, TE85, MG63, and FOB Cells; Oligonucleotide Delivery into A549 Cells | Protein Delivery Oligonucleotide Delivery |
| P3 [45] | VGIGFHLQIY | human FGF5 | 95~104 | 1~1000 mmol/L (cells) 5 μg/subject (animals) | Linear monomer | In Vitro Model: 3T3 Cell Proliferation Assay In Vivo Model: Depilated Mouse Model | Hair Follicle Repair |
| KGFp [46] | KELILENHYNTYA | human FGF7 | 140~152 | 1~100 ng/mL (cells) | Linear monomer conjugated to a 3D porous scaffold | In Vitro Model: Migration and Differentiation of Human Bone Marrow Mesenchymal Stem Cells In Vivo Model: Chronic Wound Model in Type 2 Diabetic Mice | Tissue Repair |
| 8b-13 [47,48,49] | PNFTQHVREQSLV | human FGF8 | 30~42 | 1~125 nmol/L | Linear monomer | In Vitro Model: Proliferation Assay of PC-3 and DU-145 Prostate Cancer Cells | Tumor Suppression |
| Hexafin 8 [20] | TGLYICMNKKGKLIAK | human FGF8 | 104~119 | 0~10 μmol/L | Tetramer | In Vitro Model: Neurite Outgrowth Assay of Cerebellar Granule Neurons | Neural Functional Recovery |
| Hexafin 9 [20] | SGLYLGMNEKGELYGS | human FGF9 | 112~127 | 0~100 μmol/L | Tetramer | In Vitro Model: Neurite Outgrowth Assay of Cerebellar Granule Neurons | Neural Functional Recovery |
| Hexafin 10 [20] | SNYYLAMNKKGKLYGS | human FGF10 | 128~143 | 0~10 μmol/L | Tetramer | In Vitro Model: Neurite Outgrowth Assay of Cerebellar Granule Neurons | Neural Functional Recovery |
| Hexafin 17 [20] | SEKYICMNKRGKLIGK | human FGF17 | 93~108 | 0~10 μmol/L | Tetramer | In Vitro Model: Neurite Outgrowth Assay of Cerebellar Granule Neurons | Neural Functional Recovery |
| FGF23 Peptide [50] | AEDDSERDPLNVLKPRARMTPAPAS | human FGF23 | 181~205 | 0.2 nmol/L | Linear monomer | In Vivo Model: Hyperphosphatemia in Fgf23−/− Mice | Metabolic Regulation |
| FAP1 [51] | RERNEVNHYRTY | Computational Design of Peptide Derivatives Targeting Human FGFR1c | 1~100 ng/mL (cells) 1~10 mg/kg (animals) | Linear monomer | In Vitro Model: Proliferation and Migration Assays Using NIH 3T3 Cells In Vivo Model: Diabetic Mouse Wound Healing Model | Tissue Repair | |
| C19jun [52] | AESGDDYCVLVFTDSAWTKICDWSHFRN | Phage Display Technology for Screening FGFR1c-Binding Peptide Derivatives | 0~10 nmol/L | fused with human c-Jun residues for expression Dimer | In Vitro Model: Swiss 3T3 Cell Proliferation and Neurite Outgrowth in Neuronal Cells | Tissue Repair | |
| F8 [53] | ACSLNHTVNC | Phage Display Technology for Screening FGFR1c-Binding Peptide Derivatives | 0~10 μmol/L | Cyclic peptide | In Vitro Model: BA/F3 Cell Proliferation Assay | Tumor Suppression | |
| F91-8A07 [54] | LPGRTCREYPDLWWVRCY | Phage Display Technology for Screening FGFR1c/β-Klotho-Binding Peptide Derivatives | 0~1000 μmol/L (cells) 0~1000 nmol/kg (animals) | Dimer | In Vitro Model: Primary Human Adipocyte Model In Vivo Model: Mouse Model | Metabolic Regulation | |
| CH02 [55] | GPANVET | Phage Display Technology for Screening FGFR2c-Binding Peptide Derivatives | 0~40 μmol/L | Linear monomer | In Vitro Model: Neurite Outgrowth Assay of Dorsal Root Ganglion (DRG) Neurons In Vivo Model: Rat Dorsal Root Compression Injury Model | Neural Functional Recovery | |
| H1 [56] | SNFLHLG | Phage Display Technology for Screening FGFR2c-Binding Peptide Derivatives | 0~20 μmol/L (cells) 0~1000 μmol/L (animals) | Linear monomer | In Vitro Model: 3T3 Cell Proliferation and Migration Assay In Vivo Model: Full-Thickness Excisional Wound Model | Tissue Repair | |
| P3 [57] | VSPPLTLGQLLS | Phage Display Technology for Screening FGFR3-Binding Peptide Derivatives | 0~50 μmol/L (cells) 100 μg·kg−1·d−1 (animals) | Linear monomer | In Vitro Model: ATDC5 Cell Proliferation and Chondrogenic Differentiation Model In Vivo Model: TDII Mouse Lethal Phenotype Model | Bone Regeneration | |
| peptide [58] | MQLPLAT | Phage Display Technology for Screening FGFR-Binding Peptide Derivatives | 10–20 μg/mL (conjugation dose) | Conjugation with PEI-PEG | In Vitro Model: B16F10 Cell Transfection Model | Cell Transfection | |
| 23-b6 [59] | SSPPKSP | Phage Display Technology for Screening FGFR-Klotho-Binding Peptide Derivatives | 0~0.1 μmol/L | Linear monomer | In Vitro Model: Phosphate Uptake Assay in Renal Proximal Tubule Cells | Metabolic Regulation | |
| Pro-Ile [60] | PI | Functional Screening of Human FGFR-Binding Peptide Derivatives Using Bacterial Conditioned Medium | 0~1 mmol/L | Linear monomer | In Vitro Model: Keratinocyte Proliferation Assay | Hair Follicle Repair | |
| AP8 [61] | AGNWTPI | Phage Display Technology for Screening FGF1-Binding Peptide Derivatives | 0~16 μmol/L | Linear monomer | In Vitro Model: Proliferation Assay of Breast Cancer Cells and Human Umbilical Vein Endothelial Cells | Tumor Suppression | |
| P7 [62,63,64] | PLLQATLGGGS | Phage Display Technology for Screening FGF2-Binding Peptide Derivatives | 0~16 μmol/L (cells) | Linear monomer | In Vitro Model: Proliferation and Migration Assay of MDA-MB-231 Breast Cancer Cells | Tumor Suppression | |
| P7Δ [65] | PLLQATL | Phage Display Technology for Screening FGF2-Binding Peptide Derivatives | 0~16 μmol/L (cells) 1 μmol/L (animals) | Linear monomer | In Vitro Model: Proliferation Assay of BALB/c 3T3 Cells In Vivo Model: Chick Embryo Chorioallantoic Membrane (CAM) Assay | Tumor Suppression | |
| FP16 [66] | VLWLKNR | Phage Display Technology for Screening FGF3-Binding Peptide Derivatives | 0~16 μmol/L | Linear monomer | In Vitro Model: Proliferation Assay of MDA-MB-231 and T47D Breast Cancer Cells | Tumor Suppression | |
| P12 [67] | HSQAAVP | Phage Display Technology for Screening FGF8b-Binding Peptide Derivatives | 0~16 μmol/L | Linear monomer | In Vitro Model: Proliferation Assay of PC-3 and HUVECs | Tumor Suppression | |
| P4 [68] | NVFTVSP | Phage Display Technology for Screening FGF9-Binding Peptide Derivatives | 0~16 μmol/L | Linear monomer | In Vitro Model: Proliferation Assay of SGC-7901 Gastric Cancer Cells and RT-112 Bladder Cancer Cells | Tumor Suppression | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, C.-K.; Shi, Z.-Y.; Chen, C.-B.; Li, X.-K.; Su, Z.-J. Fibroblast Growth Factor-Derived Peptides: Sources, Functions, and Applications. Bioengineering 2025, 12, 1019. https://doi.org/10.3390/bioengineering12101019
Cao C-K, Shi Z-Y, Chen C-B, Li X-K, Su Z-J. Fibroblast Growth Factor-Derived Peptides: Sources, Functions, and Applications. Bioengineering. 2025; 12(10):1019. https://doi.org/10.3390/bioengineering12101019
Chicago/Turabian StyleCao, Cheng-Kun, Zhong-Yuan Shi, Chuan-Bang Chen, Xiao-Kun Li, and Zhi-Jian Su. 2025. "Fibroblast Growth Factor-Derived Peptides: Sources, Functions, and Applications" Bioengineering 12, no. 10: 1019. https://doi.org/10.3390/bioengineering12101019
APA StyleCao, C.-K., Shi, Z.-Y., Chen, C.-B., Li, X.-K., & Su, Z.-J. (2025). Fibroblast Growth Factor-Derived Peptides: Sources, Functions, and Applications. Bioengineering, 12(10), 1019. https://doi.org/10.3390/bioengineering12101019







